Abstract
Considerable progress has been made in identifying the molecular composition of complex signaling networks controlling cell proliferation, differentiation, and survival. However, to discover general building principles and predict the dynamic behavior of signaling networks, it is necessary to develop quantitative models based on experimental observations. Here we report a mathematical model of the core module of the Janus family of kinases (JAK)–signal transducer and activator of transcription (STAT) signaling pathway based on time-resolved measurements of receptor and STAT5 phosphorylation. Applying the fitted model, we can determine the quantitative behavior of STAT5 populations not accessible to experimental measurement. By in silico investigations, we identify the parameters of nuclear shuttling as the most sensitive to perturbations and verify experimentally the model prediction that inhibition of nuclear export results in a reduced transcriptional yield. The model reveals that STAT5 undergoes rapid nucleocytoplasmic cycles, continuously coupling receptor activation and target gene transcription, thereby forming a remote sensor between nucleus and receptor. Thus, dynamic modeling of signaling pathways can promote functional understanding at the systems level.
Signaling pathways form intracellular networks that control proliferation, differentiation, and survival. Complexity arises from the large number of molecules involved in signal processing and the various interactions between them. Although considerable progress has been made in identifying the components of signaling networks, the regulation of information processing is poorly understood. To understand these networks at a system level and predict the outcome of perturbations, the dynamic interactions of individual components have to be simultaneously examined, which is facilitated by quantitative mathematical modeling (1). Previous attempts to model signaling pathways have been primarily based on qualitative data reflecting the interactions between the components and on simulations with ad hoc fixed parameters (2–4). However, if discrepancies between the mathematical model and experimental observations occur, it is not possible with this approach to decide whether the underlying biological model is incorrect or whether the parameters have been ill chosen. To resolve the dilemma and quantitatively predict the dynamic behavior of signaling pathways, databased models are required (5, 6).
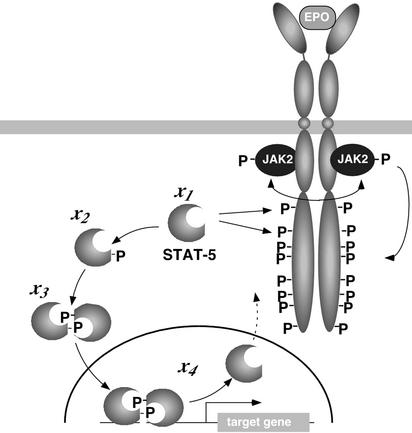
To introduce meaningful simplifications and establish a mechanism-based model, detailed qualitative knowledge of a signaling pathway is necessary. A signaling pathway that has been studied in great detail is the Janus family of kinases (JAK)–signal transducer and activator of transcription (STAT) pathway (7). This pathway is involved in signaling through multiple cell surface receptors, including receptor tyrosine kinases (8), G-protein-coupled receptors (9), and in particular hematopoietic cytokine receptors such as the erythropoietin receptor (EpoR) (10). Signal transduction through the EpoR is essential for proliferation and differentiation of erythroid progenitor cells. Binding of the hormone Epo to the receptor activates the receptor-bound tyrosine kinase JAK2 and results in tyrosine phosphorylation of the EpoR cytoplasmic domain, thereby creating docking sites for downstream signaling molecules, including the latent transcription factor STAT5 (11). The core module of the JAK-STAT pathway mediating rapid signal transduction from the cell surface receptor to the nucleus (Fig. 1) is represented by STAT5, which is tyrosine phosphorylated on recruitment to the activated receptor, dimerizes, and migrates to the nucleus (7), where it stimulates the transcription of target genes as, e.g., the gene encoding cytokine inducible SH2 domain-containing protein (CIS; ref. 12). As a general shutoff mechanism for the JAK-STAT pathway, proteasome-dependent or ubiquitin-associated degradation of STATs in the nucleus has been proposed (13), whereas evidence by others suggests that STATs are relocated back to the cytoplasm after dephosphorylation in the nucleus (14).
Fig 1.
The core module of the JAK-STAT signaling pathway. Hormone (Epo) binding to the EpoR results in transphosphorylation (double-headed arrow) of JAK2 and subsequently in tyrosine phosphorylation (P) of JAK2 and the EpoR cytoplasmic domain. Phosphotyrosine residues 343 and 401 in the EpoR mediate recruitment of monomeric STAT5 (x1). Upon receptor recruitment, monomeric STAT5 is tyrosine phosphorylated (x2), dimerizes (x3), and migrates to the nucleus (x4), where it binds to the promoter of target genes and is dephosphorylated and exported to the cytoplasm. The arrow with the dashed line indicates the possibility for STAT5 to reenter the cytoplasm.
To establish a databased mathematical model of the JAK-STAT signaling pathway, we translated the individual steps of the JAK-STAT core module into four coupled differential equations. We based the structure of the equations on biochemical knowledge and compared models reflecting the assumption of a feed-forward cascade versus a model capturing the cycling capacity of STAT5. The dynamical parameters of the equations were estimated based on time-course experiments, simultaneously measuring changes in phosphorylation of STAT5 and EpoR as well as the total amount of STAT5 in the cytoplasm by quantitative immunoblotting. By mathematical modeling, we identify nucleocytoplasmic cycling as an essential feature of the JAK-STAT core module. We predict that the steps of nuclear import and export are most sensitive to perturbations of the system and experimentally verify this by inhibiting nuclear export.
Materials and Methods
Time-Course Experiments.
The wild-type EpoR was cloned into the retroviral expression vector pMOWS (15) and introduced into BaF3 cells by retroviral transduction as described (16). Cell lines stably expressing the EpoR (BaF3-EpoR) were selected in the presence of puromycin and maintained as described (16). BaF3-EpoR cells were starved in RPMI 1640/1 mg/ml BSA for 3 h and were left unstimulated or were stimulated with 5 units/ml Epo (Cilag–Jansen, Bad Homburg, Germany). For each time point, 107 cells were taken from the pool of cells and lysed by the addition of 2× Nonidet P-40 lysis buffer to terminate the reaction (16). For immunoprecipitations, cytosolic lysates were first incubated with anti-EpoR antiserum (Santa Cruz Biotechnology), and subsequently the supernatant was subjected to immunoprecipitation with anti-STAT5 antiserum (Santa Cruz Biotechnology). For treatment with leptomycin B (LMB), starved BaF3-EpoR cells were incubated for 30 min with 10 ng/ml LMB before Epo addition. Nuclear extracts were prepared by standard procedures (details can be found in the Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org) and were used for immunoprecipitation with anti-STAT5 antiserum. The immunoprecipitates were analyzed by 15% SDS/PAGE followed by immunoblotting analysis using the antiphosphotyrosine monoclonal antibody 4G10 (Upstate Biotechnology, Lake Placid, NY). For reprobing with anti-STAT5 antiserum, the blots were treated with β-mercaptoethanol and SDS, as described (16). The immunoblots were performed under standardized conditions, incubated with enhanced chemiluminescence substrate (Amersham Pharmacia) for 1 min, and exposed for 10 min on a LumiImager (Roche Diagnostics). For quantifications, lumianalyst software (Roche Diagnostics) was used.
β-Galactosidase Assay.
The reporter plasmid pSac-CIS was generated by amplifying the CIS promoter from genomic DNA and inserting the SacI/XhoI subfragment, comprising the two STAT5-binding sites and the authentic transcriptional start site, into pGL2 basic (Promega) and transferring the promoter cassette via KpnI and XhoI into pCMVβ (CLONTECH) using the EcoRI and XhoI restriction sites in the multiple cloning site. By overlap extension, both consensus STAT5-binding sites were altered to AAxxxTTxxxxAAxxxTT, resulting in pSac-CIS-STAT−. For transient transfection, BaF3-EpoR cells were starved in RPMI 1640/1% FCS for 3 h and used for electroporation applying 2 pmol of β-galactosidase reporter plasmids pSac-CIS and pSac-CIS-STAT− and 42 μg of carrier plasmid. Electroporated cells were either left untreated or were pretreated with 10 ng/ml LMB and subsequently stimulated with 5 units/ml Epo. After 120 min, whole-cell extracts were prepared and analyzed for β-galactosidase activity. The β-galactosidase activity determined by duplicate measurements was normalized to the protein content of the lysates determined by Bradford assay and is displayed in relative light units.
Numerical Analysis.
Ordinary differential equations were integrated by 4th order Runge–Kutta (17), and integration of the delay differential equation was performed by using an integration routine similar to the RETARD algorithm (18). For parameter estimation, we used the maximum likelihood estimator with an optimization routine of the Gauss–Newton type by minimizing χ2(x1(t = 0), k⇀) = ∑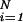 ∑
∑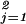 [(y
[(y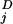 (ti) − y
(ti) − y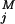 (ti))2/σ
(ti))2/σ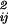 ], where y
], where y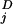 (ti) are the observed data for component j at time ti, and y
(ti) are the observed data for component j at time ti, and y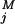 (ti, x1(t = 0), k⇀) is the model trajectory with starting value x1(t = 0) and the parameters k⇀; note that x2(t = 0) = x3(t = 0) = x4(t = 0) = 0. σij were estimated as follows: Residuals at the individual time point were calculated by Sawitzki–Golay filtering (17, 19). Because the measurement process represents a Poisson process, we apply a relative error model and estimate the proportionality constant from the rescaled residuals of duplicated measurements. Derivatives of χ2 with respect to starting values and parameters were computed by using an extrapolation scheme for finite differences, as implemented by Ridder (20). Confidence intervals were computed with help of likelihood contours (17).
(ti, x1(t = 0), k⇀) is the model trajectory with starting value x1(t = 0) and the parameters k⇀; note that x2(t = 0) = x3(t = 0) = x4(t = 0) = 0. σij were estimated as follows: Residuals at the individual time point were calculated by Sawitzki–Golay filtering (17, 19). Because the measurement process represents a Poisson process, we apply a relative error model and estimate the proportionality constant from the rescaled residuals of duplicated measurements. Derivatives of χ2 with respect to starting values and parameters were computed by using an extrapolation scheme for finite differences, as implemented by Ridder (20). Confidence intervals were computed with help of likelihood contours (17).
Results
Deriving a Mathematical Model of the Core Module of the JAK-STAT Signaling Pathway.
To model the pathway, we started with a parameterized model of the JAK-STAT core module reflecting the feed-forward information transfer from the cell surface to the nucleus and monitored ligand induced activation of STAT5 mediated by the EpoR. The individual steps of the core module (Fig. 1) were translated into four coupled differential equations describing the dynamics of the four different STAT5 populations over time
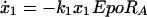 |
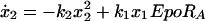 |
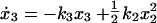 |
 |
where the pool of cytoplasmic STAT5 is represented by x1 (unphosphorylated STAT5), x2 (tyrosine phosphorylated monomeric STAT5), x3 (tyrosine phosphorylated dimeric STAT5), and x4 is the nuclear STAT5. Eqs. 1–4 assume mass-action kinetics for the individual reaction steps. Although this holds for the dimerization process, it represents an approximation for STAT5 phosphorylation and nuclear translocation, assuming that the tyrosine kinase, phosphorylating STAT5 monomers, and the nuclear import machinery are not rate limiting.
Because individual STAT5 populations are experimentally difficult to access, we measured in the cytoplasm the amount of tyrosine phosphorylated STAT5 (y1 = k5(x2 +2x3)) and the total amount of STAT5 (y2 = k6(x1 + x2 + 2x3)). The scaling parameters k5 and k6 were introduced because only relative protein amounts are measured by our quantitative experiments. As input function that determines the STAT5 response, Epo-induced tyrosine phosphorylation of the EpoR (EpoRA, Fig. 1 A and B) was quantified (y3 = k7EpoRA). Due to the mathematical structure of the differential equations, the scaling factor k7 cannot be disentangled from the rate constant k1, and k2 is coupled to x1(0) (a detailed discussion of the identifiable parameter combinations is published as supporting information on the PNAS web site). The identifiable parameter combinations were simultaneously estimated based on results from three independent experiments, whereas the nuisance parameters k5, k6 and k7 were determined separately for each experiment.
Determination of the Dynamic Parameters Based on Time-Course Experiments.
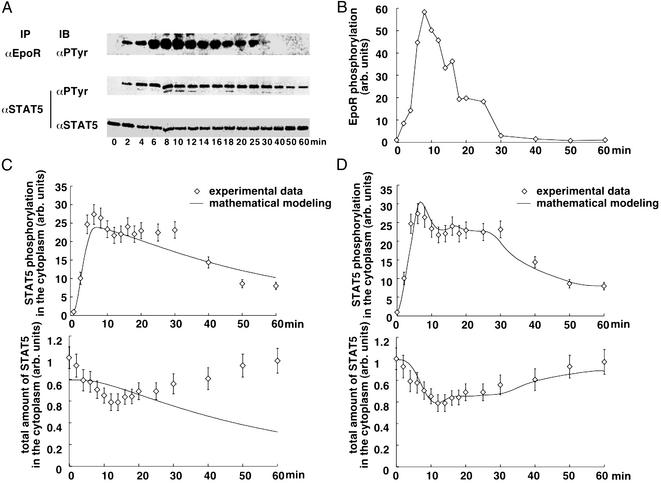
To estimate the parameters, time-course experiments were performed with BaF3 cells expressing the wild-type EpoR (BaF3-EpoR). This model system was chosen because processing (21) and surface expression (S. S. Watowich, personal communication) of the EpoR in BaF3 cells are comparable to erythroid progenitors cells derived from murine fetal liver. Furthermore, it was observed that in these cells, the JAK-STAT pathway is efficiently activated in response to Epo binding (11). Starved BaF3-EpoR cells were stimulated with 5 units/ml Epo for 2–60 min. The amount of tyrosine-phosphorylated EpoR and STAT5 present in the cytoplasmic lysates of these cells was determined by quantitative immunoblotting (Fig. 2A) (details regarding the quantification method can be found in the Supporting Text and Fig. 5, which are published as supporting information on the PNAS web site). The measurements shown in Fig. 2 revealed that both tyrosine phosphorylation of the EpoR and STAT5 increased rapidly upon Epo addition. Whereas after 10–15 min of Epo addition, receptor tyrosine phosphorylation started to decline, STAT5 tyrosine phosphorylation remained at almost maximum levels until receptor phosphorylation dropped below threshold levels after 30 min, resulting in a plateau of STAT5 phosphorylation between 10 and 30 min. The total amount of STAT5 in the cytoplasm initially rapidly declined in response to Epo stimulation but increased again after prolonged incubation times. On the basis of three independent experiments, the parameters were estimated, and the resulting time courses are displayed in Fig. 2C, showing that this model is not able to describe the experimental data.
Fig 2.
Time course (points with error bars) and mathematical modeling (solid lines) of the STAT5 nucleocytoplasmic cycle. (A) Transient activation of EpoR and STAT5. Starved BaF3-EpoR cells were left unstimulated (0) or were stimulated with 5 units/ml Epo for the time indicated. Cytoplasmic extracts were subjected to immunoprecipitation (IP) with anti-EpoR and anti-STAT5 antiserum, and were analyzed by immunoblotting (IB) with the antiphosphotyrosine (PTyr) antibody or reprobed with anti-STAT5 antiserum, followed by enhanced chemiluminescence and detection using a LumiImager. The LumiImager files are displayed. (B) Linear interpolation of EpoR tyrosine phosphorylation as input function for the four-dimensional differential equation. The time course of EpoR tyrosine phosphorylation in response to Epo stimulation was quantified with LUMIANALYST software and is displayed in arbitrary units. C and D show for the cytoplasmic tyrosine phosphorylated STAT5 and the total STAT5 pool in the cytoplasm the measured data in arbitrary units and the corresponding fit obtained with the linear model (C) and the model including nucleocytoplasmic cycling (D).
Because it has been reported that dephosphorylated nuclear STAT molecules can be relocated back into the cytoplasm (22, 23), we changed Eqs. 1 and 4 of the ordinary differential equation model into delay differential equations to include nuclear export (a detailed discussion of model derivation, including tests for other models, can be found in the Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site).
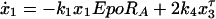 |
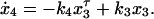 |
The superscript parameter τ represents the time STAT5 molecules reside in the nucleus. It is composed of the time STAT5 is bound to DNA and the time until dephosphorylated STAT5 leaves the nucleus (x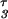 = x3(t − τ)). To preserve mass conservation, the condition k3 ≥ k4 has to hold. If the export rate k4 is smaller than the import rate k3, STAT5 accumulates in the nucleus. The time courses of the fitted model are displayed in Fig. 2D. They describe all features of the experimental data, including the plateau of phosphorylated STAT5 in the cytoplasm between 10 and 30 min. The parameters and their 1σ confidence intervals are: k1 = 0.021 min−1 (+0.004/−0.003)k7, k2 = 2.46min−1mol−1(+1.7/−1.0)/x1(0), k3 = 0.1066min−1 (+0.03/−0.022), k4 = 0.10658 min−1 (+0.0016/−0.0024) and τ = 6.4 min (+0.5/−2.6). As judged from the delay time τ, on average, a single STAT5 molecule remains in the nucleus for ≈6 min. Using these dynamical parameters and tyrosine phosphorylation of the EpoR as input function enabled us to predict the development of cytoplasmic STAT5 in an independent experiment (details can be found in the Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site).
= x3(t − τ)). To preserve mass conservation, the condition k3 ≥ k4 has to hold. If the export rate k4 is smaller than the import rate k3, STAT5 accumulates in the nucleus. The time courses of the fitted model are displayed in Fig. 2D. They describe all features of the experimental data, including the plateau of phosphorylated STAT5 in the cytoplasm between 10 and 30 min. The parameters and their 1σ confidence intervals are: k1 = 0.021 min−1 (+0.004/−0.003)k7, k2 = 2.46min−1mol−1(+1.7/−1.0)/x1(0), k3 = 0.1066min−1 (+0.03/−0.022), k4 = 0.10658 min−1 (+0.0016/−0.0024) and τ = 6.4 min (+0.5/−2.6). As judged from the delay time τ, on average, a single STAT5 molecule remains in the nucleus for ≈6 min. Using these dynamical parameters and tyrosine phosphorylation of the EpoR as input function enabled us to predict the development of cytoplasmic STAT5 in an independent experiment (details can be found in the Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site).
Dynamic Behavior of Unobserved STAT5 Populations.
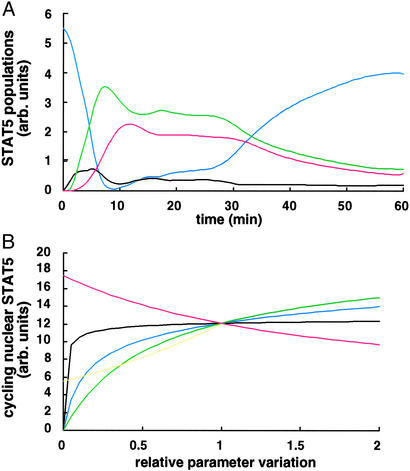
On the basis of the dynamic model, the quantitative behavior of STAT5 populations that are difficult to access experimentally can be estimated (Fig. 3A). The simulation of the fitted model shows that, on activation of the system, tyrosine phosphorylated STAT5 monomers are formed but rapidly converted into STAT5 dimers, providing an explanation for the technical difficulty of detecting monomeric tyrosine phosphorylated STAT5 experimentally. In the cytoplasm, the amount of tyrosine phosphorylated STAT5 dimers reaches an initial maximum after ≈7 min and then declines due to nuclear translocation. Dephosphorylated STAT5 monomers are subsequently exported from the nucleus and rapidly rephosphorylated at the activated receptor, thereby resulting in a second maximum of tyrosine phosphorylated STAT5 dimers in the cytoplasm after ≈17 min. Therefore, by mathematical modeling, we can reveal that the experimentally observed plateau of tyrosine phosphorylated STAT5 in the cytoplasm between 10 and 30 min is the result of repetitive reactivation facilitated by nucleocytoplasmic cycling of STAT5. Fig. 3A shows that unphosphorylated STAT5 in the cytoplasm is limited and approaches zero ≈9 min after Epo stimulation. If in a simulation the nuclear delay time is increased to 30 min, it is not possible to describe the plateau of tyrosine phosphorylated STAT5 in the cytoplasm, because STAT5 re-enters the pool of cytoplasmic STAT5 molecules when the EpoR is already in an inactive state (data not shown). Therefore, the overall nuclear sojourn time of activated STAT5 mediating target gene activation depends on efficient nuclear export and on the presence of activated EpoR. Databased mathematical modeling identifies nucleocytoplasmic cycling as an essential behavior of the JAK-STAT core module.
Fig 3.
In silico investigations. (A) Time courses of unobserved individual STAT5 populations. Depicted is the predicted quantitative behavior of unphosphorylated STAT5 (blue line), tyrosine phosphorylated STAT5 monomers (black line) and dimers (green line) in the cytoplasm, and cycling activated STAT5 molecules in the nucleus (red line). (B) Predicted effect of parameter variations on target gene activation. As an indirect indicator for target gene activation, the amount of nuclear activated STAT5 involved in cycling was determined by calculating the area under the red curve in A. The effect of relative changes of the dynamical parameters k1 (black line), k2 (blue line), k3 (green line), k4 (yellow line), and τ (red line) on the integrated area is shown.
In Silico Determination of Steps Most Sensitive to Perturbations.
On the basis of our fitted model, we evaluated the effect of relative parameter changes on the amount of nuclear tyrosine phosphorylated STAT5 involved in cycling that should be proportional to target gene activation. This in silico investigation allows determination of which step of the STAT5 nucleocytoplasmic cycle is most sensitive to perturbations. As indicated by the slope of the curves in Fig. 3B, changing the parameters of nuclear cycling (k3, k4, and τ) had the most significant effect on the amount of STAT5 in the nucleus, whereas changes in the rate of monomer–dimer interconversion had a minor impact. By setting the nuclear delay time τ to infinity or alternatively k4= 0, thus suppressing nuclear export, we could estimate that a single STAT-activation cycle contributes only 45% of activated STAT5 in the nucleus compared with successive STAT nucleocytoplasmic cycles.
Experimental Verification of the Importance of Nucleocytoplasmic Cycling.
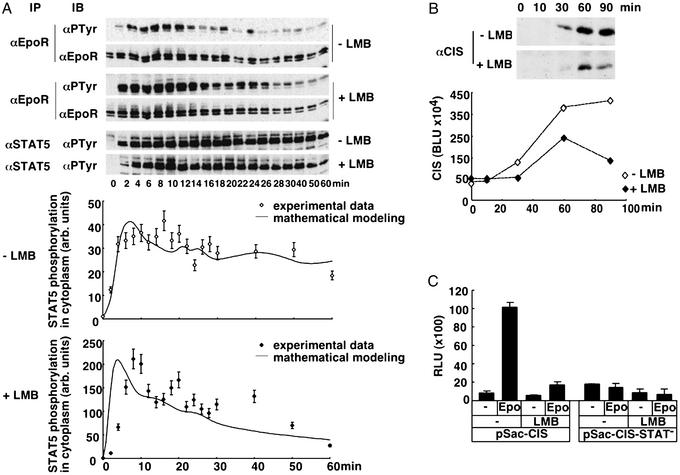
In accordance with the prediction, the experimental data presented in Fig. 4A show that in the presence of the nuclear export inhibitor LMB, the amount of tyrosine phosphorylated STAT5 in the cytoplasm declines more rapidly than in the absence of the inhibitor. The time course of EpoR tyrosine phosphorylation was not reduced by the addition of LMB but rather, for unexplained reasons, enhanced. Mathematical analysis of the experimental data revealed that the presence of LMB decreased the nuclear export rate (k4) by 60% (Fig. 4A). As judged from the relative parameter variation shown in Fig. 4B, this results within 60 min in a 40% reduction of activated STAT5 in the nucleus involved in cycling and thus should reduce the output of the system significantly. To experimentally test our prediction, we analyzed the induction of the endogenous STAT5 target gene CIS in the absence or presence of LMB. To minimize nonspecific effects of LMB, the incubation times were limited to 30-min preincubation and maximally 90-min experimental measurements. As shown in Fig. 4B, CIS was induced in BaF3-EpoR cells on Epo stimulation and reached maximum levels after 60–90 min. In agreement with our prediction, the presence of LMB resulted in a 37% reduction of CIS after 60 min of Epo addition, and this increased to a 66% reduction at 90 min. To confirm that this effect was mediated by a reduced efficiency of the JAK-STAT pathway, we performed reporter gene assays (Fig. 4C), comparing the Epo induced induction of β-galactosidase by reporter gene vectors harboring the CIS promoter limited to the STAT5-binding sites (pSac-CIS) or harboring point mutations that inactivate the respective sites (pSac-CIS-STAT−). In the absence of LMB, the addition of Epo for 120 min resulted in a 12-fold increase in β-galactosidase activity produced by the reporter gene vector pSac-CIS, whereas Epo had no effect on the reporter gene vector pSac-CIS-STAT− that lacks the STAT5-binding sites. The presence of LMB resulted in a general 2-fold reduction of β-galactosidase activity in the absence of Epo- or STAT5-binding sites, whereas the STAT5-specific yield of Epo induced β-galactosidase activity determined after 120 min was reduced by 70%. Thus, as predicted by our model, impaired nuclear export of STAT5 results in a reduced yield of the system, and the effect increases over time.
Fig 4.
Effect of impaired nuclear export on the amount of activated STAT5 in the cytoplasm and the transcriptional yield. (A) Time courses of cytoplasmic STAT5 and EpoR phosphorylation as LumiImager files (Upper) and corresponding quantification of STAT5 phosphorylation. Starved BaF3-EpoR cells untreated or pretreated for 30 min with 10 ng/ml LMB were stimulated with 5 units/ml Epo for the time indicated. Cytoplasmic extracts were subjected to immunoprecipitation with anti-STAT5 antiserum followed by immunoblotting analysis with an antiphosphotyrosine antibody. The delayed onset of signal activation in the presence of LMB is possibly due to the mild detergent function of LMB. The experimental data quantified with LUMIANALYST software is displayed in arbitrary units. (B) Reduced target gene production upon impaired nucleocytoplasmic cycling of STAT5. Starved BaF3-EpoR cells were left untreated or were pretreated with LMB and stimulated with 5 units/ml Epo for the time indicated. Cytoplasmic extracts were subjected to immunoprecipitation with anti-CIS antiserum raised against a GST-CIS fusion protein and analyzed by immunoblotting with anti-CIS antiserum followed by chemiluminescence and LumiImager detection. The quantification of a representative experiment performed using the LUMIANALYST software is shown in Boehringer Light Units (BLU). (C) Effect of nuclear export inhibition on activation of a STAT5 reporter gene. The STAT5 reporter construct pSac-CIS and as a control pSac-CIS-STAT− lacking the STAT5-binding sites were introduced by electroporation into starved BaF3-EpoR cells. The cells either were left untreated or were pretreated with LMB and then unstimulated or stimulated with 5 units/ml Epo. After 120 min, cell lysates were prepared and used for the determination of β-galactosidase activity. The results are displayed in relative light units (RLU) and represent the mean of triplicate measurements ± SD normalized to protein content.
Discussion
In summary, we demonstrate that a databased quantitative model of a signaling pathway can be formulated and applied to determine the dynamic behavior of the system in response to external and internal changes.
So far, the analysis of signaling pathways by mathematical models has been performed in the framework of simulations. For example, in the case of the mitogen-activated protein kinase, three different models were investigated by simulations (4, 24, 25). A comparison of the first two models (26) revealed that these models exhibit different properties, thus emphasizing the necessity for experimental data to enable a decision between the different models. In this type of study, the structure of the equations is based on assumptions about the underlying biology, and the parameters are taken from published data often obtained under different conditions or even from different organisms. This poses a fundamental problem to infer systemic properties of signaling networks: It is difficult to decide whether the results of the simulations reflect the underlying biology or the specific choice of the parameters. Therefore, quantitative time-resolved measurements for simultaneous estimation of the dynamical parameters are critical to achieve models of high predictive power (24).
To establish a databased dynamic model, it is advantageous to introduce simplifications limiting the number of equations and parameters. In our databased mathematical model, we used tyrosine phosphorylation of the EpoR as input function, because it summarizes the effect of JAK2 activation as well as the impact of negative regulatory molecules including SOCS family members (27) and SHP-1 (16). Our studies were performed in the model cell line BaF3, which can, in principle, express an order of magnitude more molecules EpoR than erythroid progenitor cells. However, the model should be applicable to signal transduction in primary cells, because the number of receptors present on the cell surface and therefore involved in signal transduction is comparable in both systems. Not included in our current model is the protein inhibitor of activated STAT (PIAS) and potential cytoplasmic STAT dephosphorylation, because the nucleus has been identified as the major compartment for STAT dephosphorylation (22). Furthermore, we summarize by the fixed nuclear delay translocation the on/off interaction of STAT5 with nuclear DNA-binding sites as well as the nuclear dephosphorylation reaction and export. Despite the necessary simplifications, the trajectories of the fitted model closely reproduce the experimental data, hence supporting our dynamic model.
The analysis of the JAK-STAT pathway has focused on the unidirectional flow of information from the cell surface to the nucleus, and proteasome-dependent or ubiquitin-associated degradation of STATs has been proposed as a shutoff mechanism (13). In contrast, we demonstrate by databased mathematical modeling that the yield of the system is determined by successive nucleocytoplasmic cycles, depending on the presence of the input signal. This is supported by recent biochemical and imaging studies showing that STAT5B can shuttle between the cytoplasm and the nucleus in a cytokine-dependent manner (28). Nuclear import and export of the closely related transcription factor STAT1 have been studied in detail, showing that STAT1 is dephosphorylated in the nucleus (22), exported to the cytoplasm, and potentially enters additional reactivation cycles (22, 23, 29). A recent report on STAT6 signaling provides biochemical evidence that continuous cycling of STAT6 is required, and that the estimated half-life of activated STAT6 is ≈5 min (30). In agreement with this observation, we show by mathematical modeling that the nuclear sojourn time of STAT5 is ≈6 min.
By in silico investigations simulating the effects of parameter changes on the amount of nuclear activated STAT5 involved in cycling, we identify the parameters for nuclear import and export as most sensitive for perturbation of the system. To experimentally verify this prediction, we used the antibiotic LMB that inhibits CRM1-dependent nuclear export (31). In STAT1, a leucine-rich nuclear export signal recognized by CRM1 has been located near the DNA-binding domain, and imaging studies revealed that the presence of LMB inhibited STAT1 relocalization to the cytoplasm (23). Reporter gene assays showed that the presence of LMB had an effect similar to the use of a STAT1 mutant impaired in nuclear export and resulted in a reduced yield of reporter gene activation (29). Moreover, recent reports demonstrate that prolonged treatment of cells with LMB leads to reduced levels of nuclear STAT6-binding activity in response to IL-4 stimulation (30), and that the nuclear export of STAT5B by cytokine depletion is inhibited by LMB (28). In keeping with our in silico prediction, we show that in the presence of LMB, the amount of tyrosine phosphorylated STAT5 present in the cytoplasm decays more rapidly and results in a reduced transcriptional yield.
Applying our databased mathematical model, we can quantify, to our knowledge for the first time, the contribution of successive nucleocytoplasmic cycles and predict that the first STAT5 activation cycle contributes only to ≈45% to the transcriptional yield, indicating that repetitive activation cycles are required for effective signal transmission. Because the functional STAT transactivation complex assembles in the nucleus unable to directly detect fluctuations in extracellular signals (32), rapid cycling between receptor and nucleus allows STAT proteins to tightly couple target gene activation to the changes in extracellular hormone levels. Thereby STATs form a remote sensor to monitor the activation status of the system identifying nucleocytoplasmic cycling as an important systems property of the JAK-STAT signaling pathway. Thus, our functional analysis of a signaling pathway by mathematical modeling in comparison with experimental data leads from a static description of signaling events to a dynamic understanding.
Because in a variety of human tumors oncogenic tyrosine kinases or autocrine loops result in constitutively activated STATs contributing to malignant transformation and tumor progression, the in silico identification of targets for efficient disruption of the pathway is of great medical interest. By analyzing the effect of parameter changes on the behavior of the system, we show that, in contrast to the previous focus of research, the steps of nuclear export and import are targets for effective intervention of the JAK-STAT signal pathway.
Supplementary Material
Acknowledgments
We thank R. Kehlenbach and B. Stenkamp for helpful discussions and suggestions. We are also grateful to L. C. Cantley and H.-P. Herzel for critically reading the manuscript. We thank U. Baumann, A. Walker, and A. Geist for technical help and F. U. Reuss for technical advice. For the LMB, we are indebted to M. Yoshida (University of Tokyo, Tokyo).
Abbreviations
STAT, signal transducer and activator of transcription
JAK, Janus family of kinases
LMB, leptomycin B
Epo, erythropoietin
EpoR, Epo receptor
CIS, cytokine inducible SH2 domain-containing protein
References
- 1.Kitano H. (2002) Science 295, 1662-1664. [DOI] [PubMed] [Google Scholar]
- 2.Fussenegger M., Bailey, J. E. & Varner, J. (2000) Nat. Biotechnol. 18, 768-774. [DOI] [PubMed] [Google Scholar]
- 3.Bhalla U. S. & Iyengar, R. (1999) Science 283, 381-387. [DOI] [PubMed] [Google Scholar]
- 4.Huang C. Y. & Ferrell, J. E. (1996) Proc. Natl. Acad. Sci. USA 93, 10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endy D. & Brent, R. (2001) Nature 409, 391-395. [DOI] [PubMed] [Google Scholar]
- 6.Timmer J., Rust, H., Horbelt, W. & Voss, H. U. (2000) Phys. Lett. A 274, 123-134. [Google Scholar]
- 7.Horvath C. M. (2000) Trends Biochem. Sci. 25, 496-502. [DOI] [PubMed] [Google Scholar]
- 8.Schlessinger J. (2000) Cell 103, 211-225. [DOI] [PubMed] [Google Scholar]
- 9.Williams J. G. (1999) Trends Biochem. Sci. 24, 333-334. [DOI] [PubMed] [Google Scholar]
- 10.Constantinescu S. N., Ghaffari, S. & Lodish, H. F. (1999) Trends Endocrinol. Metab. 10, 18-23. [DOI] [PubMed] [Google Scholar]
- 11.Klingmüller U., Bergelson, S., Hsiao, J. G. & Lodish, H. F. (1996) Proc. Natl. Acad. Sci. USA 93, 8324-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura A., Ohkubo, T., Kiguchi, T., Jenkins, N. A., Gilbert, D. J., Copeland, N. G., Hara, T. & Miyajima, A. (1995) EMBO J. 14, 2816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim T. K. & Maniatis, T. (1996) Science 273, 1717-1719. [DOI] [PubMed] [Google Scholar]
- 14.Koster M. & Hauser, H. (1999) Eur. J. Biochem. 260, 137-144. [DOI] [PubMed] [Google Scholar]
- 15.Ketteler R., Glaser, S., Sandra, O., Martens, U. M. & Klingmüller, U. (2002) Gene Ther. 9, 477-487. [DOI] [PubMed] [Google Scholar]
- 16.Klingmüller U., Lorenz, U., Cantley, L. C., Neel, B. G. & Lodish, H. F. (1995) Cell 80, 729-738. [DOI] [PubMed] [Google Scholar]
- 17.Press W. H., Teukolsky, S. A., Vetterling, W. T. & Flannery, B. P., (1992) Numerical Recipes (Cambridge Univ. Press, Cambridge, U.K.).
- 18.Hairer E., Norsett, S. P. & Wanner, G., (1993) Solving Ordinary Differential Equations (Springer, Berlin).
- 19.Sawitzky A. & Golay, M. J. E. (1964) Anal. Chem. 36, 1627-1639. [Google Scholar]
- 20.Ridder C. J. F. (1982) Adv. Eng. Software 4, 75-76. [Google Scholar]
- 21.Cohen J., Altaratz, H., Zick, Y., Klingmüller, U. & Neumann, D. (1997) Biochem. J. 327, 391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haspel R. L. & Darnell, J. E., Jr. (1999) Proc. Natl. Acad. Sci. USA 96, 10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride K. M., McDonald, C. & Reich, N. C. (2000) EMBO J. 19, 6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhalla U. S., Ram, P. T. & Iyengar, R. (2002) Science 297, 1018-1023. [DOI] [PubMed] [Google Scholar]
- 25.Asthagiri A. R. & Lauffenburger, D. A. (2000) Annu. Rev. Biomed. Eng. 2, 31-53. [DOI] [PubMed] [Google Scholar]
- 26.Blüthgen N. & Herzel, H., (2001) 2nd Workshop on Computation of Biochemical Pathways and Genetic Networks. (Logos, Berlin), pp. 55–62.
- 27.Starr R. & Hilton, D. J. (1999) BioEssays 21, 47-52. [DOI] [PubMed] [Google Scholar]
- 28.Zeng R., Aoki, Y., Yoshida, M., Arai, K. & Watanabe, S. (2002) J. Immunol. 168, 4567-4575. [DOI] [PubMed] [Google Scholar]
- 29.Begitt A., Meyer, T., van Rossum, M. & Vinkemeier, U. (2000) Proc. Natl. Acad. Sci. USA 97, 10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews R. P., Ericksen, M. B., Cunningham, C. M., Daines, M. O. & Hershey, G. K. (2002) J. Biol. Chem. 277, 36563-3659. [DOI] [PubMed] [Google Scholar]
- 31.Kudo N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E. P., Wolff, B., Yoshida, M. & Horinouchi, S. (1999) Proc. Natl. Acad. Sci. USA 96, 9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeman B. C. & Yamamoto, K. R. (2001) Trends Biochem. Sci. 26, 285-290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.