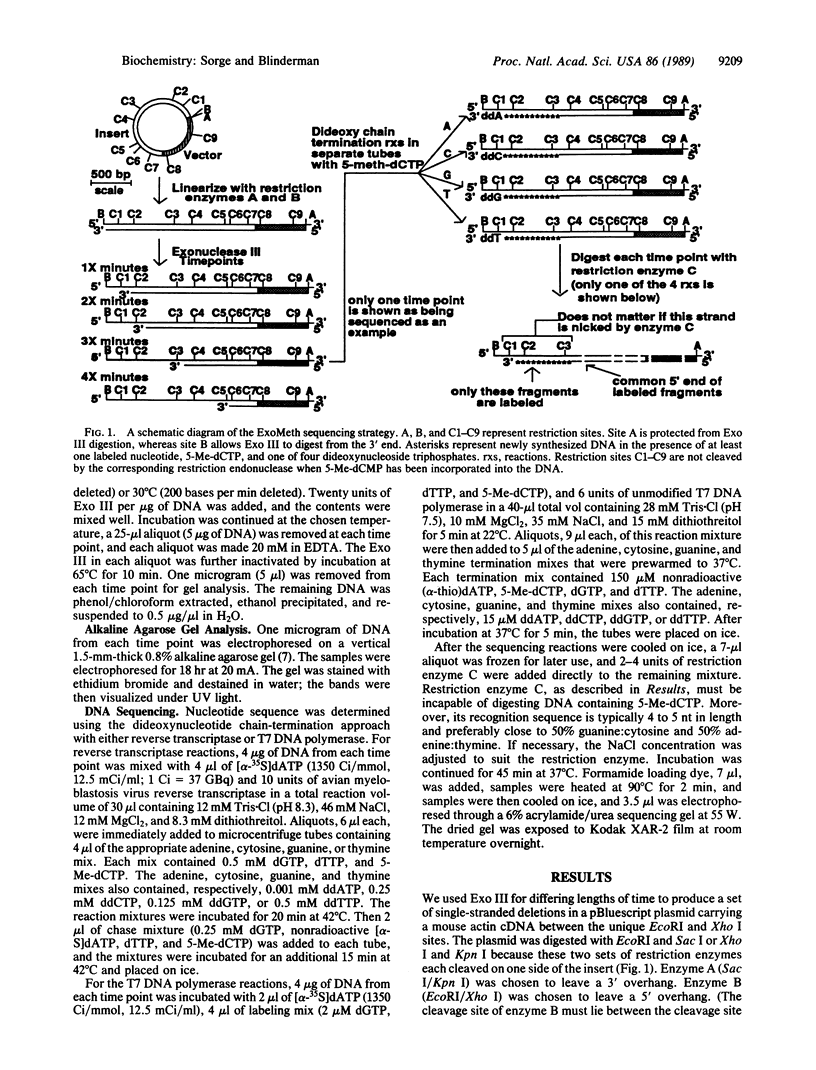
Abstract
A method is reported for sequencing DNA based on exonuclease III digestion and strand protection by using modified nucleoside triphosphates. Up to 10 kilobases of sequence information may be obtained from each strand of a given template without subcloning. Prior knowledge of the restriction map is not important; prior knowledge of any of the sequence is not required. Nor are oligonucleotide primers needed. Double-stranded cosmids, plasmids, lambda phage, or linear molecules (including amplified molecules) may be used as starting material. The method creates a single-stranded template from these starting molecules, thus generating high-quality sequence ladders. Most commonly used DNA polymerases may be utilized, including reverse transcriptase and T7 DNA polymerase. The approach is "ordered", so little time is wasted on redundant sequencing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beutler E., Gelbart T., Han J. H., Koziol J. A., Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc Natl Acad Sci U S A. 1989 Jan;86(1):192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. H., Wu R. Exonuclease III: use for DNA sequence analysis and in specific deletions of nucleotides. Methods Enzymol. 1983;100:60–96. doi: 10.1016/0076-6879(83)00046-4. [DOI] [PubMed] [Google Scholar]
- Guo L. H., Wu R. New rapid methods for DNA sequencing based in exonuclease III digestion followed by repair synthesis. Nucleic Acids Res. 1982 Mar 25;10(6):2065–2084. doi: 10.1093/nar/10.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Waterman M. S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988 Apr;2(3):231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Leader D. P., Gall I., Campbell P., Frischauf A. M. Isolation and characterization of cDNA clones from mouse skeletal muscle actin mRNA. DNA. 1986 Jun;5(3):235–238. doi: 10.1089/dna.1986.5.235. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Sanders J. Z., Kaiser R. J., Hughes P., Dodd C., Connell C. R., Heiner C., Kent S. B., Hood L. E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986 Jun 12;321(6071):674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- Zimmermann J., Voss H., Schwager C., Stegemann J., Ansorge W. Automated Sanger dideoxy sequencing reaction protocol. FEBS Lett. 1988 Jun 20;233(2):432–436. doi: 10.1016/0014-5793(88)80477-0. [DOI] [PubMed] [Google Scholar]