Abstract
Single strands of DNA serve, in rare instances, as promoters for transcription; duplex DNA promoters with individual strands that also have a promoter capacity/function have not been described. We show that the nontranscribed strand of the Saccharomyces cerevisiae U6 snRNA gene directs transcription initiation factor IIIB-requiring and accurately initiating transcription by RNA polymerase III. The nontranscribed strand promoter is much more extended than its duplex DNA counterpart, comprising the U6 gene TATA box, a downstream T7 tract, and an upstream-lying segment. A requirement for placement of the 3′ end of the transcribed (template) strand within the confines of the transcription bubble is seen as indicating that the nontranscribed strand provides a scaffold for RNA polymerase recruitment but is deficient at a subsequent step of transcription initiation factor IIIB's direct involvement in promoter opening.
Budding yeast (Saccharomyces cerevisiae) RNA polymerase III (pol III) is brought to its promoters by its transcription factor (TF)-IIIB, which is composed of three subunits, TATA-binding protein (TBP), Brf1, and Bdp1. S. cerevisiae TFIIIB, in turn, is brought to its binding site upstream of the transcriptional start by TFIIIC, but also can be directed to the promoter by TBP subunit-directed binding to a TATA box (1–3). Only a few budding yeast pol III genes, among them the U6 snRNA gene, have sufficiently strong TATA boxes to allow TFIIIC-independent and solely TFIIIB-directed transcription of DNA (4). In contrast, fission yeast (Saccharomyces pombe) genes transcribed by pol III have TATA boxes as absolutely required promoter elements (4, 5).
Our recent analysis of the functions of TFIIIB in initiation of transcription has exploited certain deletion mutants of Brf1 and Bdp1 that generate distinctive defects of promoter opening in linear DNA. The search for conditions under which activity would be restored to these defective TFIIIB assemblies has involved the exploitation of DNA templates with partially opened promoters or with breaks in either DNA strand. Exploring the architecture of the pol III promoter complex by manipulating its DNA scaffold in this manner has provided insights into the mechanism of promoter opening, including its general upstream → downstream polarity, and into the selective role of the transcribed (template) strand in specifying/measuring the distance from the TATA box to the transcriptional start site (6–8).
That analysis also has led (by a path that is specified below) to the surprising finding that is elaborated in this work: the nontranscribed strand of the U6 promoter suffices for TFIIIB-dependent and accurately initiating transcription by pol III. We show that the single-stranded U6 promoter includes a TATA element, with which TFIIIB interacts in a distinctive way, and also involves additional promoter elements that are required to be single-stranded but are not part of the duplex DNA promoter.
Methods
DNA Templates.
Transcription templates for this work are based on plasmid pU6RboxB (9), with a single G → A substitution at base pair −22 reversing the original A → G mutation. Fully duplex DNA (base pairs −60 to +138 relative to the transcriptional start site) was generated as described (10). The 5′ overhang templates and gap templates were generated by annealing the purified single-stranded components and purified by native gel electrophoresis and, when necessary, electrophoresis through gel-immobilized oligonucleotides to trap and separate partially annealed and/or unannealed contaminants (8).
Proteins.
Purification and quantification of wild-type TFIIIB subunits and TBPm3 have been described (6, 9). TBP and Bdp1 were estimated to be nearly 100%, and Brf1 was 20% active in formation of heparin-resistant promoter complexes. Bdp1(Δ355–372) was purified under native conditions as described for Bdp1(138–594) (11). NΔ68Brf1 was purified as described (10). TFIIIC and pol III were purified as described; quantities of pol III are specified as fmol of enzyme active for specific transcription (12, 13).
Transcription and 5′ End Mapping.
TFIIIB–DNA complexes were formed for 40 min at 20°C in 20 μl of reaction buffer containing 40 mM Tris⋅HCl (pH 8.0), 7 mM MgCl2, 3 mM DTT, 100 μg/ml BSA, 5 μg/ml poly(dG-dC), 5–7% (vol/vol) glycerol, 80–90 mM NaCl with 20 fmol DNA template, 52 fmol TFIIIC (as indicated), 200 fmol TBP (or TBPm3), 100 fmol Brf1 (or Brf1NΔ68), and 150 fmol Bdp1 (or Bdp1Δ355–372). Two microliters (5 fmol) of pol III were added for an additional 20 min. Multiple rounds of transcription at 20°C were started by adding 5 μl of reaction buffer containing 1 mM ATP, 1 mM CTP, 1 mM GTP, and 125 μM [α-32P]UTP and stopped after 30 min by adding 155 μl of stop solution (10 mM Tris⋅HCl/3 mM EDTA/0.2% SDS). Samples were precipitated, processed for denaturing gel electrophoresis, and quantified by phosphorimage plate analysis. Primer extension (reverse transcription) analysis of unlabeled transcripts to determine transcription start sites was carried out as described (10).
Results
The analysis that is reported here has its origin in an observation made during the course of experiments on the effects of DNA strand breaks at defined locations on transcription by RNA pol III (7, 8). The duplex DNA for this analysis was assembled from three purified oligonucleotides, and partial assemblies containing single-stranded tails were removed carefully. Control DNA preparations with long 5′ and 3′ overhanging tails (assembled with only two oligonucleotides) were made to assess inhibition of transcription by attached single-stranded DNA. Finding that one of these partially duplex DNAs, presenting only the nontranscribed strand of its essential TATA box, could serve as a template for factor-dependent and specifically initiating transcription prompted the further exploration that is recounted below.
TFIIIB-Dependent Transcription with Partially Single-Stranded Promoters.
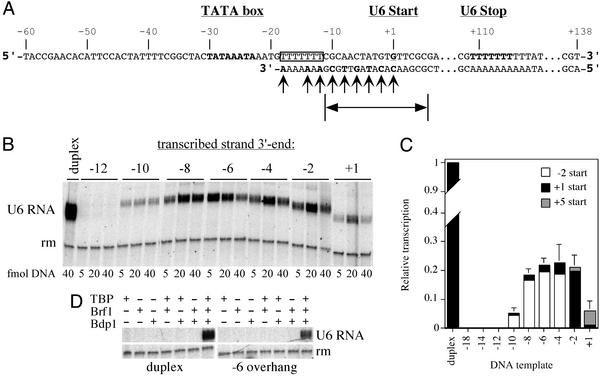
We constructed partially single-stranded templates derived from the yeast U6 snRNA (SNR6) gene, presenting only the 5′ overhanging nontranscribed (Fig. 1A Upper) strand of the upstream promoter region. The 3′ end of the transcribed (Fig. 1A Lower) strand was placed at different positions relative to the normal transcription start site, and activity was monitored in multiple rounds of transcription. Only templates with transcribed strand 3′ ends placed downstream of base pair −12 (relative to the normal transcription start site as +1) were found to be active (Fig. 1B and data not shown). At the optimal concentration (Fig. 1B), 5′ overhang DNA was seen to yield approximately one-fifth as many transcripts as duplex DNA (Fig. 1C). Excess DNA diminished activity (perhaps by nonspecifically and unproductively sequestering TBP or pol III). TFIIIC did not restore transcription activity to the inactive −12 5′ overhang (o/h) construct (data not shown). Transcription from these 5′ overhang templates strictly required each TFIIIB subunit, including Bdp1 (Fig. 1D). Transcriptional start sites, mapped by primer extension, were shifted slightly upstream to base pair −2 for constructs −10 o/h to −4 o/h, remained at base pair +1 for construct −2 o/h, and were shifted downstream to base pair +5 for construct +1 o/h (Fig. 1C and data not shown). Similar shifts of start site have been noted in transcription of duplex DNA with breaks at these locations and have been attributed to the removal of a constraint that is imposed by continuity of the transcribed strand between the TATA box and base pair +1 (8). That constraint is, of course, also removed in these constructs.
Figure 1.
(A) The single-stranded SNR6 promoter. The 198-nt nontranscribed (Upper) strand comprises the U6 snRNA gene sequence from −60 to +138 relative to the normal transcription start site (bold/underlined; designated as +1). The TATA box and the terminator sequence are in bold letters; the T7-stretch is boxed; 3′ ends of the transcribed strand are indicated by arrows. The extent of the transcription bubble is noted at the bottom. (B) Transcription activity of the single-stranded promoter. Pol III transcription was carried out in the presence of TFIIIB on duplex DNA or 5′ overhang templates, with transcribed-strand 3′ ends located as indicated. Because of the potential inhibitory effect of single-stranded DNA, templates were titrated, as indicated below each lane. (C) Transcriptional activity at 20 fmol 5′ overhang DNA relative to duplex DNA. Error bars are SEM of at least five independent experiments. The distribution of transcriptional start sites, as determined by primer extension, is specified for each construct: open boxes, −2; black, +1; gray, +5. Relative activities of promoters with transcribed strand 3′ ends at base pairs −18, −14, and −12, regarded as indistinguishable from background, were 0.008, 0.008, and 0.009, respectively. (D) TFIIIB dependence of transcription. Transcription was assessed on duplex DNA and the −6 overhang template in the presence of single, any two, or all three TFIIIB subunits. rm, recovery marker.
The preceding experiment yields the surprising result that TFIIIB can assemble on a single-stranded version of its DNA-binding site, accurately recruiting pol III to the transcriptional start site. The ability of the resulting peculiar TFIIIB–pol III-promoter complex to produce transcripts is restricted to 5′ overhang constructs placing the template strand's 3′ end downstream of the U6 gene's T7 segment (T7:A7 in duplex DNA), that is, within the bounds of the transcription bubble. The experiments that follow explore the promoter sequence requirements of these constructs and also exploit previously analyzed TBP, Bdp1, and Brf1 mutations to gain further insight into the mode of action of TFIIIB in this special context.
Transcriptional Activity of Partially Single-Stranded Promoters Is Sequence-Dependent.
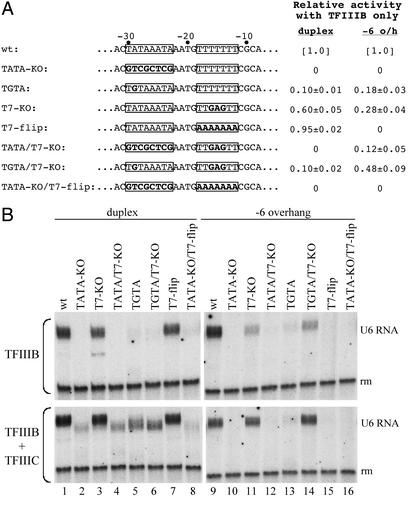
Mutations in the TATA box and the T7:A7 stretch were introduced into the transcriptionally active 5′ overhang construct −6 o/h, the inactive construct −12 o/h, and the corresponding duplex DNA (Fig. 2A) and tested for transcriptional activity with TFIIIB and pol III (Fig. 2B). Destruction of the TATA box eliminated transcription of duplex DNA (Fig. 2B, lanes 2, 4, and 8), and A → G at base pair −29 (TATA → TGTA) greatly diminished transcription (lanes 5 and 6), whereas changes in the T7:A7 stretch only moderately affected transcription (lanes 3 and 7). This is consistent with prior observations that transcription of the U6 gene in vitro tolerates multiple substitutions in the T7:A7 stretch (14), although the latter becomes an essential promoter element of the U6 gene under circumstances altering chromatin structure in vivo (15). The same mutations introduced into the −6 o/h construct showed that transcriptional activity remained sequence-dependent, but with additional elaborations. Transcription activity was eliminated by destroying the TATA box (Fig. 2B, lane 10) and reduced strongly by the TGTA mutation (lane 13), but the changes in the T7 stretch also strongly reduced or entirely eliminated transcriptional activity (lanes 11 and 15). To our surprise, the T7-KO mutation appeared to diminish the effects of both TATA box mutations (compare lanes 10 and 13 with lanes 12 and 14, respectively; the difference between lanes 10 and 12 is seen clearly in the original data and was observed consistently).
Figure 2.
Mutating the nontranscribed strand U6 promoter. (A) Changes of the TATA box and T7-stretch (boxed) are highlighted in bold letters. Percent transcriptional activity with TFIIIB only (i.e., no TFIIIC) relative to the respective wild-type reference (wt; set to 1.0) is shown at the right (the numbers represent averages ± SEM of at least five independent experiments). (B) Transcription assay. The effects of mutations were assessed for transcription of duplex DNA (lanes 1–8) and the −6 o/h template (lanes 9–16) in the presence of TFIIIB only (Upper) or TFIIIB and TFIIIC (Lower).
TFIIIC makes TFIIIB assembly on the promoter much less dependent on specific DNA sequence (4, 16), and TFIIIC mitigated the effects of these TATA box mutations on transcription of duplex DNA (generating 2- to 10-fold-increased transcription), as expected (Fig. 2B, lanes 2, 4–6, and 8). In contrast, TFIIIC did not restore transcription of completely inactive −6 o/h constructs (e.g., TATA-KO; lane 10) and only slightly stimulated transcription of constructs retaining weak activity with TFIIIB alone (e.g., TGTA; lane 13). The −12 o/h construct versions of these promoter mutants were all transcriptionally inactive (in the presence and absence of TFIIIC).
Two properties of the promoter in 5′ overhang DNA that are seen in this experiment have no counterpart in the linear duplex DNA: (i) an interaction between the T7 site and the TATA box that allows the T7-KO change to restore some activity to TATA box mutants instead of compounding inactivity and (ii) an inability of TFIIIC to mitigate the detrimental effects of TATA box mutations on transcription. These differences imply that assembling TFIIIB on the U6 promoter as duplex DNA and as its nontranscribed strand produces complexes with different properties arising from different protein–DNA interactions and, possibly, DNA secondary structures.
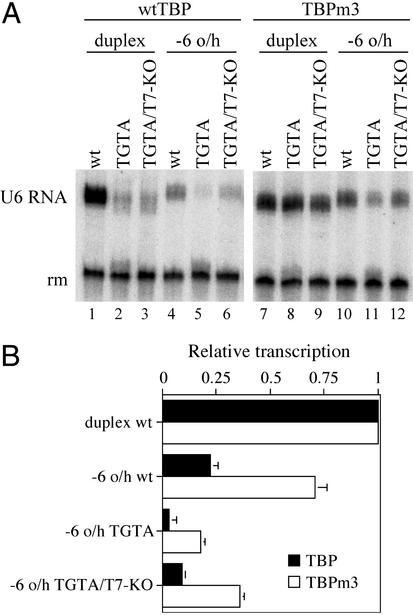
Rescue of the TGTA Mutation by TBPm3.
Three mutations changing amino acid side chains that are located close to DNA in the TBP–DNA complex confer relaxed TATA box recognition on TBPm3 (17–19). In particular, TBPm3 restores pol III transcription activity to the U6 A-29 → G (TATA → TGTA) mutant promoter (9). TFIIIB assembled with TBPm3 was examined for the ability to rescue transcription of TATA box and other mutations in the 5′ overhang context (Fig. 3). TBPm3 restored full transcriptional activity to the mutant TGTA element in duplex DNA (Fig. 3, compare lanes 1–3 with lanes 7–9) and generally increased transcription of −6 o/h constructs relative to the corresponding duplex DNA (for example, compare lane 10 relative to lane 7 with lane 4 relative to lane 1). However, substituting TBPm3 for TBP consistently had only a limited effect on transcription of the TGTA and TGTA/T7-KO mutant −6 o/h constructs relative to the corresponding “wild-type” promoter (compare lanes 11 and 12 relative to lane 10 with lanes 5 and 6 relative to lane 4; data not shown). Mitigation of the effect of TATA → TGTA on transcription by the T7 stretch mutation T7-KO in the 5′ overhang construct was retained with TFIIIB(TBPm3) (compare lanes 11 and 12).
Figure 3.
Effect of TBPm3 on transcription. (A) Transcriptional activity of TGTA-containing duplex and −6 o/h promoters was tested in the presence of TFIIIB assembled with wild-type TBP (lanes 1–6) or TBPm3 (lanes 7–12). Template nomenclature follows Fig. 2A. (B) Transcriptional activity in the presence of TBP (filled bars) or TBPm3 (open bars) relative to duplex DNA. Error bars are SEM of at least three independent experiments.
In summary, TBPm3 reduces the stringency of recognition of the single-stranded (nontranscribed strand) U6 promoter but exerts a smaller specific effect on the TGTA/TATA discrimination.
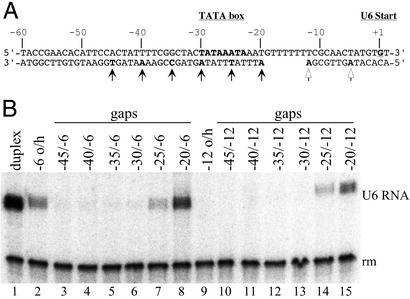
The Extent of the Single-Stranded Promoter.
Two approaches were taken to determine the upstream extent of single-stranded DNA required for activity of the U6 promoter as 5′ overhanging DNA. One group of constructs explored whether making segments of upstream DNA double-stranded would interfere with transcription of −6 o/h DNA or restore activity to the TFIIIB–pol III complex assembled on −12 o/h DNA (Fig. 4A). Possible problems of inhibition of transcription that might have been caused by contaminating short template strand DNA fragments were minimized by gel purification of these gapped DNA constructs (see Methods) and by DNA titration. Activity of the −6 o/h series of gap templates was lost when the DNA segment upstream of nucleotide −25 was double-stranded. Even conversion of the 5′ terminal 16 nucleotides to duplex DNA (construct −45/−6) inactivated transcription, although the base pair −60/−45 segment of the U6 promoter is not required for transcription of, or TFIIIB complex formation on, fully duplex DNA (data not shown). Transcriptional activity of the −12 o/h construct was restored by making the base pairs −60 to −20 segment double-stranded (the −20/−12 gap template) and also partially recovered in the −25/−12 construct. The 5′ ends of these transcripts were shifted predominantly to base pair −8, whereas the start sites on the active −6 o/h gap constructs (−25/−6 and −20/−6) remained at base pair −2 (data not shown). These locations of start sites have been noted for transcription of duplex DNA with simple transcribed strand breaks at base pairs −12 and −6, respectively (7).
Figure 4.
Transcription activity of gapped promoters. (A) Gapped promoter templates. The upstream edge of the single-stranded gap is indicated by a solid arrow for each construct; open arrows indicate the downstream edge of the gap for the −12 o/h and −6 o/h series, respectively. (B) Transcription with gapped promoter templates. TFIIIB-dependent transcription was carried out with the gapped promoters (lanes 3–8 and 10–15) and with duplex DNA and −12 o/h and −6 o/h constructs (lanes 1, 2, and 9, respectively) as controls/references. The nomenclature follows A: e.g., −35/−6 is the construct with a transcribed strand gap extending from nucleotide −34 to nucleotide −7. (The transcription start site of the active −6 gap constructs is at base pair −2; the start site for the active −12 gap constructs is at base pair −8.)
The other approach taken was to progressively truncate the nontranscribed strand from its 5′ end. Even the truncation to nucleotide −45 eliminated TFIIIB-specific transcription, and more extensive truncations, to nucleotides −40, −35, −30, and −25, also were completely inactive (data not shown). Transcription activity of duplex DNA was retained substantially even for deletion to base pair −30 (data not shown); evidently, the Bdp1 contact with duplex DNA upstream of the TATA box (21) and stable TFIIIB–DNA complex formation (22) are not activity-limiting under these assay conditions.
These experiments taken together specify that the architecture of the U6 promoter is changed entirely when the double-stranded DNA segment comprising its TATA box and neighboring sequence is replaced by its nontranscribed single strand.
A 5′ Overhang Promoter Restores Activity to Transcription-Defective Bdp1 and Brf1 Mutants.
The requirement of the 5′ overhang U6 promoter for a double-strand–single-strand junction downstream of base pair −12 (Fig. 1) is compatible with two interpretations that are not necessarily mutually exclusive: (i) the essential T7 segment (nucleotide −18 to −12) of the 5′ overhang promoter (Fig. 2) has to be single-stranded, and (ii) TFIIIB assembled on the nontranscribed strand of the 5′ overhang promoter is defective in promoter opening and placing the 3′ end of the transcribed strand within the boundaries of the transcription bubble (Fig. 1) restores transcription by facilitating promoter opening. The next experiments pursue the latter possibility.
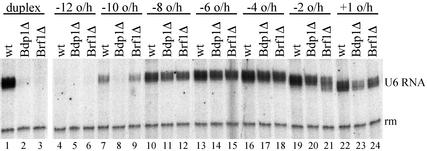
TFIIIB assembled with certain Brf1 and Bdp1 deletion proteins recruits pol III to the promoter but does not transcribe linear DNA because of a failure in promoter opening (ref. 20; Fig. 5, lanes 1–3). Transcription can be restored by unpairing short segments of the transcription bubble (6) and also by placing breaks in the transcribed strand segment that is ultimately converted to the transcription bubble (7). The 5′ overhang constructs shown in Fig. 1A were examined for the ability to rescue the transcription activity of TFIIIB assembled with Bdp1Δ355–372 or Brf1NΔ68.
Figure 5.
Transcription with mutant TFIIIB subunits. Transcription of the indicated 5′ overhang templates and duplex DNA was carried out in the presence of wild-type TFIIIB (wt), TFIIIB assembled with Bdp1(Δ355–372) substituting for Bdp1 (Bdp1Δ), or Brf1NΔ68 substituting for Brf1 (Brf1Δ). Lanes 1–3 and 4–24 are from separate experiments.
Fig. 5 shows that defects of TFIIIB assembled with either of these deletion proteins were rescued specifically by 5′ overhang constructs with transcribed 3′ ends between base pairs −10 and +1, resembling their requirements for transcription with wild-type TFIIIB (Fig. 1C). Transcription required each TFIIIB subunit (data not shown), as is the case for wild-type TFIIIB (Fig. 1D), and start sites for transcription remained (essentially) as for wild-type TFIIIB-dependent transcription of the same constructs (data not shown). We conclude that placing the transcribed strand 3′ end between base pairs −10 and +1 in these 5′ overhang constructs does facilitate promoter opening. The results also suggest that TFIIIB contributes pol III recruitment and postrecruitment functions in transcription of these templates with their (partially) single-stranded promoter just as it does in transcription of duplex DNA.
Discussion
The primary focus and interest of this work is in revealing an unsuspected DNA–structural context for TFIIIB-requiring and accurately initiating pol III transcription and, as we discuss below, suggesting an architectural role for DNA in supporting the postrecruitment functions of TFIIIB in promoter opening.
An upstream segment of the U6 gene nontranscribed strand serves as a promoter. Single-stranded promoters also are known to function in vivo. The best-studied single-stranded promoters direct transcription of N4 virus early genes by the virion RNA polymerase, which is injected into the (Escherichia coli) host cell along with the viral genome at infection. N4 early promoters require specific DNA sequence within a secondary structure consisting of a 5-bp stem and 3-nt loop (23, 24). The N4 virion RNA polymerase is a highly diverged member of the single-subunit RNA polymerase family, of which the phage T7 polymerase is the prototype (25). In eukaryotes, single-subunit RNA polymerases, encoded by nuclear genes, are located in mitochondria and chloroplasts (26). Single-stranded promoters also are recognized by the multisubunit E. coli RNA polymerase during conjugal DNA-strand transfer and direct RNA primer synthesis in rolling circle DNA replication, as well as transcription of minicircle single-stranded DNA (27–30). Even single-stranded RNA (hepatitis delta virus negative strand) may serve as a promoter (of eukaryotic RNA polymerase II) (31).
The promoter elements of the U6 nontranscribed strand and of the standard (duplex DNA) U6 gene differ. The TATA element remains essential, and the TATA → TGTA mutation debilitates the nontranscribed strand promoter, just as it does on the duplex DNA promoter. However, the T7 segment is also an essential promoter element: it cannot be “inverted” (to A7), and changing three Ts (T-16/T-14) to purine (GAG) more severely weakens the nontranscribed strand promoter than it does the duplex DNA promoter. Most strikingly, a far-upstream-lying segment (nucleotides −60 to −45) is an essential component only of the nontranscribed strand promoter. Converting this nucleotide −60/−45 segment to duplex DNA completely inactivates the nontranscribed strand promoter. Extending the double-strand conversion into the TATA box restores activity (Fig. 4) but transforms the promoter to its standard (duplex DNA) form, as indicated by the fact that the transcribed strand 3′ end is no longer constrained to the characteristic nucleotide −10/−1 band of the nontranscribed strand promoter (Fig. 4). TATA box and T7 stretch changes are individually inactivating but compensatory when combined in the nontranscribed strand promoter, indicating a linkage between these two elements.
These differences specify an entirely distinctive interaction of the nontranscribed strand promoter with TFIIIB, despite a retained involvement of the TATA box. We see two additional manifestations of this exceptionality. (i) TBPm3, which relaxes the requirement for A at TATA, generally elevates the activity of nontranscribed strand promoters (relative to duplex DNA), including the promoter with wild-type U6 gene sequence. However, TBPm3 does not selectively rescue TATA → TGTA in the nontranscribed strand promoter above this generally elevated background, although it clearly restores activity to the duplex DNA (standard) TGTA promoter. (ii) The nontranscribed strand promoter does not discriminate against TFIIIB assembled with deletion variants of Brf1 and Bdp1 that inactivate transcription of linear duplex DNA.
What is the structure of the nontranscribed strand promoter? Its two salient characteristics, (i) its extent, with essential components located between nucleotides −60 and −45, at the TATA box, and in the downstream T7 segment between nucleotides −18 and −12, and (ii) its mutual compensation, instead of additive effect of separately deleterious mutations, suggests that the nontranscribed strand promoter may fold into a secondary structure. Alternative secondary structure models of the nontranscribed strand promoter can be constructed, but are only modestly stable (calculated ΔG0 −6 to −10 kcal/mol at 0°C). No model converts the entire TATA box to a double-stranded form. Of course, it is the stability of any putative secondary structure in a TFIIIB or TFIIIB/pol III complex that is functionally significant, which means that secondary structure features will need to be elucidated empirically.
The ability of TBPm3 to generally elevate transcription at the nontranscribed strand U6 promoter also suggests that TATA box–TBP interactions of the nontranscribed strand and duplex DNA promoters differ. The three point mutations in TBPm3, which create enough space to accommodate the C2-NH2 group of G pointing into the minor groove of the TGTA mutant TATA box, may also favor TFIIIB binding to the nontranscribed strand promoter by relaxing other constraints on TBP–DNA interaction. It has been argued that TBP principally recognizes the differential deformability of the TATA box in duplex DNA (32). TBP also makes approximately equal numbers of contacts with the sugar–phosphate backbone and the floor of the minor groove in its TATA box complexes. There is even an isolated report of considerable affinity of TBP for a TATA box in single-stranded DNA (33).
We suggest that TFIIIB-dependent transcription supported by the nontranscribed strand U6 promoter (at least in part) reflects the ability of TBP to interact specifically with noncanonical DNA structure. Whether the nontranscribed strand of the TATA box is recognized by TBP in TFIIIB as single-stranded or, as we suspect, in a form with partial secondary structure remains to be determined.
The nontranscribed strand promoter requires a specifically located single-stranded–double-stranded DNA junction, with the 3′ end of the transcribed strand placed within the confines of the transcription bubble. This suggests two interpretations that are not mutually exclusive.
(i) If the T7 promoter element of the nontranscribed strand promoter must be single-stranded, encroachment by the transcribed strand 3′ end upstream of base pair −10 would be prohibited. In contrast, the T7 segment of the standard promoter does not have to be double-stranded, as evidenced by activity of the −20/−12 construct (Fig. 4).
(ii) The nontranscribed strand promoter may fail to support the role of TFIIIB in promoter opening (6). Placing a transcribed strand 3′ end within the base pair −10/−1 segment (corresponding to the upstream and central segments of the transcription bubble) facilitates promoter opening by TFIIIB assembled with BrfNΔ68 or Bdp1Δ355–372, which is defective in opening this promoter in linear duplex DNA (7). We suggest that the required transcribed strand 3′ end placement in the base pair −10/−1 segment signifies that the nontranscribed strand U6 promoter has an interesting defect: it provides a scaffold for TFIIIB placement that suffices for pol III recruitment but fails at a subsequent step of the reaction sequence that leads to initiation of transcription.
Acknowledgments
We thank L. B. Rothman-Denes and J. T. Kadonaga for helpful comments on the manuscript and the National Institute of General Medical Sciences for support of this research.
Abbreviations
- TFIIIB
transcription factor IIIB
- TBP
TATA-binding protein
- pol III
RNA polymerase III
- o/h
5′ overhang
References
- 1.White R J. RNA Polymerase III Transcription. New York: Springer; 1998. [Google Scholar]
- 2.Geiduschek E P, Kassavetis G A. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 3.Schramm L, Hernandez N. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 4.Dieci G, Percudani R, Giuliodori S, Bottarelli L, Ottonello S. J Mol Biol. 2000;299:601–613. doi: 10.1006/jmbi.2000.3783. [DOI] [PubMed] [Google Scholar]
- 5.Hamada M, Huang Y, Lowe T M, Maraia R J. Mol Cell Biol. 2001;21:6870–6881. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassavetis G A, Letts G A, Geiduschek E P. EMBO J. 2001;20:2823–2834. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassavetis G A, Grove A, Geiduschek E P. EMBO J. 2002;21:5508–5515. doi: 10.1093/emboj/cdf533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grove A, Adessa M S, Geiduschek E P, Kassavetis G A. EMBO J. 2002;21:704–714. doi: 10.1093/emboj/21.4.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehall S K, Kassavetis G A, Geiduschek E P. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 10.Kassavetis G A, Letts G A, Geiduschek E P. EMBO J. 1999;18:5042–5051. doi: 10.1093/emboj/18.18.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Kassavetis G A, Geiduschek E P, Hambalko M, Brent C J. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 13.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlach V L, Whitehall S K, Geiduschek E P, Brow D A. Mol Cell Biol. 1995;15:1455–1466. doi: 10.1128/mcb.15.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M P, Gerlach V L, Brow D A. Mol Cell Biol. 2001;21:6429–6439. doi: 10.1128/MCB.21.19.6429-6439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joazeiro C A, Kassavetis G A, Geiduschek E P. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 17.Strubin M, Struhl K. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 18.Kim J L, Nikolov D B, Burley S K. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 20.Kassavetis G A, Kumar A, Letts G A, Geiduschek E P. Proc Natl Acad Sci USA. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah S M, Kumar A, Geiduschek E P, Kassavetis G A. J Biol Chem. 1999;274:28736–28744. doi: 10.1074/jbc.274.40.28736. [DOI] [PubMed] [Google Scholar]
- 22.Colbert T, Lee S, Schimmack G, Hahn S. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X, Rothman-Denes L B. Genes Dev. 1998;12:2782–2790. doi: 10.1101/gad.12.17.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glucksmann M A, Markiewicz P, Malone C, Rothman-Denes L B. Cell. 1992;70:491–500. doi: 10.1016/0092-8674(92)90173-a. [DOI] [PubMed] [Google Scholar]
- 25.Kazmierczak K M, Davydova E K, Mustaev A A, Rothman-Denes L B. EMBO J. 2002;21:5815–5823. doi: 10.1093/emboj/cdf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cermakian N, Ikeda T M, Miramontes P, Lang B F, Gray M W, Cedergren R. J Mol Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 27.Masai H, Arai K. Cell. 1997;89:897–907. doi: 10.1016/s0092-8674(00)80275-5. [DOI] [PubMed] [Google Scholar]
- 28.Ohmichi T, Maki A, Kool E T. Proc Natl Acad Sci USA. 2002;99:54–59. doi: 10.1073/pnas.012589099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Althorpe N J, Chilley P M, Thomas A T, Brammar W J, Wilkins B M. Mol Microbiol. 1999;31:133–142. doi: 10.1046/j.1365-2958.1999.01153.x. [DOI] [PubMed] [Google Scholar]
- 30.Higashitani A, Higashitani N, Horiuchi K. Proc Natl Acad Sci USA. 1997;94:2909–2914. doi: 10.1073/pnas.94.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modahl L E, Macnaughton T B, Zhu N, Johnson D L, Lai M M. Mol Cell Biol. 2000;20:6030–6039. doi: 10.1128/mcb.20.16.6030-6039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 33.Sokolenko A A, Sandomirskii I I, Savinkova L K. Mol Biol (Moscow) 1996;30:279–285. [PubMed] [Google Scholar]