Abstract
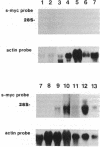
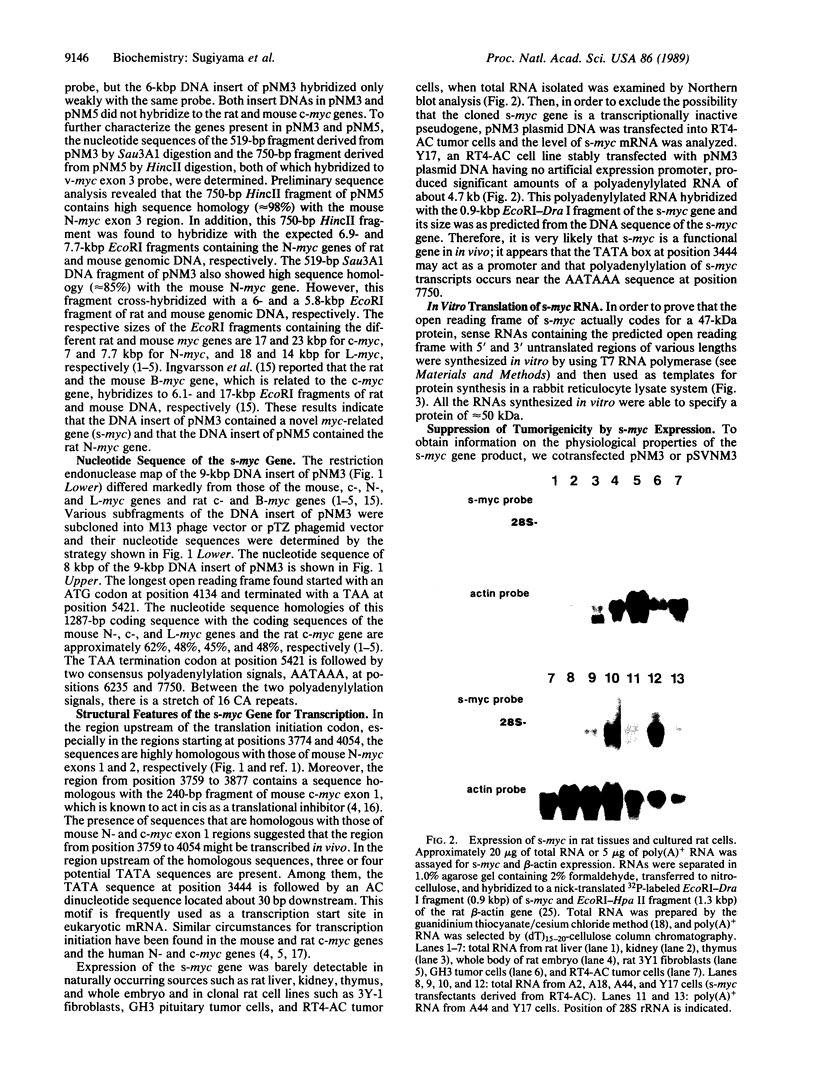
A 9-kilobase-pair clone containing a myc-related gene that we have designated s-myc was isolated from a rat genomic library. The entire nucleotide sequence of the cloned DNA was determined, showing that the s-myc gene contains an open reading frame consisting of 1287 base pairs without introns. In vitro transcription-translation analysis of s-myc indicated that this gene produces a protein of approximately 50 kDa. The amino acid sequence predicted from the DNA sequence showed that the s-Myc protein is closely related to the murine N-Myc protein but lacks an acidic amino acid-rich sequence commonly present in the Myc-family proteins. Studies on transfection of s-myc into rat RT4-AC tumor cells revealed that the gene produces a polyadenylylated transcript of approximately 4.7 kilobases and that its high-level expression suppresses the tumor-igenicity of RT4-AC tumor cells in nude mice.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988 Oct;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge J., Webster T., Smith T. F., Paucha E. Prediction of similar transforming regions in simian virus 40 large T, adenovirus E1A, and myc oncoproteins. J Virol. 1988 May;62(5):1814–1818. doi: 10.1128/jvi.62.5.1814-1818.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. R., Sueoka N. Induction and segregation of glial intermediate filament expression in the RT4 family of peripheral nervous system cell lines. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5808–5812. doi: 10.1073/pnas.84.16.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Makino R., Kawamura H., Arisawa A., Yoneda K. Characterization of rat c-myc and adjacent regions. Nucleic Acids Res. 1987 Aug 25;15(16):6419–6436. doi: 10.1093/nar/15.16.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Cooper J. A. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- Ingvarsson S., Asker C., Axelson H., Klein G., Sümegi J. Structure and expression of B-myc, a new member of the myc gene family. Mol Cell Biol. 1988 Aug;8(8):3168–3174. doi: 10.1128/mcb.8.8.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Nemoto K., Kawai S., Nishimura S. Activation of c-myc gene transcription by Rous sarcoma virus infection. Jpn J Cancer Res. 1985 Feb;76(2):75–78. [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel U., Zakut R., Shani M., Neuman S., Levy Z., Yaffe D. The nucleotide sequence of the rat cytoplasmic beta-actin gene. Nucleic Acids Res. 1983 Mar 25;11(6):1759–1771. doi: 10.1093/nar/11.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N., Darveau A., Nicholson R., Sonenberg N. cis-acting translational effects of the 5' noncoding region of c-myc mRNA. Mol Cell Biol. 1988 Jul;8(7):2875–2883. doi: 10.1128/mcb.8.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Robertson M. Molecular associations and conceptual connections. Nature. 1988 Jul 14;334(6178):100–102. doi: 10.1038/334100a0. [DOI] [PubMed] [Google Scholar]
- Sargent T. D., Wu J. R., Sala-Trepat J. M., Wallace R. B., Reyes A. A., Bonner J. The rat serum albumin gene: analysis of cloned sequences. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3256–3260. doi: 10.1073/pnas.76.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stanton L. W., Schwab M., Bishop J. M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashjian A. H., Jr, Yasumura Y., Levine L., Sato G. H., Parker M. L. Establishment of clonal strains of rat pituitary tumor cells that secrete growth hormone. Endocrinology. 1968 Feb;82(2):342–352. doi: 10.1210/endo-82-2-342. [DOI] [PubMed] [Google Scholar]
- Taya Y., Mizusawa S., Nishimura S. Nucleotide sequence of the coding region of the mouse N-myc gene. EMBO J. 1986 Jun;5(6):1215–1219. doi: 10.1002/j.1460-2075.1986.tb04349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennström B., Moscovici C., Goodman H. M., Bishop J. M. Molecular cloning of the avian myelocytomatosis virus genome and recovery of infectious virus by transfection of chicken cells. J Virol. 1981 Aug;39(2):625–631. doi: 10.1128/jvi.39.2.625-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]