Abstract
Reversible protein phosphorylation plays an important role in many cellular processes. However, a simple and reliable method to measure changes in the extent of phosphorylation is lacking. Here, we present a method to quantitate the changes in phosphorylation occurring in a protein in response to a stimulus. The method consists of three steps: (i) enzymatic digestion in H216O or isotopically enriched H218O to label individual pools of differentially phosphorylated proteins; (ii) affinity selection of phosphopeptides from the combined digests by immobilized metal-affinity chromatography; and (iii) dephosphorylation with alkaline phosphatase to allow for quantitation of changes of phosphorylation by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. We applied this strategy to the analysis of the yeast nitrogen permease reactivator protein kinase involved in the target of rapamycin signaling pathway. Alteration in the extent of phosphorylation at Ser-353 and Ser-357 could be easily assessed and quantitated both in wild-type yeast cells treated with rapamycin and in cells lacking the SIT4 phosphatase responsible for dephosphorylating nitrogen permease reactivator protein. The method described here is simple and allows quantitation of relative changes in the level of phosphorylation in signaling proteins, thus yielding information critical for understanding the regulation of complex protein phosphorylation cascades.
Phosphorylation is one of the most frequently occurring posttranslational modifications in proteins. It plays an essential role in transferring signals from the outside to the inside of a cell, and it regulates many diverse cellular processes such as growth, metabolism, proliferation, motility and differentiation. Regulation of these processes is achieved through changes in phosphorylation of receptors, adaptor proteins, and protein kinases. Thus, it is important to obtain a complete image of phosphorylation occurring in such proteins, and to achieve this, rapid and sensitive methods to track and quantitate changes of phosphorylation occurring in proteins are necessary. Although mass spectrometry has been a valuable tool in elucidating sites of phosphorylation in proteins (1), problems persist because of technical difficulties in measuring the changes in the extent of phosphorylation at these sites in response to defined stimuli.
In the simplest case, phosphorylation stoichiometries can be quantified by dividing integrated peak intensities for a given phosphopeptide by the sum of intensities of its phosphorylated and unphosphorylated forms. However, this procedure often results in an underestimation of the phosphorylation stoichiometry, as phosphopeptides sometimes ionize with lower efficiency than their corresponding unphosphorylated peptides (2, 3). Quantitation in site-specific phosphorylation has been achieved by the use of stable isotope labeling (4) and by phosphoprotein-specific isotope-coded affinity tagging (5). However, tracking changes in the extent of phosphorylation in low-abundance proteins, such as signaling molecules, has not been demonstrated with the above methods.
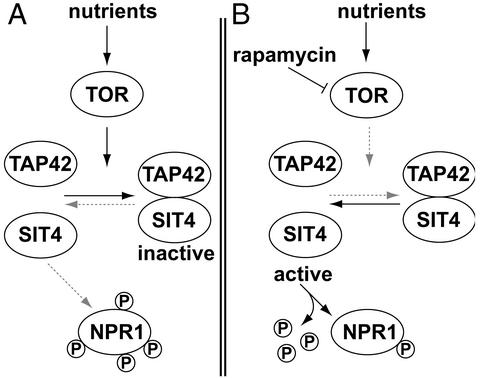
One of the major growth control elements in the yeast Saccharomyces cerevisiae is the target of rapamycin (TOR) proteins (6). The TOR proteins are highly conserved phosphatidylinositol kinase-related kinases that control cell growth through an extensive signaling network (6). Inhibition of TOR by treatment of yeast cells with the immunosuppressant and anti-cancer drug rapamycin leads to a number of responses resembling nutrient deprivation, indicating that TOR controls cell growth in response to nutrients. The starvation program of rapamycin-treated cells includes a severe decrease in protein synthesis (7), arrest in the G1 phase of the cell cycle (8, 9), down-regulation of transcription of genes encoding rRNA and ribosomal proteins (10–13), glycogen accumulation (7), autophagy (14), and induction of processes involved in scavenging secondary nitrogen sources (12, 13, 15). In response to the nitrogen source, TOR also influences the cell's repertoire of amino acid transporters. Upon nitrogen starvation, high-affinity transporters such as the tryptophan permease TAT2 are rapidly targeted to the vacuole and degraded, whereas the broad specificity amino acid transporter GAP1 is up-regulated and stabilized (16). The reprogramming of permeases in response to nutrients is controlled via the Ser/Thr kinase nitrogen permease reactivator protein (NPR1), which is maintained phosphorylated by the TOR signaling pathway (17). TOR promotes the association between the phosphatase SIT4 and the regulatory subunit TAP42 (18). When associated, SIT4 is inactive because of the inhibitory effect of TAP42, and NPR1 is kept in a hyperphosphorylated form (Fig. 1). When cells are treated with rapamycin, or grown in the presence of poor nitrogen sources, TOR is inactivated, leading to the dissociation of the SIT4-TAP42 complex and, thus, to the activation of SIT4. As a consequence, NPR1 becomes dephosphorylated (Fig. 1; refs. 17 and 19).
Figure 1.
Regulation of NPR1 phosphorylation by the rapamycin-sensitive TOR signaling pathway. (A) Under good nutrient conditions, TOR promotes the association of TAP42 and SIT4, thereby inactivating the phosphatase activity of SIT4. NPR1 is, therefore, maintained in a highly phosphorylated state. (B) Rapamycin treatment leads to inactivation of TOR and to dissociation of the regulatory subunit TAP42 from SIT4. In turn, activated SIT4 dephosphorylates NPR1. Arrows indicate activation; bars indicate inhibition.
To characterize the rapamycin-sensitive phosphorylation sites in NPR1 (SwissProt database accession no. P22211), a mass spectrometric analysis was initiated. To determine which of the phosphorylation sites in NPR1 is most sensitive to rapamycin treatment, a method was needed to follow the dephosphorylation of the protein quantitatively. In this paper, we present a simple method to quantitate changes in phosphorylation of proteins by a combination of stable isotope labeling, affinity selection, and matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) mass spectrometry.
Materials and Methods
Yeast Strains and Media.
The plasmid harboring GST-NPR1 was constructed in two steps. For the plasmid pBS1, a 0.8-kb Xbal-HindIII fragment coding for the GST part was cloned into a YEplac195 (2μ, URA3) plasmid containing the promoter region of NPR1 (pAS103 derived; ref. 17). Next, a 2.4-kb HindIII fragment containing the NPR1 ORF was cloned into the plasmid pBS1, yielding plasmid pBS2. Yeast strains used in this study were isogenic derivatives of TB50a (ura3–52 leu2–3,112 trp1 his3 rme1 HMLa). The strains JC19-1A (TB50a npr1∷HIS3MX6) and JC28-1B (TB50a npr1∷HIS3MX6 sit4∷kanMX4) were transformed with the plasmid pBS2 according to standard procedures. By using an npr1 mutant background results in the plasmid (pBS2)-encoded Gst-NPR1 as the sole cellular copy of NPR1. Thus, strain JC19–1A transformed with pBS2 is referred to as wild-type strain. Cells were grown in synthetic medium (yeast nitrogen base, supplemented with amino acids) containing 2% glucose to an optical density of 0.7. Rapamycin treatment was done at an optical density of 0.7 for 15 min. by adding the drug from a stock solution (1 mg/ml in 90% ethanol/10% Tween-20) to a final concentration of 200 ng/ml. Cells were harvested by centrifugation and washed with water, and the cell pellets were stored at −80°C until use.
Purification of GST-NPR1.
The cell pellet obtained from a 6-liter culture was resuspended in 50 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.0, 100 mM NaCl/1% Nonidet P-40/1 mM EDTA/10 mM sodium azide/1 mM benzamidine/1 μg/ml each of pepstatin, leupeptin, aprotinin, and 10 mM each of p-nitrophenylphosphate, sodium pyrophosphate, and β-glycerophosphate). Cells were broken with glass beads (500 μm) in a bead beater (Biospec Products, Bartlesville, OK) with seven bursts for 20 sec. Cell debris was removed by centrifugation at 3,000 × g. The supernatant was passed through a glutathione Sepharose 4B column (1.5 × 2 cm; Amersham Pharmacia), which had been equilibrated with lysis buffer. After washing the column with lysis buffer and 100 mM Tris⋅HCl, pH 8.0/150 mM NaCl, GST-NPR1 was eluted with 10 mM reduced glutathione (Sigma) in 100 mM Tris⋅HCl, pH 8.0/150 mM NaCl.
Peptide Isotope Labeling.
Twenty microliters of reduced and carboxamidomethylated chicken ovalbumin (0.2 μg/μl in 100 mM Tris⋅HCl, pH 8.0; Sigma) or GST-NPR1 (in 100 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.2 μg/μl protein concentration) were dried and dissolved either in 20 μl of H216O or (if derived from rapamycin-treated or mutant cells) in 20 μl of isotopically enriched H218O (>95% pure; CAMPRO, Berlin, Germany). The protein was digested with trypsin (Promega) at an enzyme/substrate ratio of 1:25 at 37°C for 2 h. The reaction was stopped by adding trifluoroacetic acid (TFA) to a final concentration of 0.1%. For desalting, peptides were adsorbed on C18 ZipTips (Millipore), washed with 0.1% TFA, and desorbed with 5 μl of 80% acetonitrile/0.1% TFA.
Selection of Phosphopeptides.
Five microliters of phosphopeptide solution (ZipTip-desalted) was incubated with ≈200 nl of Chelating Sepharose Fast Flow beads (Amersham Pharmacia) charged with FeCl3 for 15 min at room temperature. After washing the beads, phosphopeptides were released with either 5 μl of 100 mM Tris⋅HCl, pH 8.0 or with 5 μl of 100 mM Tris⋅HCl, pH 8.0 containing 0.5 units of calf intestinal alkaline phosphatase (CIP, Roche Diagnostics, Mannheim, Germany). Dephosphorylation was allowed to proceed for 15 min at 37°C. The beads were removed by centrifugation, and the supernatant was acidified with 0.5 μl of 10% (vol/vol) TFA. For MALDI measurements, the peptides were desalted on C18 ZipTips as described above, except that elution was done with 80% (wt/vol) AcCN, 0.1% TFA, containing 1 μg/μl α-cyano-4-hydroxycinnamic acid (CHCA, Aldrich).
MALDI-TOF Mass Spectrometry.
Five hundred nanoliters of phosphopeptides was deposited onto anchor spots of a Scout 400 μm per 36 sample support (Bruker Daltonik, Bremen, Germany), and the droplet was left to dry at room temperature. Mass spectra were recorded on a Bruker Reflex III instrument (Bruker Daltonik). Phosphopeptides eluted from the immobilized metal affinity chromatography (IMAC) beads with Tris buffer were analyzed in linear mode, and peptides eluted from the beads in the presence of CIP were analyzed in reflector mode by using delayed ion extraction with a total acceleration voltage of 23 kV. Assignments of tryptic peptides of GST-NPR1 were done by comparing the measured masses to those predicted by the protein sequence of NPR1.
Peptide Nomenclature.
Peptides generated by trypsin cleavage are labeled with T. The peptides are numbered sequentially according to their position based on the N-terminal methionine of NPR1. The cloning procedure resulted in a fusion protein in which the sequence of the GST from Schistosoma japonicum is fused via the linker peptide SDLSGGGGGKL to the N-terminal methionine of NPR1. The calculated masses are given as mass to charge ratios of the singly protonated peptides.
Data Analysis.
To quantitate changes in the extent of phosphorylation, the ratio R of paired sets of isotope clusters 4-Da apart was calculated. Firstly, the sequences of the tryptic peptides were used to calculate the theoretical isotope peak distribution by using the MS-isotope pattern calculator (http://prospector.ucsf.edu). Secondly, the predicted intensity of the fifth isotope peak from the peptide digested in H216O was subtracted from the fifth peak of the isotope cluster of the mix of unlabeled and 18O-labeled peptide. Thirdly, to correct for incomplete incorporation of 18O into the C terminus of the peptides, the difference between the measured intensity of the third peak of the isotope envelope and the predicted intensity was added to the intensity of the fifth peak of the isotope envelope. The equation used for all calculations is the following: R = I0/[(I4 − v4 × I0 + (I2 − v2 × I0)], where I0, I2, and I4 are the measured peak intensities for the monoisotopic peak for peptides digested in H216O, the peak with masses 2 Da higher, and the peak with 4 Da higher, respectively; v2, and v4 are the percent maximum of the isotopes 2 and 4 Da higher, respectively.
Results
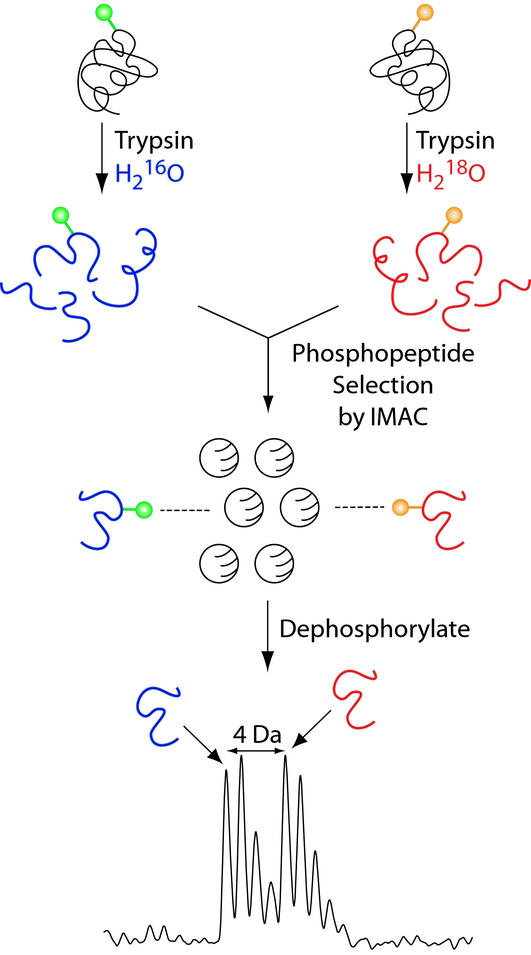
One of the major difficulties in using mass spectrometry to measure precise stoichiometries of phosphorylation is that the pair consisting of the phosphopeptide and the corresponding nonphosphorylated peptide differ in their behavior during the ionization process. It is well known that during electrospray ionization phosphopeptides ionize less efficiently than the corresponding nonphosphorylated peptides (2, 3). In MALDI-TOF analysis, dominant metastable decomposition of phosphopeptides occurs, which complicates quantitation (20). Therefore, we sought a method to remove the phosphate such that information indicating the extent of phosphorylation of a specific site is preserved. To this end, we chose the C termini of peptides as targets for stable isotope tagging during proteolysis (21). In short, a protein obtained from one particular phosphorylation state (e.g., hyperphosphorylated) is digested with trypsin in the presence of H216O, whereas the same protein differing only in its phosphorylation state (e.g., hypophosphorylated) is digested in the presence of H218O. The two peptide pools are then mixed, and phosphopeptides are selected by IMAC-beads (22). If, for example, two phosphopeptides containing the same phosphorylation site have different levels of modification, the relative amounts of phosphopeptides bound to the IMAC matrix should reflect their relative levels of modification. Consequently, peptides that undergo a change in their level of modification yield pairs of MS peaks with intensity ratios that reflect these changes. Because of metastable loss of phosphoric acid from serine or threonine residues in MALDI MS (20), phosphopeptides have to be detected in linear mode to be able to quantitate relative peak intensities. However, the resolution achievable in linear mode is not sufficient to separate completely the two sets of isotope clusters. Therefore, to be able to measure for the highest possible resolution in reflector mode, bound phosphopeptides are eluted from the IMAC-beads with alkaline phosphatase. Although the phosphate group is lost upon this treatment, the extent of phosphorylation is reflected by the relative abundance of their monoisotopic clusters, and hence, their relative peak areas can be used to quantitate the change in phosphorylation that occurred in a particular peptide (Fig. 2).
Figure 2.
Outline of the strategy. Two protein pools differing in their extent of phosphorylation are digested with trypsin either in H216O or in H218O to obtain differential mass labeling. Equal amounts of the two pools are mixed, and phosphopeptides are selected with IMAC beads charged with Fe3+. To enable mass determinations of the bound peptides with high accuracy, the peptides are eluted from the IMAC beads with alkaline phosphatase. The two peptide pools can be distinguished by a shift of 4 Da in the isotope cluster, and the difference in the extent of phosphorylation is reflected by the peak areas of the two monoisotopic peaks.
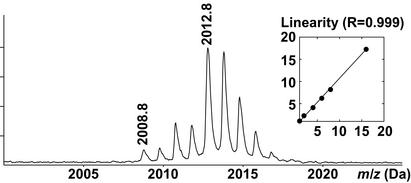
To test the above strategy, we used ovalbumin as a model phosphoprotein. Ovalbumin contains two phosphorylated serines at positions 68 and 344. Tryptic cleavage of ovalbumin releases two phosphopeptides, T8 (LPGFGDpSIEAQCGTSVNVHSSLR) and T31 (EVVGpSAEAGVDAASVSEEFR) with expected masses of 2,354.11 Da and 2,088.91 Da, respectively. To simulate two proteins of different levels of phosphorylation, peptides from tryptic digests of ovalbumin performed in H216O or in isotopically enriched H218O were mixed in different ratios and phosphopeptides were selected with IMAC beads and eluted with alkaline phosphatase. Measuring the areas of the 2,008.8-Da and 2,012.8-Da signal, corresponding to the dephosphorylated T31 peptide (2,088.91 Da or 2,092.91 Da minus 80 Da for the phosphate group), the procedure consisting of 18O-labeling, IMAC selection, and alkaline phosphatase treatment was found to be linear within mixing ratios of between 1:1 and 1:16 (Fig. 3 Inset). It is important to note that accurate quantitation depends critically on equal amounts of each of the protein pools being subjected to proteolysis. Comparison of the intensities of the monoisotopic peaks of the two sets of isotope clusters indicates if unequal amounts of protein were subjected to enzymatic digestion. If so, a correction factor can be calculated from any nonphosphorylated peptide to compensate for the relative amounts of the two protein pools.
Figure 3.
Linearity assay. To simulate two protein pools with differing extent of phosphorylation, the relative amounts of ovalbumin digested in H218O or H216O was set to 16:1, and a MALDI spectrum was acquired for the T31 phosphopeptide after IMAC selection and elution with alkaline phosphatase. (Inset) The plot of the ratios of the theoretical vs. the experimentally measured areas of the T31 (2,008.8 Da) and the 18O-labeled T31 (2,012.8 Da).
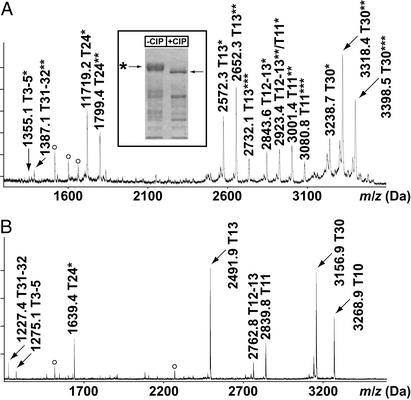
Next, the method was applied to assess rapamycin-induced changes in phosphorylation of the yeast NPR1 kinase. Purification of GST-tagged NPR1 from untreated yeast cells yields a hyperphosphorylated protein migrating as a diffuse band of ≈120 kDa (Fig. 4 Inset; ref. 17). Analysis of a tryptic digest of GST-NPR1 by IMAC selection and MALDI-TOF analysis in linear mode reveals seven candidate phosphopeptides, carrying a total of 14 phosphates (Fig. 4A, for a compilation of all phosphopeptides of GST-NPR1; also, see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). When elution of the IMAC beads is done in the presence of alkaline phosphatase, the mass of the seven peptides decreases by 80 Da, or multiples thereof (Fig. 4B), indicating that the peptides selected by the IMAC beads are indeed phosphorylated. Also, one phosphopeptide (T10) which was difficult to detect in linear mode became easily detectable after treatment with alkaline phosphatase (Fig. 4). In addition, tryptic peptides whose molecular masses remained unaltered upon treatment with alkaline phosphatase were also found to bind unspecifically to the IMAC beads. These include the peptides AEISMLEGAVLDIR and LLLEYLEEKYEEHLYER (residues 90–103 and 19–35) of the GST moiety, and T70 (GNIEEIMEDPWIR, residues 731–743) of NPR1.
Figure 4.
IMAC phosphopeptide selection from a tryptic digest of GST-NPR1. (A) MALDI-TOF spectrum of phosphopeptides in linear mode. *, Number of phosphates associated with the peptide. White circles mark unspecifically bound peptides. (B) MALDI-TOF spectrum of the phosphopeptides eluted from the IMAC beads in the presence of alkaline phosphatase. The spectrum was acquired in reflector mode. (Inset) Coomassie blue-stained SDS gel of purified GST-NPR1 without (−CIP) or with (+CIP) alkaline phosphatase treatment.
However, attempts to deduce alterations in the extent of phosphorylation by directly comparing affinity-selected phosphopeptides of GST-NPR1 from untreated- and rapamycin-treated cells in linear MALDI-TOF were largely unsuccessful because of variations in relative signal intensities of the phosphopeptides (results not shown). Therefore, stable isotope labeling combined with affinity selection was used to track changes in the extent of phosphorylation of individual phosphopeptides of NPR1 occurring upon rapamycin treatment.
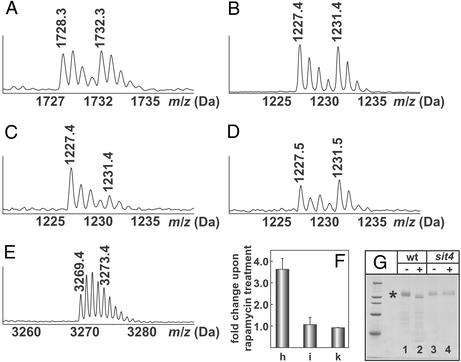
Treating wild-type cells with rapamycin for 15 min results in increased mobility of GST-NPR1 because of decreased phosphorylation of the protein (Fig. 5G, lanes 1 and 2). To track changes in the extent of phosphorylation of NPR1 upon rapamycin treatment of wild-type cells, equal amounts of GST-NPR1 from untreated or rapamycin-treated cells were subjected to tryptic digestion in H216O or in H218O, respectively. The digests were mixed, and a MALDI-TOF spectrum was recorded in reflector mode. The signal intensity of the isotope cluster for the unphosphorylated peptide T68 (residues 707–721 of GST-NPR1; calculated mass = 1,728.94 Da) digested in H216O or in H218O indicates equal amounts of the two protein pools (Fig. 5A). Also, equal amounts of the phosphopeptide T31–32 (SQHSSIGDLKR, residues 353–362; calculated mass = 1,227.64 Da, doubly phosphorylated on Ser-353 and Ser-357) were selected from a H216O- and a H218O-digest of GST-NPR1 isolated from untreated cells (Fig. 5B). However, when equal amounts of GST-NPR1 digests from untreated (trypsinized in H216O) and rapamycin-treated cells (trypsinized in H218O) were selected by IMAC, the intensity of the monoisotopic signal of the 18O-labeled T31–32 phosphopeptide was strongly reduced (Fig. 5C). Peak integration showed that rapamycin treatment induces a 3.6 ± 0.5-fold dephosphorylation of this peptide (Fig. 5F, column h).
Figure 5.
MALDI-TOF spectra of tryptic peptides T68 (A), T31–32 (B–D), and T10 (E) of GST-NPR1. (A and B) The nonphosphorylated peptide T68 (1,728.3 Da and 1,732.3 Da H216O/H218O digest) and the doubly phosphorylated T31–32 peptide from untreated cells (1,227.4 Da and 1,231.4 Da H216O/H218O digest) indicate that equal amounts of protein were digested and equal amounts of phosphopeptides were selected. (C and D) T31–32 phosphopeptide from untreated (1,227.4 Da) and from rapamycin-treated (1,231.4 Da) wild-type cells (C) or from sit4 mutant cells (D). (E) Phosphopeptide T10 from untreated (3,269.4 Da) or rapamycin-treated (3,273.4 Da) wild-type cells. (F) Ratios between the isotope clusters of the T31–32 (column h), the T10 (column i) peptide from the wild-type strain treated with rapamycin, and the T31–32 peptide from the sit4 mutant JC28-1B treated with rapamycin (column k) digested in H216O or H218O. Error bars represent the mean of three independent experiments. (G) Coomassie blue-stained SDS gel of GST-NPR1 (labeled with an asterisk) from wild-type cells (JC19-1A, wt, lanes 1 and 2) or sit4 mutant cells (JC28-1B, sit4, lanes 3 and 4), either untreated (−) or treated (+) with 200 nM rapamycin for 15 min.
Preliminary data indicated that some of the phosphorylation sites of GST-NPR1 were not equally sensitive to rapamycin treatment. In particular, selecting phospho-T10 (spanning amino acids 63–94; calculated mass = 3,269.5 Da for the nonphosphorylated peptide) with IMAC beads from either untreated (digested in H216O) or rapamycin-treated GST-NPR1 (digested in H218O) and subjecting the peptide to MALDI-TOF analysis after CIP treatment indicated almost equal signal intensities of the peptide derived from untreated and rapamycin-treated cells (Fig. 5E). After subtracting the contribution of the signal of the peptide with two 16O atoms from the signal of the monoisotopic peak of the peptide with two 18O atoms and correcting for incomplete labeling, a value of 1.1 ± 0.34 (Fig. 5F, column i) was obtained, indicating that the phosphorylation site contained in T10 is indeed unaffected by the drug treatment.
TOR promotes the association of the regulatory subunit TAP42 with the phosphatase SIT4 when cells are grown in medium containing a good nitrogen source (such as ammonium or glutamine; see Fig. 1). Nitrogen starvation or rapamycin treatment leads to dissociation of the regulatory subunit and consequently to activation of SIT4, ultimately resulting in dephosphorylation of NPR1 (Fig. 1). Purification of GST-NPR1 from a sit4 deletion mutant yields a protein whose migration during SDS gel electrophoresis is identical to GST-NPR1 from the wild-type strain (Fig. 5G, lane 3). However, GST-NPR1 in the sit4 mutant strain is not dephosphorylated upon rapamycin treatment, as evidenced by its unaltered mobility when compared with GST-NPR1 from untreated sit4 cells (Fig. 5G, lane 4; ref. 19). To see whether the newly developed assay correlates with the results obtained by SDS gel electrophoresis, GST-NPR1 was isolated from the wild-type strain, digested in H216O, whereas the protein from the sit4 mutant treated with rapamycin was trypsinized in H218O. The digests were mixed in equal amounts, affinity-selected with IMAC beads, and the bound phosphopeptides were eluted in the presence of alkaline phosphatase. The signal intensity of the two isotope clusters for the phosphopeptide T31–32 indicates no effect of rapamycin treatment on the extent of phosphorylation of Ser-353 and Ser-357 contained in the T31–32 peptide (Fig. 5D). Calculating the ratio of the 1,227.5 and 1,231.5 Da peak areas yielded a value of 0.88 ± 0.03 (Fig. 5F, column k). Therefore, the present method is consistent with previous results that NPR1 is resistant to the SIT4-mediated dephosphorylation upon rapamycin treatment (19).
Discussion
There is an ever growing demand for rapid and reliable methods to assess protein phosphorylation. Here, we devised a method which consists of a combination of simple steps: proteolytic digestion in H216O or H218O to tag two different phosphoprotein pools, affinity selection of the phosphopeptides from the combined pools by IMAC, and dephosphorylation with alkaline phosphatase to allow for quantitation by mass spectrometry of the changes of modification occurring in a given phosphopeptide. The main advantage of the method presented here is that phosphopeptides originating from protein pools with varying extent of phosphorylation are measured simultaneously within the same matrix crystal, therefore avoiding fluctuations in relative signal intensities, which frequently occur in independent measurements.
Although the overall yield of the method was found to be excellent, it is worth commenting on the individual steps of the procedure. First, the basis of the present method is the application of stable isotope labeling of peptides during trypsinolysis. When highly enriched H218O (>95%) is used, products with two atoms of 18O are predominant, thereby shifting the mass of a peptide 4 Da higher (21, 23). Ideally, to be of general use for quantitative studies, incorporation of two 18O atoms should proceed to 100%. In our studies, we found that in all peptides investigated so far, the incorporation of two 18O atoms proceeded to ≈90%, whereas 10% had only one atom of 18O incorporated after two h of trypsinolysis. Neither prolonging the incubation time up to 36 h, nor adding more trypsin was able to bring about 100% labeling (results not shown). Nevertheless, because the proportion of peptides labeled with two atoms of 18O was found to be 90% for all peptides investigated, reliable correction factors can be introduced to compensate for less than quantitative labeling.
Secondly, phosphopeptide selection with IMAC beads is essentially quantitative as long as the maximal binding capacity is not exceeded (≈20 pmol/μl resin), and dephosphorylation with alkaline phosphatase goes to completion when working at the picomole level of phosphopeptides. Furthermore, the use of alkaline phosphatase introduces an additional element of specificity because it is well known that, in addition to phosphopeptides, histidine- and cysteine-containing peptides, as well as peptides containing multiple carboxylic acid groups (24), also bind to IMAC beads. By using alkaline phosphatase, the mass shift of 80 Da caused by the removal of the phosphate allows unambiguous determination whether a given peptide is phosphorylated or not (Fig. 4).
The incorporation of two atoms of 18O shifts the mass of a peptide by 4 Da. With the resolution obtainable by MALDI-TOF instruments operated in reflector mode, isotopic resolution can be achieved up to 2,000 Da. For larger peptides, proper correction factors have to be introduced to compensate for the overlap of the two sets of isotopes. Alternatively, the low-specificity protease elastase can be used to produce smaller phosphopeptides with masses ranging from 0.5 to 1.5 kDa (25). Such peptides would be ideally suited for the method presented here. Also, the large number of peptides produced by elastase digestion is likely to result in peptides carrying only one phosphate, so that site-specific regulation of phosphorylation can be studied. However, because of the low specificity of elastase, peptide assignment by mass alone cannot be achieved with a MALDI-TOF instrument, unless high-resolution mass spectrometers are used (25).
Only recently, mass spectrometric methods for quantification of protein modification have been developed. For example, by using differential labeling of yeast cells grown in normal or in 15N-enriched medium, changes in the extent of phosphorylation of the STE20 protein kinase produced under the control of the inducible GAL1 promoter have been monitored (4). Although the method is conceptually well suited to track phosphorylation dynamics at individual sites, it is limited to the analysis of proteins expressed at relatively high levels. To be able to track changes in the extent of phosphorylation of low-abundance proteins involved in signaling events, large cell cultures are required, making the use of isotopically enriched medium impractical. Recently, a method has been presented that uses β-elimination of the O-phosphate followed by the addition of 1,2-ethanedithiol containing either four alkyl hydrogen or deuterium atoms for differential labeling of two phosphoprotein pools (PhIAT method; ref. 5). The PhIAT method can isolate phosphopeptides from such complex mixtures as digests of whole-cell lysates from S. cerevisiae; however, its ability to quantitate changes in the extent of phosphorylation of proteins has yet to be demonstrated. Another method to isolate and quantitate phosphopeptides uses a series of chemical reactions equally applicable to P-Ser-, P-Thr-, and P-Tyr-containing peptides (26, 27). Although the method is not intended for direct quantitation of changes in the extent of phosphorylation, the blocking step potentially allows for incorporation of stable isotope tags that can be differentiated and quantitated by mass spectrometry. Likewise, the method proposed by Ficarro et al. (24), which uses conversion of carboxylate groups to methyl ester for increased specificity of the IMAC selection process, can be modified to allow for quantitation of phosphopeptides obtained from two different samples.
In short, we have devised a simple and efficient method which allows quantitation of changes of phosphorylation of proteins isolated from two different cellular states. It is conceivable that the method described here is applicable to processes controlling growth or diseases. Furthermore, future work should be carried out to see whether the method can be used in model organisms such as the yeast S. cerevisiae on a proteome-wide level.
Supplementary Material
Acknowledgments
We thank Dr. S. Helliwell for critical reading of the manuscript. This work was supported by grants from the Swiss National Foundation (to P.J. and M.N.H.). J.L.C. is a recipient of a Long Term Fellowship of the Federation of European Biochemical Societies, and E.J. is a recipient of a fellowship of the American Cancer Research Institute.
Abbreviations
- TOR
target of rapamycin
- NPR1
nitrogen permease reactivator protein
- MALDI-TOF
matrix-assisted laser desorption ionization/time-of-flight
- TFA
trifluoroacetic acid
- CIP
calf intestinal alkaline phosphatase
- IMAC
immobilized metal affinity chromatography
References
- 1.McLachlin D T, Chait B T. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 2.Radimerski T, Mini T, Schneider U, Wettenhall R E, Thomas G, Jenoe P. Biochemistry. 2000;39:5766–5774. doi: 10.1021/bi9927484. [DOI] [PubMed] [Google Scholar]
- 3.Resing K A, Ahn N G. Methods Enzymol. 1997;283:29–44. doi: 10.1016/s0076-6879(97)83005-4. [DOI] [PubMed] [Google Scholar]
- 4.Oda Y, Huang K, Cross F R, Cowburn D, Chait B T. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goshe M B, Conrads T P, Panisko E A, Angell N H, Veenstra T D, Smith R D. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 6.Schmelzle T, Hall M N. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 7.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heitman J, Movva N R, Hall M N. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 9.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 10.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers T, Walter P. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 15.Beck T, Hall M N. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 16.Beck T, Schmidt A, Hall M N. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Como C J, Arndt K T. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 19.Jacinto E, Guo B, Arndt K T, Schmelzle T, Hall M N. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 20.Annan R S, Carr S A. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 21.Schnolzer M, Jedrzejewski P, Lehmann W D. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 22.Posewitz M C, Tempst P. Anal Chem. 1999;14:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 23.Yao X, Freas A, Ramirez J, Demirev P A, Fenselau C. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 24.Ficarro S B, McCleland M L, Stukenberg P T, Burke D J, Ross M M, Shabanowitz J, Hunt D F, White F M. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 25.Schlosser A, Pipkorn R, Bossemeyer D, Lehmann W D. Anal Chem. 2001;73:170–176. doi: 10.1021/ac000826j. [DOI] [PubMed] [Google Scholar]
- 26.Zhou H, Watts J D, Aebersold R. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 27.Oda Y, Nagasu T, Chait B T. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.