Abstract
The presence of a genetic component in longevity is well known. Here, the association of a mtDNA mutation with a prolonged life span in humans was investigated. Large-scale screening of the mtDNA main control region in leukocytes from subjects of an Italian population revealed a homoplasmic C150T transition near an origin of heavy mtDNA-strand synthesis in ≈17% of 52 subjects 99–106 years old, but, in contrast, in only 3.4% of 117 younger individuals (P = 0.0035). Evidence was obtained for the contribution of somatic events, under probable nuclear genetic control, to the striking selective accumulation of the mutation in centenarians. In another study, among leukocyte mtDNA samples from 20 monozygotic and 18 dizygotic twins, 60–75 years old, 30% (P = 0.0007) and 22% (P = 0.011), respectively, of the individuals involved exhibited the homoplasmic C150T mutation. In a different system, i.e., in five human fibroblast longitudinal studies, convincing evidence for the aging-related somatic expansion of the C150T mutation, up to homoplasmy, was obtained. Most significantly, 5′ end analysis of nascent heavy mtDNA strands consistently revealed a new replication origin at position 149, substituting for that at 151, only in C150T mutation-carrying samples of fibroblasts or immortalized lymphocytes. Considering the aging-related health risks that the centenarians have survived and the developmental risks of twin gestations, it is proposed that selection for a remodeled replication origin, inherited or somatically acquired, provides a survival advantage and underlies the observed high incidence of the C150T mutation in centenarians and twins.
Recently we reported a large aging-dependent accumulation of tissue-specific point mutations at critical control sites for mtDNA replication in human skin fibroblasts and skeletal muscle (1–3). The T414G transversion within the promoter for light (L)-strand transcription and for synthesis of the RNA primer of heavy (H)-strand synthesis (4, 5) (Fig. 1) was found in a generally high proportion (up to 50%) of mtDNA molecules in skin fibroblast cultures from 8 of 14 normal individuals above 65 years of age, but was absent in fibroblast cultures from 13 younger individuals (1). The age distribution and the results of two longitudinal studies indicated clearly that the T414G mutation was not inherited (1). A search for possible point mutations in the main mtDNA control region of skeletal muscle revealed, surprisingly, the presence of two mutations that had not been observed in fibroblast mtDNA (2). In particular, an A189G transition, very close to the main origin of H-strand synthesis (position 191), in 11–64% of the mtDNA and a T408A transversion, within the promoter for the RNA primer of H-strand synthesis (Fig. 1), in 2–16% of the mtDNA were found in the muscle from the majority of 27 normal individuals above 53 years old, while being absent or marginally present in the muscle from 19 individuals younger than 34 years. Both the fibroblast T414G mtDNA mutation and the muscle A189G and T408A mutations showed a striking tissue specificity, being absent in heart, liver, lymph nodes, and spleen (2).
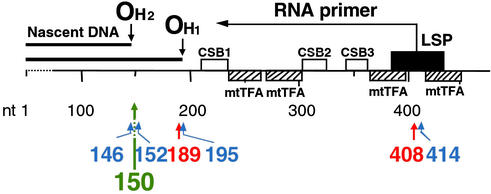
Figure 1.
Positions of the tissue-specific aging-dependent somatic mutations identified in human mtDNA main control region (1, 2) and of the C150T transition. OH1 and OH2, primary and secondary origin of H-strand synthesis; LSP, promoter for transcription of L-strand and synthesis of primer for H-strand synthesis (4, 5); CSB1, CSB2, and CSB3, conserved sequence blocks 1, 2, and 3 (6). The positions of binding of mitochondrial transcription factor A (mtTFA; the densely hatched rectangle indicates a position of high-affinity binding) are shown (1). Blue arrows and numbers indicate fibroblast-specific mutations (1), red arrows and numbers indicate skeletal muscle-specific mutations (2), and the green arrow and number indicate the C150T transition.
The occurrence of the fibroblast-specific T414G transversion in four of six centenarians (1, 3) raised the question of whether aging-dependent mutations may play a role in longevity. The availability of leukocytes from a large group of centenarians and control subjects of an Italian population offered us the opportunity of testing this possibility. In this paper, we report that a homoplasmic C150T transition, very close to a replication origin in the main mtDNA control region (Fig. 1), occurs at a much higher frequency in leukocytes from centenarians and from twins than in leukocytes from the rest of the population. Evidence was obtained that this mutation causes a remodeling of the replication origin at position 151, and that both maternal inheritance and somatic events play a role in this phenomenon. Furthermore, the aging-dependent somatic accumulation of the same mutation in skin fibroblasts was also demonstrated.
Materials and Methods
Source of Tissue Samples.
Blood leukocyte samples from a total of 207 subjects from Northern, Central, and Southern Italy (all of Italian origin) were analyzed (see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org, for a detailed list). Control groups were made up of 117 individuals (18- to 98-year age range; median age 54 years; 63 females and 54 males), of which 94 were from Northern/Central and 23 from Southern Italy. The groups of centenarians from the same Italian population included 52 subjects (37 females and 15 males), of which 33 were from Northern/Central and 19 from Southern Italy. Both the controls and the centenarians were unrelated to each other, and except that C497 and C498, and C692 and C693 were brothers, and C472 and C473, and C508 and C509 were sisters. The twin groups (60–75 years old), from the Northern/Central Italian population, included 20 monozygotic and 18 dizygotic individuals (all males). All the subjects included in this study were free of any clinically manifest pathologies.
The skin fibroblast cultures from 43 individuals of varying ages from fetal to 103 years old were obtained either from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ) or from the NIA Aging Cell Culture Repository (Camden, NJ), or were derived from biopsies of old individuals (all from retirement homes) at the Clinical Neurology Institute of the University of Milan (Italy). See Table 3 for a detailed list. All individuals were free of any neurological or muscular pathology.
Lympho-Monocyte and Granulocyte Isolation.
For the isolation of lymphocyte and monocytes–macrophages, hereafter referred to as “lympho-monocytes,” and granulocytes, the blood sample, mixed with 0.1% EDTA as anticoagulant, was centrifuged at 800 × g for 20 min through a Ficoll/metrizoate gradient (Isoprep, Biochrom, Berlin), made with a solution having a density of 1.077 g/ml (7–9). The band at the interface between the sample and Isoprep, consisting of ≈95% mononuclear cells, was removed and the cells washed three times with PBS by centrifuging at 250 × g for 10 min. The granulocytes were recovered from the upper layer of the erythrocyte fraction pelleted at the bottom of the tube and washed with PBS. The buffy coat was isolated by diluting the blood sample with an equal volume of 10 mM Tris⋅HCl, pH 7.4 (25°C)/100 mM NaCl/1 mM EDTA, centrifuging it for 15 min at 400 × g, and recovering the intermediate layer. Immortalization of lymphocytes was carried out as described in ref. 10.
Fibroblast Culture Conditions.
Human skin fibroblasts were cultured in DMEM supplemented with 20% FBS. In general, the fibroblast cultures were used within two or three passages after receipt.
Isolation of mtDNA Fragments.
Total cell DNA was isolated by the standard proteinase K/SDS treatment, followed by phenol/chloroform extraction. PCR amplification and cloning of the large mtDNA fragment comprised between positions 21 and 719 in the Cambridge sequence (11), which contains all the initiation sites for transcription and replication of mtDNA (Init-Tra-Rep), and secondary PCR amplification and purification of the DLP4 and DLP6 subfragments (Fig. 6, which is published as supporting information on the PNAS web site), which were chosen for analysis in this work, were carried out as previously described (2, 12).
Allele-Specific Termination of Primer Extension.
Quantification of the specific point mutations was carried out by allele-specific termination of primer extension by using the large Init-Tra-Rep mtDNA fragment as a template, with minor modifications of the original protocol (13), as detailed in refs. 1, 2, and 12. The combinations of oligodeoxynucleotide primer and of deoxynucleoside and dideoxynucleoside triphosphates used for the quantifications of the T414G, A189G, and T408A mutations were previously specified (1, 2), and those for the quantification of the C150T and G228A mutations are specified below:
C150T, 5′-GCAGTATCTGTCTTTGATTCCTGCCTC (positions 121–147 in Cambridge sequence); dA, dT, ddC.
G228A, 5′-GACATTCAATTGTTATTATTATGT (positions 254–230); dC, ddT.
Identification of 5′ Ends of mtDNA H-Strand Nascent Chains.
The analysis of the distribution of the 5′ ends of the nascent mtDNA H-strands and their identification were carried out by the following approach. The first step in this approach was a primer extension carried out on all nascent H-strand chains of mtDNA contained in a total cell DNA sample by using Vent DNA polymerase and a purified 5′-32P-labeled L-strand primer corresponding to a segment of the D-loop. This step was followed by separation of the extended products by 7 M urea/10% PAGE and analysis of the dried gel in a Phosphor-Imager or by autoradiography. For the identification of the 5′ ends of the nascent chains, an undried gel was exposed for autoradiography, the bands of interest were excised, and the DNA eluted and subjected to circularization by intramolecular ligation of the 5′ and 3′ ends of each extended primer with T4 RNA ligase. The circularized molecules were subjected to PCR amplification by using two adjacent primers divergent from a position about one-third or one-half of the way around the circle from the 3′ end of the extended primers, and the PCR products were separated by electrophoresis in a native polyacrylamide gel, cloned in Escherichia coli by using a TA vector, and 25–50 plasmids derived from each PCR product were then sequenced to identify the 5′–3′ end junctions. See supporting information on the PNAS web site for more detailed information.
Results
Markedly Increased Frequency of C150T Transition in the Replication Control Region of Leukocyte mtDNA of Human Centenarians.
In a preliminary large-scale screening, mtDNA of lympho-monocytes and granulocytes from 43 centenarians aged 99–106 years and 78 control subjects 18–98 years old of the same Italian population (see Table 3) were analyzed by allele-specific termination of primer extension for the possible presence of the main aging-dependent mutations previously identified in fibroblasts and skeletal muscle by allele-specific termination of primer extension. This analysis (data not shown) revealed the absence of the T414G and T408A mutations from all 231 leukocyte mtDNA samples and the rare presence of the A189G polymorphism (in <2.5% of centenarians and ≈5% of controls).
To investigate the possibility that some point mutations accumulate with aging in leukocytes at different nucleotide positions from those previously found in fibroblasts and skeletal muscle, the Init-Tra-Rep fragments (Fig. 6), PCR-amplified from mtDNA of lympho-monocytes and granulocytes from two subjects 72 years old (72-1 y and 72-2 y) and four centenarians 100 or 102 years old (100-6 y, 100-10 y, 102-4 y, and 102-5 y) were cloned in E. coli, and 50 plasmids were isolated from each source following a previously described protocol (1, 2, 12). The DNAs of these plasmids were subjected to PCR amplification of the two Init-Tra-Rep subfragments DLP4 and DLP6, which harbor all aging-dependent mutations so far identified in the main control region (1, 2) (Fig. 6). These PCR products were then analyzed by denaturing gradient gel electrophoresis and sequencing, as previously described (1, 12). As shown in Table 4, which is published as supporting information on the PNAS web site, the most significant observation was that the mtDNA DLP4 fragment from lympho-monocytes of the two centenarians 100-6 y and 100-10 y exhibited transitions C150T and G228A in homoplasmic or near-homoplasmic form, whereas the DLP4 from the corresponding granulocytes lacked them or carried them at a marginal level. This observation was important, in that it excluded a pseudogene origin of these transitions, while pointing to their possible somatic derivation.
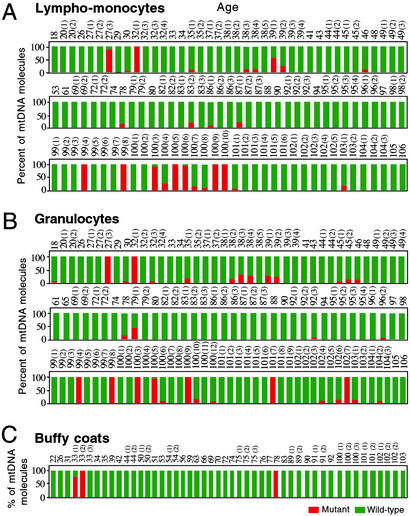
A large-scale screening by allele-specific termination of primer extension (1, 2, 12) of the available leukocyte mtDNA samples for the presence of the two transitions mentioned above failed to show a significant increase in incidence of the G228A mutation in the samples from centenarians, as compared with those from younger subjects, and, by contrast, revealed in the same samples a striking increase in frequency of the C150T transition. The C150T mutation was particularly interesting because of its location near the site of the secondary origin of mtDNA replication (sequence 146–151) (4, 5, 14, 15). As shown in Fig. 2A, the C150T mutation was observed in homoplasmic or nearly homoplasmic form in seven lympho-monocyte mtDNA samples (19.4%) among those derived from 36 subjects 99–106 years old, and, in striking contrast, in only two samples (from the 27-3 y and 32-1 y individuals) (2.6%) among those derived from 76 individuals 18–98 years old (P = 0.0048). Similarly, as shown in Fig. 2B, the homoplasmic or nearly homoplasmic C150T transition was found in seven granulocyte samples (16.2%) among those derived from 43 subjects 99–106 years in age, and, by contrast, in only two samples (from the same 27-3 y and 32-1 y individuals) (2.6%) among those derived from 76 subjects 18–98 years old (P = 0.0107). The homoplasmic C150T mutation was also found in two buffy coat samples (from the 33-2 y and 78 y individuals). Very surprisingly, for both lympho-monocytes and granulocytes, there was no aging-related trend, below the donor's age of 99 years, in the accumulation of the homoplasmic C150T transition. This observation clearly pointed to a role of selection in the high frequency of the mutation in centenarians.
Figure 2.
Bar graph summarizing the age distribution and frequency of the C150T transition (as determined by primer extension analysis) in mtDNA from lympho-monocytes (A), granulocytes (B), and buffy coats (C) of differently aged individuals. The number above each bar indicates the age in years of the individual analyzed. Red bars, C150T mutation-carrying mtDNA; green bars, wild-type mtDNA.
A complete analysis of the data presented in Fig. 2 is presented in Table 1. Because no significant difference was found between the frequencies of the male and female centenarians exhibiting this mutation (20% and 16%, respectively), male and female individuals were considered together in the above analysis. As shown in Table 1 Upper, the overall frequency of the homoplasmic C150T transition among the centenarians (17.3%) is very significantly higher than that among the younger subjects of the same population (3.4%) (P = 0.0035).
Table 1.
Distribution of mtDNA C150T homoplasmic transition in leukocytes from an Italian population
| Overall sample donors
|
G samples
|
L-M samples
|
||||
|---|---|---|---|---|---|---|
| n | C150T (%)* | n | C150T (%)* | n | C150T (%)* | |
| Centenarians,† 99–106 y | 52 | 9 (17.3) | 43 | 7 (16.2) | 36 | 7 (19.4) |
| Controls,‡ | 117 | 4 (3.4) | 76 | 2 (2.6) | 76 | 2 (2.6) |
| 18–98 y | P = 0.0035 | P = 0.0107 | P = 0.0048 | |||
| Monozygotic
|
Dizygotic
|
|||
|---|---|---|---|---|
| n | C150T (%) | n | C150T (%) | |
| Twins, 60–75 y | 20 | 6 (30) | 18 | 4 (22) |
| Controls, | 117 | 4 (3.4) | 117 | 4 (3.4) |
| 18–98 y | P = 0.0007 | P = 0.0113 | ||
G, granulocytes; L-M, lympho-monocytes. The P values for the difference in frequency of the C150T homoplasmic transition between centenarians and controls and between monozygotic or dizygotic twins and controls were calculated by Fisher's exact test.
The numbers and percentages shown represent the absolute and relative frequencies of the C150T homoplasmic transition.
These include 43 donors of G and/or L-M samples and donors of buffy coats.
These include 78 donors of G and/or L-M samples and 39 donors of buffy coats. The apparent discrepancy between the number of overall G and L-M sample donors (78) and the number of G samples and L-M samples is due to the fact that, for a few of the donors, only the G sample or the L-M sample was available (74 shared donors).
Distribution of C150T Transition in Lympho-Monocytes and Granulocytes from the Same Individuals.
Five of the seven centenarian subjects carrying the C150T mutation in lympho-monocytes also exhibited it in granulocytes, as expected for an inherited mutation. On the contrary, the other two subjects (100-6 y and 100-10 y) carried in their granulocytes a heteroplasmic C150T mutation, and even this at a low level (7% and 17%, respectively), substantially corroborating the previously mentioned result obtained by the cloning–denaturing gradient gel electrophoresis-sequencing approach (see Table 4).
The C150T transition occurred in heteroplasmic form in mtDNA from lympho-monocytes and granulocytes at similar frequencies in centenarians and younger subjects (≈14–15% for lympho-monocytes and ≈16–21% for granulocytes) (Fig. 2 A and B). Most interestingly, a comparison between the distribution of the heteroplasmic C150T transition in the lympho-monocyte mtDNA samples and in the corresponding granulocyte samples showed an increase with age in the frequency of discordance of its presence in the two types of leukocytes. In particular, the discordance reached 89% in the centenarians (Table 2). In view of the fact that the frequency of somatic mutations is expected to increase with age, the observations reported above strongly suggested a contribution of aging-dependent somatic events to the observed distribution of the transition.
Table 2.
Distribution of homoplasmic and heteroplasmic C150T transition in lympho-monocytes (L-M) and granulocytes (G) from the same individuals
| Age range, years | Number of shared donors for L-M and G | Homoplasmic C150T
|
Heteroplasmic C150T
|
||
|---|---|---|---|---|---|
| A
|
B
|
C
|
D
|
||
| Pairs of L-M and G with both samples homoplasmic* | Pairs with only one sample homoplasmic and the other wild-type or heteroplasmic* | Pairs of L-M and G with both samples heteroplasmic* | Pairs with only one sample heteroplasmic and the other wild-type or homoplasmic* | ||
| 18–49 | 39 | 2 (100%) | 0 | 7 (58%) | 5 (42%) |
| 53–98 | 35 | 0 | 0 | 1 (14%) | 6 (86%) |
| 99–106 | 36 | 5 (71%) | 2 (29%) | 1 (11%) | 8 (89%)** |
For each age group, the numbers of pairs in columns A and B and in columns C and D are summed. Each value in parentheses gives the proportion of the pair sums which is concordant (A or C) or discordant (B or D) in exhibiting the indicated mutation.
This group includes the two pairs listed in B.
The above findings raised the question of the degree of nucleotide selectivity in this distribution. Sequencing of the mtDNA fragment between positions 100 and 200 in the Cambridge sequence in four lympho-monocyte and four granulocyte samples from centenarians carrying the homoplasmic C150T transition revealed only one granulocyte sample exhibiting, in the 70-nt stretch surrounding the C150T position, a homoplasmic T146C and a homoplasmic T152C transition (Fig. 7, which is published as supporting information on the PNAS web site). Similarly, among the six lympho-monocyte and five granulocyte samples from centenarians lacking or exhibiting a marginal level of the C150T transition, only six carried in the same stretch the homoplasmic T146C mutation or the homoplasmic T152C mutation or a few random mutations. The T146C and T152C transitions are well known polymorphisms (ref. 16). These results showed the striking nucleotide selectivity of the C150T transition, indicating at the same time positions 146 and 152 as preferential sites of secondary mutations.
Aging-Dependent Accumulation of C150T Transition in Fibroblasts.
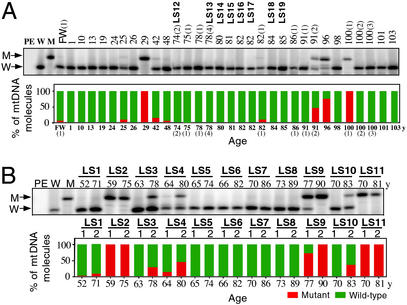
The previous detection of a homoplasmic C150T transition in fibroblast mtDNA from one of five centenarians analyzed (1) prompted us to carry out a large-scale screening of fibroblast mtDNA from 32 subjects, 20-week fetal to 103 years in age, belonging to a different set of individuals from those used for leukocyte analysis (see Table 3). As shown in Fig. 3A, the C150T transition was observed in homoplasmic or heteroplasmic form in 28% of the samples, with an apparent tendency to increase with age. Furthermore, the availability of longitudinal fibroblast samples, i.e., samples taken twice from the same individuals at time intervals ranging from 9 to 19 years, offered the unique opportunity for testing the inherited or somatic origin of this transition in this cell type. As shown in Fig. 3B, in two of 11 studies [i.e., LS2 and LS11 (Table 3)], both the earlier and later samples exhibited the C150T transition in homoplasmic form, pointing to its possible inheritance. By contrast, in five other longitudinal studies (LS1, LS3, LS4, LS9, and LS10), the mutation appeared (LS3, LS10) or became more abundant (LS1, LS4, LS9) in the second sample, up to homoplasmy in LS9, providing strong evidence for the role of somatic events in this phenomenon.
Figure 3.
(A) Detection of C150T transition by allele-specific termination of primer extension, by using the PCR-amplified Init-Tra-Rep fragment as a template, in fibroblast mtDNA from the single biopsies of 32 subjects, 20-week fetal [FW (1)] and 1–103 years old, and from the second biopsies of the longitudinal studies LS12–LS19. The bar graph summarizes the age distribution and frequency of the C150T transition. (B) Detection of the C150T transition in the fibroblast mtDNA from the first and second biopsies of the longitudinal studies LS1–LS11, and age distribution and frequency of the mutation. W and M, primer extension products obtained by using as a template plasmid DNA carrying a cloned Init-Tra-Rep mtDNA fragment with wild-type and, respectively, mutant sequence; PE, primer+ sequenase only.
Many fibroblast samples carrying the C150T transition in either homoplasmic or heteroplasmic (>25% mutant mtDNA) form were also sequenced in the 100- to 200-nt mtDNA segment (Fig. 7). These fibroblast mtDNAs revealed a marked nucleotide selectivity of the C150T mutation, as observed in leukocytes, and also the frequent occurrence of the T152C polymorphism (in 6 of 13 samples). What role, if any, the latter polymorphism may play in the evolutionary or somatic establishment of the C150T transition is not known.
Remodeling of mtDNA Replication Origin Is Associated with the C150T Mutation.
The observation of a strikingly increased frequency of the homoplasmic C150T transition in leukocyte mtDNA from centenarians raised the question of the possible effect of this transition on the initiation of H-strand synthesis at or near that position. To investigate this question, the distribution of the 5′ ends of the nascent H-mtDNA strands originating in that sequence segment was analyzed by the approach illustrated in Fig. 8, which is published as supporting information on the PNAS web site and described in Materials and Methods. By this approach, primer extension was carried out on all nascent H-strand chains, i.e., those that abort at the 3′ end of the D-loop, as well as on those that extend beyond the D-loop and that are, therefore, the true replicating H-strands. Previous work had shown that the distribution and relative amounts of the true replicating H-strands starting at different positions within the D-loop in human cells are very similar to the distribution and relative amounts of the abortive nascent chains (15). A test experiment using as an artificial extended primer a 75-nt-long oligodeoxynucleotide corresponding to the L-strand sequence from position 81 to 155 of the Cambridge sequence (11), demonstrated the accuracy of determination of the 3′ end nucleotide of the extended primer (T in this case) by the method used here (Fig. 8C).
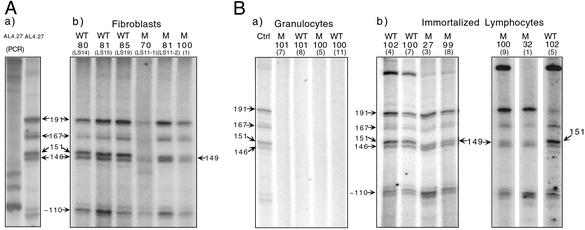
Fig. 4Aa shows the primer extension products obtained in an experiment by using mtDNA from AL4.27 (3), a cell line carrying the wild-type mtDNA sequence between positions 100 and 200. Several labeled bands were observed. As shown in Fig. 8 A and B (and data not shown), their 5′ ends mapped at or near nucleotide positions 191, 167, 146–151, and 110, which are the major initiation sites of H-strand synthesis in KB, HeLa, and Jurkat cells (4, 5, 14, 15). The 191 and 146–151 sites have been referred to in this work, respectively, as primary (OH1) and secondary (OH2) origin of H-strand synthesis. No artifactual bands corresponding in size to the primer extension products of interest were observed in a control experiment by using as a template an AccI-generated and PCR-amplified 2,818-bp mtDNA fragment encompassing the whole main control region, in which no nascent chains were expected to be present (Fig. 4Aa).
Figure 4.
Detection of nascent H-mtDNA chains and identification of their 5′ ends. (A) Primer extension products of all nascent H-strand chains, obtained by using as templates the original mtDNA from the wild-type cell line AL4.27 or its PCR-amplified ACCI 2,818-bp fragment containing the mtDNA main control region (a), or the mtDNA from three fibroblast samples lacking (WT) and three fibroblast samples carrying (M) the C150T transition (b). (B) Primer extension products of nascent H-strand chains obtained by using as templates the mtDNA from AL4.27 cells (Ctrl), or from circulating granulocytes of centenarians lacking (WT) or carrying (M) the C150T transition (a), or from immortalized lymphocytes derived from centenarians lacking or carrying the mutation (b). In A and B, the age of the donors of fibroblasts, granulocytes, and immortalized lymphocytes is indicated above each lane. The 5′ ends of the nascent H-strand chains starting at positions 191, 167, 151, 149, and 146 were determined by sequencing, whereas the 5′ end of the 110 nascent chains was estimated from comparison of their migration with that of the marker (MSP1-digested pBR322 DNA).
The approach described above was applied to identify the nascent H-strand chains and to determine their 5′ ends in fibroblast and leukocyte DNA samples. The pattern of nascent H-strand chains observed in wild-type fibroblast mtDNA from three individuals was identical to that obtained for AL4.27 mtDNA (Fig. 4Ab). By contrast, in the patterns observed in the fibroblast mtDNA from three individuals carrying the homoplasmic C150T transition, the band corresponding to nascent chains starting at position 151 was missing, and was substituted by a new band corresponding to nascent chains starting at position 149. It should be noted that the nucleotide at position 151 is a C, whereas the nucleotide at position 149 is a T, arguing against a requirement for a C in initiation.
A similar analysis carried out on granulocyte (Fig. 4Ba) and lympho-monocyte mtDNA samples (data not shown) lacking or carrying the homoplasmic C150T mutation revealed a complete absence of nascent H-strand chains. This result was not completely surprising, because blood lympho-monocytes and granulocytes are known not to proliferate and therefore presumably not to replicate their mtDNA while in circulation. In agreement with this idea, when three immortalized lymphocyte DNA samples lacking the C150T mutation were analyzed, the normal pattern of bands, including those corresponding to positions 146 and 151, was observed (Fig. 4Bb). Most significantly, when five immortalized lymphocyte DNA samples carrying the homoplasmic or nearly homoplasmic C150T mutation were analyzed, again the nascent chain pattern characteristic of mutant fibroblasts was observed (Fig. 4Bb and data not shown). It should be noted that this remodeling was also observed in the immortalized lymphocytes derived from the two young individuals (27-3 y and 32-1 y) exhibiting the homoplasmic C150T transition.
Markedly Higher Frequency of the C150T Transition in Twin Leukocytes.
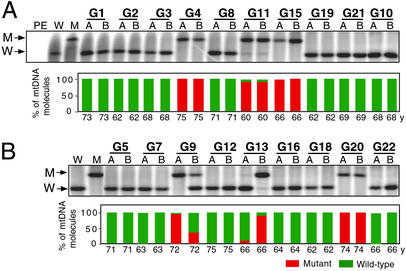
To obtain information about the inheritance of the C150T base transition, leukocyte (buffy coats) samples from 20 monozygotic (MZ) twins and 18 dizygotic (DZ) twins (all 60–75 years old) from the same Italian population investigated in the studies described above (17) were analyzed by allele-specific termination of primer extension for the presence and abundance of the C150T transition (Fig. 5, Table 1, Lower). Surprisingly, six of 20 samples from MZ twins (30%) belonging to three pairs exhibited the C150T transition in homoplasmic or near-homoplasmic form. Most interestingly, there was perfect concordance in levels of mutation in the two subjects of each of the three twin pairs. The incidence of the homoplasmic C150T transition was very significantly higher than that observed in the control population (P = 0.0007). Also among the 18 DZ twins, an unusually high number, i.e., four (22%), exhibited the C150T transition in homoplasmic or near-homoplasmic form (P = 0.0113) and two more in heteroplasmic form. However, in contrast to the MZ twins, of the three DZ pairs in which both members carried the mutation, only one revealed concordance in mutation level between the two twins.
Figure 5.
Detection of the C150T transition by allele-specific termination of primer extension and its distribution and abundance in buffy coat mtDNA from MZ (A) and DZ (B) twins. Abbreviations are as in Fig. 3.
Discussion
The C150T transition has been previously identified as a polymorphism in the mtDNA control region of individuals from Europe, Asia, and Africa (16). However, no relationship of its occurrence to aging, longevity, or twin gestations has been reported. The high incidence of the C150T transition in leukocytes from centenarians and twins has provided striking examples of a rare mutation becoming selected either through inheritance or through a somatic event(s). The aging-related increase in discordance, as concerns the presence and/or level of the C150T transition between lympho-monocytes and granulocytes from the same individuals, has clearly indicated that the observed accumulation of the mutation in centenarians has a somatic contribution. Furthermore, a nuclear genetic control of this somatic contribution is strongly suggested by the striking nucleotide selectivity of the mutation. The younger control individuals that exhibited the above-mentioned discordance between the two groups of leukocytes may be representative of the pool of subjects that have the potential of reaching a centenarian age.
The observed identity as concerns the presence and level of the C150T transition between the leukocytes from the two members of each of three MZ pairs, which were derived from a single egg of the mother, has strongly suggested a maternal inheritance of the mutation. However, it cannot be excluded that a somatic mutation, which was amplified under the control of the nuclear genotype of the twins, was involved. In fact, the phenomenon could be similar to that described for the age-related telomere shortening in MZ and DZ twins, in which a surprisingly high (≈78%) heritability of the telomere length phenotype has been determined (18). In the present work, the discordance in the level of the C150T mtDNA transition, observed between the two members of two of the DZ pairs, presumably reflected a remarkable heterogeneity in C150T mutation level among the eggs of the mother or, alternatively, a somatic event(s). Because the somatic appearance and expansion of mtDNA mutations would require mtDNA replication, the proposed contribution of somatic events to the increased frequency of the C150T transition in the peripheral lympho-monocytes and granulocytes of centenarians and twins would have to occur in the precursors of these cells and, possibly, in stem cells. This possibility is now open to investigation, with implications for the possible role of the C150T transition in cancer and other blood cell diseases.
Strong support for the conclusion of a contribution of somatic events to the phenomenon investigated here has come from the longitudinal studies of fibroblasts, which have provided convincing evidence that the mutation can arise during life or change level in the same individual during aging. Furthermore, in these cells as well, the nucleotide selectivity of the mutation has reinforced the suggestion that the postulated somatic event(s), induced by an environmental or internal stimulus, is under nuclear genetic control.
In conclusion, the data obtained in the present study on the C150T mutation in both leukocytes and fibroblasts have indicated that this mutation can be inherited or can arise de novo during life, its level changing with age to a different extent in different individuals, most probably under genetic control.
A property common to centenarians and twins is survival under unfavorable circumstances. In the case of centenarians, they have survived the aging-related decline in critical functions and the increasing risks of diseases. In twins, the unfavorable circumstances occur at a different time, i.e., during gestation and in the early phase of their lifespan. This is documented by the fetal growth restriction and the 5- to 10-fold higher rates of perinatal mortality of twin gestations, as compared with singleton gestations (19–21), and by the higher rate of postnatal morbidity in twins (21). On the basis of the arguments presented above, it is a plausible hypothesis that the observed marked increase in frequency of the C150T transition in the centenarians and twin “survivors” results from a selection process favoring the individuals that have inherited it from their mother and those who have accumulated it during life.
Further work is needed to establish whether the postulated survival advantage provided to the centenarians and twins and, in general, to aged individuals by the remodeling of an mtDNA replication origin observed here in mitotic lymphocytes and fibroblasts is related to the regulation of mtDNA replication and, in particular, to the copy number control mechanism. This mechanism, which is still not understood, ensures that there is on average one replication event per cell cycle (22). Relevant in this connection is our earlier observation that, during recovery from an ethidium bromide-induced depletion of HeLa cell mtDNA, the average number of replication events per cell cycle rises from 1 to 2.5 (23). A similar relaxation of the copy number control, in this case produced by an inherited or somatic C150T mutation, may conceivably operate to accelerate mtDNA replication in centenarians and twins. It should be mentioned that a very significant decline with the donor's age in mtDNA content has been previously demonstrated in transformants constructed by transfer of mitochondria from fibroblasts of differently aged individuals into human mtDNA-less cells (24). An interesting possibility is that the somatic event(s) at or near position 150 leading to the appearance and/or amplification of the C150T transition may be a part of a general remodeling of the mtDNA replication machinery, probably nuclearly controlled. This remodeling could accelerate mtDNA replication and compensate for the oxidative damage of mtDNA and its functional deterioration occurring in old age and, possibly, during or after twin gestations. The aging-dependent accumulation of tissue-specific point mutations, which we have previously identified in fibroblasts and skeletal muscle at critical control sites for mtDNA replication (1, 2), is conceivably also a part of this remodeling.
Supplementary Material
Acknowledgments
We are grateful to N. Bresolin and F. Mazzucchelli for providing some of the fibroblast cultures analyzed in the present work. We thank A. Chomyn for valuable discussions and L. Bellavia and M. Del Mar Roldan for expert technical assistance. This work was supported by National Institute on Aging (National Institutes of Health) Grant AG12117 (to G.A.), by Ellison Medical Foundation Senior Scholar Award AG-SS-0622-00 (to G.A.), and by an Associazione Italiana per la Ricerca sul Cancro grant (to C.F.).
Abbreviations
- H-strand
heavy strand
- L-strand
light strand
- Init-Tra-Rep
initiation sites for transcription and replication
- MZ
monozygotic
- DZ
dizygotic
References
- 1.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Kevin E, Yarasheski K E, Miller C A, Askanas V, Engel W K, et al. Proc Natl Acad Sci USA. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michikawa Y, Laderman K, Richter K, Attardi G. Somatic Cell Mol Genet. 2002;25:333–342. doi: 10.1023/a:1019972500785. [DOI] [PubMed] [Google Scholar]
- 4.Attardi G. Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 5.Chang D D, Clayton D A. Proc Natl Acad Sci USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walberg M W, Clayton D A. Nucleic Acids Res. 1981;9:5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyum A. Nature. 1964;204:793–794. doi: 10.1038/204793a0. [DOI] [PubMed] [Google Scholar]
- 8.Boyum A. Scand J Clin Lab Invest. 1968;21,(Suppl. 97):9–29. [PubMed] [Google Scholar]
- 9.Boyum A. Scand J Clin Lab Invest. 1968;21,(Suppl. 97):50. [PubMed] [Google Scholar]
- 10.Louie L G, King M C. Am J Hum Genet. 1991;48:637–638. [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson S, Bakier A T, Barrell B G, de-Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 12.Michikawa Y, Attardi G. In: Methods in Molecular Biology. Copeland W C, editor. Vol. 197. Totowa, NJ: Humana; 2002. pp. 75–92. [DOI] [PubMed] [Google Scholar]
- 13.Hofhaus G, Attardi G. Mol Cell Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crews S, Ojala D, Posakony J, Nichiguchi J, Attardi G. Nature. 1979;277:192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- 15.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. J Biol Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 16. MITOMAP: A Human Mitochondrial Genome Database (2002) www.mitomap.org. [DOI] [PMC free article] [PubMed]
- 17.Bonafé M, Cardelli M, Marchegiani F, Cavallone L, Giovagnetti S, Olivieri F, Lisa R, Pieri C, Franceschi C. Exp Gerontol. 2001;36:1063–1073. doi: 10.1016/s0531-5565(01)00112-7. [DOI] [PubMed] [Google Scholar]
- 18.Slagboom P E, Droog S, Boomsma D I. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 19.Spellacy W N, Handler A, Ferre C D. Obstet Gynecol. 1990;75:168–171. [PubMed] [Google Scholar]
- 20.Kiley J. Bull NY Acad Med. 1990;66:618–637. [PMC free article] [PubMed] [Google Scholar]
- 21.Kinzler W L, Ananth C V, Vintzileos A M. J Soc Gynecol Invest. 2000;7:321–327. [PubMed] [Google Scholar]
- 22.Flory P J, Jr, Vinograd J. J Mol Biol. 1973;74:81–94. doi: 10.1016/0022-2836(73)90100-9. [DOI] [PubMed] [Google Scholar]
- 23.Wiseman A, Attardi G. Mol Gen Genet. 1978;167:51–63. doi: 10.1007/BF00270321. [DOI] [PubMed] [Google Scholar]
- 24.Laderman K A, Penny J R, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. J Biol Chem. 1996;271:15891–15897. doi: 10.1074/jbc.271.27.15891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.