Abstract
Although a vast amount of life sciences data is generated in the form of images, most scientists still store images on extremely diverse and often incompatible storage media, without any type of metadata structure, and thus with no standard facility with which to conduct searches or analyses. Here we present a solution to unlock the value of scientific images. The Global Image Database (GID) is a web-based (http://www.g wer.ch/qv/gid/gid.htm) structured central repository for scientific annotated images. The GID was designed to manage images from a wide spectrum of imaging domains ranging from microscopy to automated screening. The annotations in the GID define the source experiment of the images by describing who the authors of the experiment are, when the images were created, the biological origin of the experimental sample and how the sample was processed for visualization. A collection of experimental imaging protocols provides details of the sample preparation, and labeling, or visualization procedures. In addition, the entries in the GID reference these imaging protocols with the probe sequences or antibody names used in labeling experiments. The GID annotations are searchable by field or globally. The query results are first shown as image thumbnail previews, enabling quick browsing prior to original-sized annotated image retrieval. The development of the GID continues, aiming at facilitating the management and exchange of image data in the scientific community, and at creating new query tools for mining image data.
INTRODUCTION
The nature of digital information stored in life sciences research organizations has become much more varied, and now includes image, video and 3D coordinate or volume files created on a wide range of systems using multiple formats. The vast majority of this information is currently thought to be unstructured (i.e. stored without any type of metadata structure) and thus with no standard facility with which to conduct searches or analyses.
In life sciences research, visual information has been estimated to represent as much as 70% of all data generated. Due to the development of digital image capture devices connected to personal computers, a number of academic and industrial laboratories now generate large amounts of digital biological image files. Despite this technological evolution, visual information continues to be treated like traditional films and is still stored on personal media, thus resulting in research-based organizations being increasingly swamped by scattered information, unable to create novel content-based mining tools.
These elements clearly indicate that the future of biological imaging relies on networked database systems allowing the scientific community to store, retrieve and process large amounts of visual information. Indeed, projects like BioImage (1) and the GID bring innovative solutions to the problem of managing this morass of data.
CONCEPTS
Biological image data management requires some degree of structured annotation to guarantee subsequent understanding of the visual information. To minimize the effort needed to annotate experimental images, we defined a minimal, but sufficient, set of annotations with expert input from biologists. The minimal pertinent annotations that encompass the widest range of biological image types can be summarized in four words: ‘who’, ‘when’, ‘how’ and ‘what’. By simply indicating in a fixed format who are the authors of the experiment, when did the experiment take place, how the biological specimen was processed, how the images were produced and what was the biological source of the specimen, it becomes possible to clearly define images from very diverse fields. For instance, histology, intracellular protein localization, tissue gene expression or automated high-throughput screening assay images fit perfectly in such an annotation scheme.
To limit the administration overhead required to maintain the image database, each author of an experiment personally introduces the annotations prior to an image set submission. In addition, all the server-side processing of the incoming annotated images is completely automated, including creating previews of the images, assigning accession codes and sending back a receipt.
To access the database and ensure the highest level of simplicity and interactivity, we developed a digitally signed Java applet running in common web browsers, which connects to a Java server application (Fig. 1).
Figure 1.
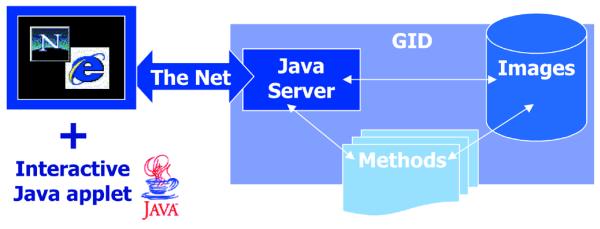
Scheme of the GID client-server system. With a web browser, a program written in Java starts on your computer from the network. This program that receives and sends information is called the client, while the central storage computer is called the server. The server contains a second Java program listening to the client’s calls. It receives the images and annotations and stores them in a temporary folder. In parallel, a program written in Perl language is started every minute to process incoming data, assign an accession number and send a receipt to communicate the accession numbers to the user. The source code of these components of the GID can be obtained by contacting the authors. In the future, the server will call quantification methods to automatically extract information from certain image types.
By promoting usage of the GID, we expect the digital imaging evolution that took place in biological research to finally not only facilitate team work and permanent storage of images, but also to enable the creation of automated image classification and novel quantitative visualization methods. Without well-defined large sets of images, the development of such methods could not be envisaged.
ACCESSING THE GID
The GID release 1.1 is available on the Web. A first connection setup procedure explaining how to open a user account and install the required Java plug-in is found at http://www.gwer.ch/qv/gid/gid.htm. This plug-in allows Java applets to run homogeneously using Netscape 3 and 4 and Internet Explorer 3, 4 and 5 on Windows, SGI-Irix, HP-UX and Solaris platforms. It is under development for the Mac (see http://java.sun.com:80/products/plugin/plugin.faq.html#pricing).
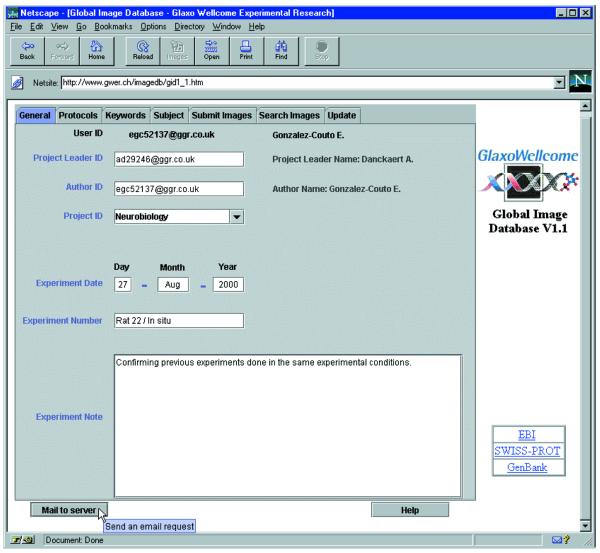
At each connection, every user has to log in, thus allowing retrieval of his profile saved during the previous submission. The GID user interface is then displayed (Fig. 2). The four panels entitled General, Protocols, Keywords and Subject are used to enter the annotations, while the Submit panel allows the selection from your local disk of the images to submit to the GID. Finally, the Search panel is used to send queries to the GID, browse the search results, including the image previews, and retrieve the original images.
Figure 2.
The GID user interface. Access to the GID takes place from a web page containing the GID user interface organized in panels. Below the panels, a mail button can be used to directly contact us and an integrated help that follows the changes of panels to guide you can also be started, in addition to the classical ToolTips that appear, as show down left, next to the Mail button, to inform you. A list of hyperlinks pointing to other biological databases of interest is located on the right side of the page.
ANNOTATING AND SUBMITTING IMAGES
The first four panels can be filled sequentially to annotate an experimental set of images (Fig. 2 and Supplementary Material). The content of most annotation fields is immediately checked for validity, with the note, or comment fields available to add personal information.
The General panel is used to indicate the author of the experiment and the project leader, together with the date of the experiment and the project’s domain. In addition to these constrained fields, an experimental note allows the user to add personal information.
The Protocols panel contains two lists of experimental protocol titles grouped by cell and tissue preparation procedures or by staining, in situ hybridization and immunostaining protocols. When a title is selected, the corresponding protocol is displayed and can be saved to the local disk. If the selected protocol is an in situ or an immunostaining protocol, an additional table is displayed to enter one or several probes, sequences or antibody names used. This information can be supplemented with accession codes to the most popular gene or protein databases. Such references are particularly valuable, since they allow searching for images showing the distribution of a mRNA, or the localization of a protein. Finally, the initial content of the protocols can be edited to ‘deviate’ from the original. This feature allows submission of in situ images with, for instance, custom hybridization conditions or stringency washes.
The Keywords panel contains a list of selectable keywords from SWISS-PROT, with the possibility to add personal keywords defining the experiment.
The Subject panel regroups the information concerning the biological specimen used in the experiment. Several experimental models with their respective strains are already listed. Besides, some comments, additional information, like the model’s disease, sex, age, weight and the type of sample, can be entered in this panel.
The Submit Images panel is designed to navigate through the local disks to view and select the image(s) corresponding to the annotations. The image formats currently accepted are GIF, animated GIF, JPG and TIF. The microscope magnification can be set individually or for all of the images. Finally, after accepting the annotation summary, clicking the Send Image(s) button allows submission of the annotated images to the GID, together with the user profile. Once the submitted data is processed on the GID server, an acknowledgement message containing the accession code(s) is sent back by email.
SEARCHING AND RETRIEVING IMAGES
The search panel is an interactive and simple tool for creating sophisticated queries without needing a complex syntax.
The GID search starts directly when selecting the Search Images panel, using the current annotations as queries on the corresponding fields in the database. It is also possible to switch to the ‘free field’ global search mode, to scan all the annotations text independent of the field. The search results are displayed in a table as the number of hits for each annotation field. Multiple selection of the lines of this table allows combination of the sets of hits to refine the initial query and browse the search results under the form of annotations (see Supplementary Material) with thumbnails. While browsing, it is possible to select the original images to retrieve from the GID.
We recently started the Internet version of the GID with more than 9000 images and 14 different experimental protocols. The database contains optical sections showing the intracellular localization of immunofluorescently labeled proteins (i.e. transferrin receptor, LGP120, TGN38, calnexin, protein disulfide isomerase) using confocal microscopy; images of Nissl-stained coronal, sagittal and horizontal sections of rat brains; and finally pictures of classical histology tissue stainings. Obviously, we encourage scientists to contribute new submissions.
CONCLUSION
Visual information is less structured and more complex to exchange through networks than text-only information. Therefore, we developed a Java client-server system to annotate and interactively access biological visual information without either the inherent limitations of HTML or the need of a specialized file transfer program.
The GID development continues with a relational version including new search features, information privacy levels and integrated access to image recognition or quantification methods associated to precise image types (2). In addition, we would like to develop query by content on well-defined data sets (3), directly access GID data from other image analysis software and let the GID become the ‘imaging core’ for more specialized biological image databases containing, for example, a brain gene expression database (4).
The profitable development of such a novel type of biological database clearly depends on how well it is populated. We therefore adopted a collaborative approach to promote the usage of the GID. Our strategy, as a BioImaging group, is to develop internal and academic collaborations aimed at creating new image visualization, quantitatification and classification tools. In such projects, the experimental images will be made available on the GID. For instance, in GlaxoWellcome the Cell Biology Department (Stevenage) has submitted images showing typical intracellular staining for Golgi body, endosomes and endoplasmic reticulum, and a scientific collaboration was started with a neurobiology group at the IBCM (University of Lausanne, Switzerland). The members of this group will submit mouse brain images to the Internet GID for us to develop a computerized mouse brain atlas. In broad terms, the GID is useful for many groups of interest in the life sciences domains where images are of central importance.
Considering the increasing number of genome-wide projects producing large amounts of visual data (5), the need for reference data sets of intracellular protein localization or tissue gene expression images, we can expect an increase in the usage of bioimaging tools. The GID should ultimately increase the intrinsic value of biological image data for the long-term benefit of scientists.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Manuel Peitsch for stimulating discussions, Mark Paulik for comments on the manuscript, our colleagues from Stevenage for sharing their expertise, Dominique Talabot-Ayer for expert technical assistance and the Medical School of the University of Geneva for the first image examples.
References
- 1.Carazo J.M. and Stelzer,E.H. (1999) The BioImage Database Project: organizing multidimensional biological images in an object-relational database. J. Struct. Biol., 125, 97–102. [DOI] [PubMed] [Google Scholar]
- 2.Boland M.V., Markey,M.K. and Murphy,R.F. (1998) Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry, 33, 366–375. [PubMed] [Google Scholar]
- 3.de Alarcon P.A., Gupta,A. and Carazo,J.M. (1999) A framework for querying a database for structural information on 3D images of macromolecules: A web-based query-by-content prototype on the BioImage macromolecular server. J. Struct. Biol., 125, 112–122. [DOI] [PubMed] [Google Scholar]
- 4.Chicurel M. (2000) Databasing the brain. Nature, 406, 822–825. [DOI] [PubMed] [Google Scholar]
- 5.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L., Heidtman,M., Nelson,F.K., Iwasaki,H., Hager,K., Gerstein,M., Miller,P., Roeder,G.S. and Snyder,M. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.