Abstract
The recombination-activating gene (RAG)1 and RAG2 proteins comprise the lymphocyte-specific components of the V(D)J recombinase and are required for the assembly of antigen-receptor variable-region genes. A mutant truncated RAG2 protein (“core” RAG2) lacking the C-terminal 144 amino acids, together with core RAG1, is able to mediate the basic biochemical steps required for V(D)J recombination in vitro and in transfected cell lines. Here we examine the effect of replacing the endogenous RAG2 locus in mice with core RAG2. These mice generate substantial numbers of B and T cells, demonstrating that the core RAG2 protein retains significant in vivo function. However, core RAG2 mice display a reduction in the total number of B and T cells, reflecting impaired lymphocyte development at the progenitor stage associated with reduced chromosomal V(D)J recombination. We discuss potential roles of the RAG2 C terminus in mediating rearrangement of endogenous antigen-receptor loci.
During B and T cell development, the Ig and T cell receptor (TCR) variable-region genes are assembled from component germ line variable (V), diversity (D), and joining (J) gene segments through a process known as V(D)J recombination (1–3). Each V, D, and J gene segment is flanked by a recombination signal sequence (RSS) composed of conserved heptamer and nonamer sequences separated by either a 12- or 23-bp spacer. V(D)J recombination is initiated by the lymphoid-specific recombination-activating gene (RAG) 1 and 2 proteins, which recognize a pair of 12 and 23 RSSs and introduce a DNA double-strand break between each RSS and flanking coding sequence. The RAG-liberated ends are held in a complex with RAG1 and RAG2 until resolved into precise recombination signal and imprecise coding joins via the generally expressed nonhomologous end-joining DNA-repair factors (1).
During lymphocyte development, V(D)J recombination is highly regulated in the contexts of lineage specificity, developmental-stage specificity, and allelic exclusion (4, 5). In addition, developing lymphocytes use strategies that require productive V(D)J rearrangements for continued developmental progression (4, 5). For example, Ig heavy chain (IgH) variable-region genes are assembled in progenitor (pro) B cells via an ordered process in which DH-to-JH rearrangement occurs on both alleles before VH-to-DJH rearrangement. Productive VH-to-DJH rearrangements produce cell-surface expression of IgH μ chains that signal expansion and differentiation of pro-B cells to the pre-B cell stage. This expression of IgH μ chains also signals the cessation of further VH-to-DJH rearrangement via feedback regulation. Pro-B cells that first assemble nonproductive VH-to-DJH rearrangements proceed to rearrange their second allele.
Both RAG1 and RAG2 are required for V(D)J recombination, because RAG1- or RAG2-deficient mice exhibit a complete block in lymphocyte development at the progenitor stage (6, 7). In addition, RAG1 and RAG2 together are sufficient to initiate V(D)J recombination in vitro (8). Nearly all in vitro studies of RAG function have used the minimal regions of RAG1 (amino acids 384–1,008 of 1,040) and RAG2 (amino acids 1–383 of 527) required for activity, because full-length RAG1 and RAG2 are largely insoluble. However, the activities of these mutant “core” proteins differ from those of the full-length RAGs when assayed with extrachromosomal substrates in transfected cells. In this context, the mutant core RAG proteins support V(D)J recombination with reduced efficiency and with different levels and types of recombination products (9–15).
Although not required for the biochemistry of V(D)J recombination, the noncore regions of RAG1 and RAG2 are conserved throughout evolution. For example, sequence conservation across the entire RAG2 protein from pufferfish to humans is ≈60%, with conservation of the noncore C-terminal region slightly higher than the core domain (16). Therefore, the noncore regions of RAG1 and RAG2 may serve important accessory and/or regulatory functions in chromosomal V(D)J recombination. Consistent with this notion, studies of Abelson murine leukemia virus-transformed pre-B cells have suggested that the noncore regions of RAG1 are important for IgH locus DH-to-JH rearrangement (17), whereas the C terminus of RAG2 is more important for VH-to-DJH rearrangement than for DH-to-JH rearrangement (18). The C terminus of RAG2 is predicted to fold into a plant homeodomain (PHD) (19, 20), a motif found in a number of chromatin-associated proteins including some that have chromatin-modifying activities (21–24).
To investigate potential in vivo function of the RAG2 noncore region in chromosomal V(D)J recombination and normal lymphocyte development, we have generated and characterized mice containing specific replacement of the full-length endogenous RAG2 gene with a gene encoding the mouse core RAG2 protein.
Materials and Methods
Generation of Core RAG2 Mice.
For information about the generation of core RAG2 mice, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Southern Blotting and PCR.
Standard methods were used for the isolation of genomic DNA and for Southern hybridization (25). IgH and TCRβ rearrangements were amplified by PCR with primers as described (18, 26).
Flow Cytometry.
Single-cell suspensions from each tissue were stained with FITC-, phycoerythrin (PE)-, and CyChrome (CyC)-conjugated antibodies (PharMingen) and analyzed by a FACScalibur or FACSVantage SE (Becton Dickinson). Data were analyzed with CELLQUEST (Becton Dickinson) software. Cell sorting was performed on a MoFlo machine (Cytomation, Fort Collins, CO).
Results
Core RAG2 Mice Exhibit a Partial Arrest in B Cell Development.
A targeting construct was used to replace the endogenous RAG2 gene with a gene encoding the mouse core RAG2 protein (Fig. 6 A–C and Supporting Results, which are published as supporting information on the PNAS web site). Expression of core RAG2 in the resulting mice was found to be similar to the levels seen with full-length RAG2 in WT mice (Fig. 6D). B cell development in core RAG2 mice was assessed by using fluorescence-activated cell sorter (FACS) analysis to examine the expression of lineage- and stage-specific cell-surface markers on bone marrow cells (Fig. 1). In this assay, cells from mice with RAG2 genotypes +/+, +/−, and +/c [WT (+), knockout (−), or core (c)] exhibited no obvious phenotypic differences and are subsequently all referred to as “WT.” Likewise, cells from mice of genotypes c/c and −/c are phenotypically identical and both referred to as “core.” Genotypes are indicated when representative results are presented.
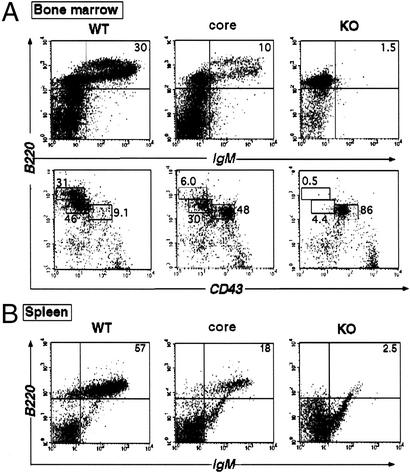
Figure 1.
B cell development of core RAG2 mice is impaired. (A) Representative FACS analysis of bone marrow cells from RAG2 WT (+/−), core (c/−), and knockout (KO, −/−) mice stained with CyC-anti-B220 and either PE-anti-IgM or FITC-anti-CD43. (B) FACS analysis of peripheral splenic B cells from RAG2 WT (+/−), core (c/−), and knockout (−/−) mice stained with CyC-anti-B220 and PE-anti-IgM. The percentage of B220+IgM+ cells of total lymphocytes is shown. The percentages of B220 and CD43 staining are calculated on the basis of the total number of B220+ cells in each sample.
Staining with IgM and B220 revealed that the number of IgM+ B cells is reduced substantially in the core RAG2 mouse (approximately one-third of the number of WT cells) (Fig. 1A). The CD43/B220 stain shows an increased percentage of CD43hi/B220lo (pro-B) cells in the core RAG2 mouse and a reduction in the percentage of CD43lo/B220int (pre-B and immature B cells) and CD43−/B220hi (mature B cells) compared with WT (Fig. 1A). The absolute numbers of pro-B cells are equivalent in core RAG2 and WT mice (data not shown). This FACS profile indicates that core RAG2 mice exhibit impaired development at the pro-B cell stage. Peripheral lymphoid compartments such as spleen and lymph nodes were also examined, and they too contain decreased numbers of IgM+ B cells in the core RAG2 mice (≈50% of WT controls) (Fig. 1B), which is consistent with the decreased rate of B cell production in bone marrow.
Reduced V(D)J Recombination of the IgH Locus.
B cell development in RAG-deficient mice is arrested completely at the pro-B cell stage with virtually no CD43lo/B220int or CD43−/B220hi cells (Fig. 1A) (6, 7). Thus, we investigated whether the impairment in B cell development in core RAG2 mice is a reflection of impaired IgH gene rearrangement. For this purpose, we first took advantage of the fact that B cell development occurs in a nearly synchronized manner in fetal liver (27, 28) such that DH-to-JH rearrangement begins around day 13 of gestation, whereas VH-to-DJH rearrangement is not detectable until day 14 or 15 (29). Thus, at earlier days we could measure the level of DH-to-JH rearrangement in the absence of VH-to-DJH rearrangement and then measure VH-to-DJH rearrangement at later times.
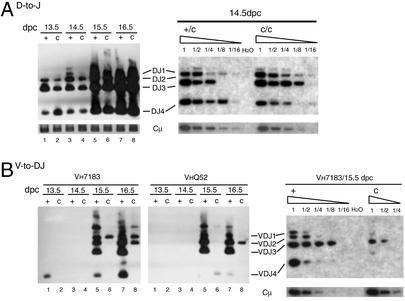
Fetal livers from WT and core RAG2 littermates were harvested at 13.5, 14.5, 15.5, and 16.5 days postconception (dpc), and genomic DNA from each sample was analyzed by PCR using standard primers that allow detection of rearrangements to JH 1–4 (Fig. 2). At days 13.5 and 14.5, when no VH-to-DJH rearrangements are detectable, DH-to-JH rearrangement can be detected in both WT and core RAG2 cells (Fig. 2). In addition, serial dilutions of 14.5-dpc samples indicate that the level of DH-to-JH rearrangement in core RAG2 mice is similar to WT at this time point (Fig. 2A). In contrast, at days 15.5 and 16.5, we see a marked reduction in the amount of VH-to-DJH rearrangement in the core RAG2 cells compared with WT (Fig. 2B). Three different VH family-specific primer pairs were tested [VH7183 (D-proximal), VHQ52, and VHJ558 (D-distal)] with similar results in all three cases (Fig. 2B and data not shown). Furthermore, direct comparison of PCR amplifications with VH7183 family primers of serially diluted DNA from multiple independent fetal livers derived from WT and core RAG2 littermates indicates that the level of VH7183-to-DJH rearrangements in WT mice is 4- to 8-fold higher than in the core RAG2 mice (see Fig. 2B Right for representative data). Therefore, expression of core RAG2 results in a more dramatic decrease in VH-to-DJH rearrangement of VH7183, VHQ52, and VHJ558 family members than DH-to-JH rearrangement in fetal liver cultures.
Figure 2.
VH-to-DJH rearrangement is reduced in developing B cells from core RAG2 mice. (A) PCR analysis of DH-to-JH rearrangement in fetal liver genomic DNA from RAG WT (+/c) and core (c/c) littermates is shown with serial dilutions from 14.5-dpc embryos (Right). (Left) PCR amplification of Cμ is included as a loading control and shown at the bottom. (B) PCR analysis of VH-to-DJH rearrangement of VH7183 and VHQ52 segments with serial dilutions for VH7183 rearrangements from 15.5-dpc embryos (Right). PCR products were probed with the JH4 probe.
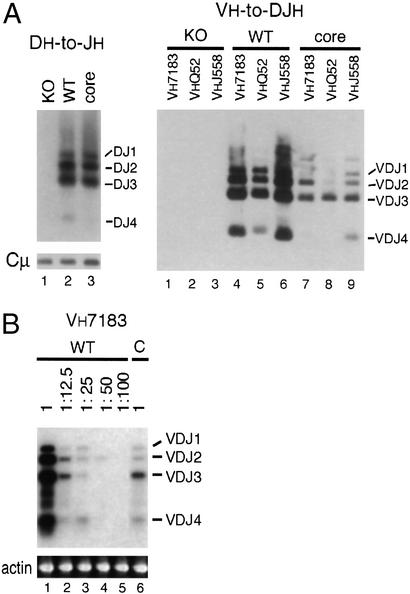
We also analyzed DH-to-JH and VH-to-DJH rearrangements at the IgH locus of unselected, sorted pro-B (CD43+B220lo) populations from the bone marrow of WT and core RAG2-expressing mice (Fig. 3). Consistent with the fetal liver experiments, the level of DH-to-JH rearrangement appeared similar in core RAG2 and WT pro-B cells (Fig. 3A), whereas the level of VH-to-DJH rearrangement of three different VH family members appeared lower in core RAG2 pro-B cells compared with WT controls (Fig. 3A). Furthermore, serial dilutions indicated that VH7183-to-DJH rearrangement was reduced ≈10- to 20-fold in the DNA from bone marrow-derived pro-B cells in the core RAG2 mice (Fig. 3B). Therefore, expression of core RAG2 also results in a more dramatic decrease in VH-to-DJH rearrangement of VH7183, VHQ52, and VHJ558 family members than DH-to-JH rearrangement in unselected pro-B cell populations.
Figure 3.
Reduced VH-to-DJH rearrangement in sorted core RAG2 pro-B cells. (A) PCR analysis of DH-to-JH rearrangement (Left) and VH-to-DJH rearrangement of VH7183, VHQ52, and VHJ558 segments (Right) in B220loCD43+ cells sorted from the bone marrow of RAG WT (+/−) and core (c/−) mice. Cμ amplification is shown (Left) as a control for input DNA. (B) Serial dilutions of WT DNA were amplified and compared with undiluted core DNA. The ratio of the mixed DNA (WT:KO) is indicated above each lane. The same dilutions were amplified with primers specific for β-actin as a control for DNA input.
Normal B cells almost always contain JH rearrangements on both JH alleles (30). In this context, in addition to their productive VH(D)JH rearrangement, ≈60% of normal B cells should contain DJH rearrangement with the remaining 40% containing nonproductive VHDJH rearrangement (30). Therefore, to analyze more quantitatively the extent to which DJH and overall VHDJH rearrangements are reduced in core RAG2 mice, we analyzed IgH gene rearrangements in B cell hybridomas. Three separate fusion experiments were analyzed (Table 1). Southern blotting with a JH probe demonstrated that ≈90% of control hybridomas contained two non-germ-line bands representing JH rearrangements (Table 1). The remainder, 10 ± 2%, contained a germ-line JH band and thus most likely represents tripartite fusions involving a nonlymphoid cell (30, 31). In contrast, 45 ± 8% of core RAG2 hybridomas contained a germ-line (nonrearranged) band (Table 1). Therefore, DH-to-JH rearrangement is reduced significantly in core RAG2 mice, and as a consequence nearly half of core RAG2 mature B cells, in contrast to normal B cells, develop with a germ-line JH allele.
Table 1.
IgH status of hybridomas derived from Rag2+/+ or Rag2c/c B cells
| Genotype | Experiment | V(D)J/DJ, % | V(D)J/V(D)J, % | V(D)J/germ line (%) | Total |
|---|---|---|---|---|---|
| RAG+/+ | Fusion 1 | 14 (61) | 7 (30) | 2 (9) | 23 |
| Fusion 2 | 7 (64) | 3 (27) | 1 (9) | 11 | |
| Fusion 3 | 5 (63) | 2 (25) | 1 (12) | 8 | |
| Total | 26 (63 ± 2%) | 12 (27 ± 3%) | 4 (10 ± 2) | 42 | |
| RAG2c/c | Fusion 1 | 8 (47) | 0 (0) | 9 (53) | 17 |
| Fusion 2 | 5 (56) | 0 (0) | 4 (44) | 9 | |
| Fusion 3 | 8 (62) | 0 (0) | 5 (38) | 13 | |
| Total | 21 (55 ± 8) | 0 (0 ± 0) | 18 (45 ± 8) | 39 |
To quantify the level of DH-to-JH versus VH-to-DJH rearrangement on the remaining nonselected alleles we conducted Southern blotting with a probe that hybridizes between the VH and DH segments. In control B cell hybridomas, 63 ± 2% and 27 ± 13% of the nonselected alleles contained DH-to-JH and VH-to-DJH rearrangements, respectively (Table 1). In core RAG2 hybridomas, 55 ± 8% of the nonselected alleles contained DH-to-JH rearrangements, but none contained VH-to-DJH rearrangements (Table 1). Therefore, the level of VH-to-DJH rearrangement on the nonselected allele is reduced dramatically in core RAG2 mice and thus impaired more substantially than DH-to-JH rearrangement.
Core RAG2 Mice Exhibit Partial Blocks in αβ and γδ T Cell Development.
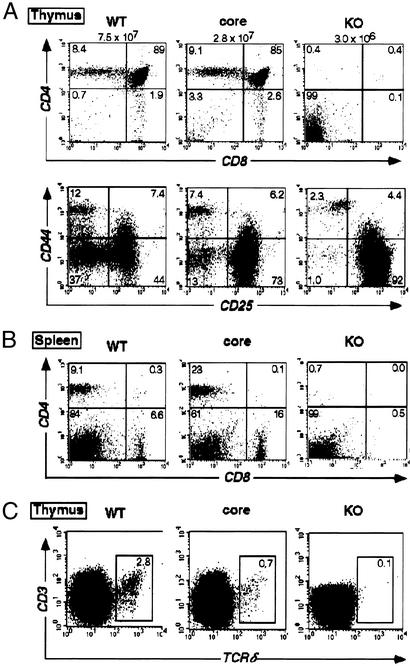
To assess the effect of core RAG2 expression on T cell development we carried out FACS analysis on cells derived from the thymus of core RAG2 (c/−) and WT (+/−) mice (Fig. 4). Core RAG2 mice contain reduced numbers of thymocytes (approximately one-third of controls) with an almost normal distribution of CD4+/CD8+ double-positive and CD4+CD8− or CD4−CD8+ single-positive thymocytes but a 4-fold increase in the percentage of cells in CD4−CD8− double-negative (DN) compartment (Fig. 4A). Further analysis demonstrated that this increase was due to a partial block in development at the CD25+CD44− (DNIII) stage (Fig. 4A). FACS analysis of splenic lymphocytes revealed that core RAG2 mice contain a higher percentage of CD4+ and CD8+ T cells because of the reduction in B cells (Fig. 4B); however, the absolute numbers of peripheral T lymphocytes is equivalent between core RAG2 and WT mice. Finally, a significant reduction (≈4-fold) in the number of thymic γδ T lymphocytes was also observed in core RAG2 mice (Fig. 4C).
Figure 4.
Analysis of T cell development in core RAG2 mice. (A Upper) FACS analysis of thymocytes from RAG2 WT (+/−), core (c/−), and knockout (KO−/−) mice using CD8-CyC and CD4-PE. (Lower) FACS analysis of thymocytes using CD44-PE and CD25-FITC after the removal of CD4+, CD8+, B220+, MAC1+, and GR1+ cells by electronic gating. (B) FACS analysis of splenic T cells using CD8-CyC and CD4-PE. (C) FACS analysis of thymocytes using TCRδ-FITC and CD3-PE after the removal of CD4+ and CD8+ cells by electronic gating.
Core RAG2 Mice Display Impaired TCRβ and TCRδ Recombination.
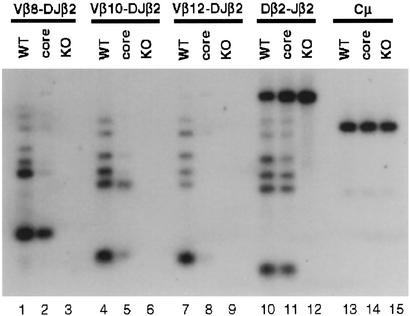
T cell development in RAG-deficient mice is arrested completely at the CD25+CD44− (DNIII) stage (Fig. 4A) (6, 7). Thus, we investigated whether the impaired T cell development in core RAG2 mice is a reflection of altered TCRβ gene rearrangement. For this purpose, the effect of core RAG2 expression on TCRβ V(D)J recombination was evaluated by PCR analysis on genomic DNA isolated from sorted, unselected CD25+CD44− DN thymocytes of WT mice and those expressing the core RAG2 protein. The levels of PCR products corresponding to Dβ2-to-Jβ2 rearrangements were reduced only mildly in the DNA from RAG2 core thymocytes (Fig. 5). However, the levels of PCR products corresponding to Vβ-to-Dβ2Jβ2 rearrangements of multiple Vβ genes showed a greater reduction, with only minimal complete VβDβ2Jβ2 rearrangements observed (Fig. 5). Similar results were obtained when Dβ1-to-Jβ1 and Vβ-to-Dβ1Jβ1 rearrangements were analyzed (data not shown). However, core RAG2 mice exhibited no significant differences in Vβ utilization (data not shown), indicating that the observed reduction in Vβ rearrangement is not biased by chromosomal location or other factors.
Figure 5.
Analysis of TCRβ rearrangement. PCR analysis of Dβ2-to-Jβ2 and Vβ-to-Dβ2Jβ2 rearrangement on DNA isolated from sorted CD44−CD25+ (DNIII) thymocytes of RAG2 WT (+/c), core (c/c), and knockout (KO, −/−) mice is shown. PCR amplification of Cμ was included as a loading control.
The reduction in number of thymic γδ T lymphocytes likely reflects impairment of TCRδ or TCRγ rearrangement. Thus, we examined the effect of core RAG2 expression on both TCRδ and TCRγ rearrangement by Southern blot analysis of thymus DNA from core RAG2 and WT control mice. We found that core RAG2 mice carry out an appreciable level of either Dδ1(Dδ2)Jδ1 or Vδ(Dδ1)Dδ2 rearrangement; however, a reduced level of complete TCRδ gene assembly was observed (Supporting Results and Fig. 7, which are published as supporting information on the PNAS web site). In contrast, no defect in TCRγ rearrangement was found (data not shown).
Discussion
The RAG2 C Terminus Is Required for Normal Lymphocyte Development.
The original finding that RAG2 retains its enzymatic activity in the absence of the C-terminal one-fourth of the protein provided a tremendous experimental convenience: a well behaved protein that could be readily overexpressed and purified for biochemical study. For this reason almost all biochemical work on RAG2 has been done with the truncated protein. Here we show that although the core RAG2 protein supports substantial lymphocyte development in vivo, removal of the C terminus is not without biological consequence. In mice expressing only core RAG2, development of the immune system is impaired. This impairment is seen in the B cell compartment as a reduction in the number of mature B cells (B220+IgM+) and pre-B and immature B cells (CD43lo/B220int) and a concomitant increase in the percentage of pro-B cells (CD43hi/B220lo) that accumulate at the developmental stage when VH-to-DJH joining should occur. Similarly, although the overall number of peripheral αβ T cells is unaffected in mice expressing core RAG2, thymic cellularity is reduced ≈3-fold with an accumulation of cells at the pro-T (CD44−CD25+) stage, during which Vβ to DJβ rearrangement occurs. Core RAG2 mice also exhibit a 4-fold reduction in the number of thymic γδ T lymphocytes.
The RAG2 C Terminus Is Required for Normal V(D)J Recombination of the Endogenous IgH, TCRβ, and TCRδ Loci.
In principle, the impaired lymphoid development in core RAG2 mice could be a result of altered V(D)J recombination or decreased cell proliferation/increased cell death or some combination of these factors. Cell cycle-regulated RAG2 expression mediated through the phosphorylation of Thr-490 (located in the deleted C terminus of RAG2) serves to restrict V(D)J recombination to G1 (32). Because the activity of the core RAG2 protein is not cell cycle-regulated in core RAG2 mice (Y.A., unpublished observations), continued expression of core RAG2 during S phase after the productive assembly and expression of IgH and TCRβ variable-region genes theoretically could lead to decreased cell proliferation/increased cell death. However, the lack of impaired lymphocyte development in heterozygous core RAG2 mice indicates that expression of the core RAG2 protein during S phase does not cause significant reduced cell proliferation/increased cell death after productive IgH and TCRβ V-to-DJ rearrangement.
Along with impaired lymphocyte development and decreased cellularity, we find that the core RAG2 protein supports a reduced level of chromosomal V(D)J recombination. Analysis of IgH gene assembly by PCR in unselected pro-B cells and Southern blotting in B cell hybridomas of core RAG2 mice indicates that both DH-to-JH and VH-to-DJH rearrangement are reduced significantly, with VH-to-DJH rearrangement dramatically more impaired. Similarly, analysis of TCRβ gene assembly by PCR in unselected pro-T cells of core RAG2 mice indicates that Vβ-to-DJβ rearrangement is reduced more substantially than Dβ-to-Jβ rearrangement. Furthermore, a reduction in the level of complete TCRδ variable-region gene assembly was observed by Southern blot analysis of core RAG2 thymus DNA. Therefore, we conclude that the observed reduction in complete V(D)J recombination of the endogenous IgH, TCRβ, and TCRδ loci is responsible for impaired lymphocyte development in core RAG2 mice.
The core RAG2 protein exhibits ≈50% activity of the full-length protein as measured with plasmid substrates in tissue culture (9–13). Therefore, on the simplest level, the observed reduction in the complete assembly of IgH, TCRβ, and TCRδ variable-region genes might reflect only a similar decrease of in vivo V(D)J recombinase activity. In this context, the 4-fold reduction in the number of γδ T cells that develop in RAG2 core mice can be explained entirely by the straightforward calculation that overall V(D)Jδ joining should be reduced on the order of 4-fold if both Dδ-to-Jδ and Vδ-to-Dδ joining were reduced ≈50%. However, our analyses clearly indicate that VH-to-DJH rearrangement was dramatically more impaired than DH-to-JH rearrangement and, in this context, cannot be accounted for by decreased recombination activity alone.
The ordered assembly of IgH genes is likely effected via developmental stage-specific DH and VH accessibility as DH-to-JH rearrangement occurs in an earlier developmental stage than VH rearrangement (30). Approximately half of core RAG2 mature B cells lack IgH rearrangements on their nonselected allele, indicating that expression of core RAG2 permits the development of pro-B cells to the stage in which VH rearrangement occurs without DH-to-JH rearrangement on both alleles. Therefore, the completion of DH-to-JH rearrangement on both alleles is not required for pro-B cells to progress to the developmental stage in which VH rearrangement occurs.
Potential Roles of the RAG2 C Terminus.
How does the C terminus of RAG2 influence IgH, and possibly TCRβ, V-to-DJ rearrangement? An intriguing possibility is that the C-terminal region of RAG-2 directly influences accessibility of antigen-receptor loci (18). Thus, the RAG2 C terminus might directly facilitate changes in chromatin structure or, more likely, facilitate assembly of the RAG1/2 complex on RSSs wrapped in chromatin. The association of other PHD-containing proteins with chromatin remodeling and modification (21–24) is consistent with such a role for RAG2 in influencing the accessibility of antigen-receptor loci. In this context, the PHD motif of RAG2 might serve a unique, undefined function in promoting accessibility of VH and Vβ segments.
Another possibility involves differential recombination efficiency of specific RSS pairs with the full-length versus core RAGs; for example, in the context of beyond 12/23 restrictions observed at the TCRβ locus (33). The 5′-Dβ1 12-RSS precisely targets rearrangement of a diverse repertoire of endogenous Vβ gene segments based on properties beyond those of simple 12/23 RSS compatibility (33). However, we find that the specific targeting of Vβ rearrangement to the 5′-Dβ1 12-RSS, versus the Jβ1.1 12-RSS, is not affected in mice expressing core RAG-2 (D.D.D., C.H.B., and F.W.A., unpublished observations). Therefore, the C terminus of RAG2 does not serve a specific role in the targeting of Vβs to the 5′-Dβ1 12-RSS via beyond 12/23-RSS restrictions. However, it is possible that the core RAG2 protein is particularly less efficient at assembling a synaptic complex or cleaving a signal pair derived from IgH and TCRβ V/D versus D/J RSS pairs. In this regard, the core and full-length RAGs catalyzed slightly different recombination patterns with regard to position of target RSS in extrachromosomal plasmid substrates (34).
Finally, other possibilities cannot be excluded. The noncore domains of RAG2 could facilitate localization of the protein to specific sites within the nucleus of progenitor lymphocytes where IgH and TCRβ V-to-DJ joining takes place. The recently noted order of synaptic complex assembly, with RAG proteins initially assembling on one RSS (preferably a 12-RSS) then searching for the second site (35, 36), might also be affected by the absence of the C terminus of RAG2 if, for example, the complex is less stable. In this context, the PHD theoretically could associate with chromatin over IgH and Vβ segments to facilitate synaptic complex assembly between V RSSs and RAG-bound 5′-D RSSs (1). Further in vivo experiments will be required to determine the one or more ways that the C terminus of RAG2 participates in chromosomal V(D)J recombination. Of particular interest will be whether IgH and TCRβ locus allelic exclusion is mediated, at least in part, through a signal that modifies the PHD motif and/or other residues of the RAG2 C terminus in progenitor lymphocytes to inhibit V-to-DJ rearrangement.
Supplementary Material
Acknowledgments
We thank Mark Schlissel for discussing unpublished data; Barry Sleckman and David Schatz for critical review of the manuscript; Laurie Davidson, Heather S. Hunt, and Yoko Akagi for technical assistance; and members of the F.W.A., M.A.O., and Y.T. laboratories for helpful discussions. C.H.B. is a Research Associate of The Howard Hughes Medical Institute. Y.A. was supported by the Japan Society for the Promotion of Science. F.W.A. is an Investigator of The Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants AI35714 (to F.W.A.), AI20047 (to F.W.A.), and GM48026 (to M.A.O.) and the Leukemia and Lymphoma Scholars Program (to M.A.O.).
Abbreviations
- TCR
T cell receptor
- RSS
recombination signal sequence
- RAG
recombination-activating gene
- IgH
Ig heavy chain
- PHD
plant homeodomain
- PE
phycoerythrin
- CyC
CyChrome
- FACS
fluorescence-activated cell sorter
- dpc
days postconception
- DN
double-negative
References
- 1.Bassing C H, Swat W, Alt F W. Cell. 2002;109,Suppl.:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.Hesslein D G, Schatz D G. Adv Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 5.Gorman J R, Alt F W. Adv Immunol. 1998;69:113–181. doi: 10.1016/s0065-2776(08)60607-0. [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 7.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 8.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 9.Cuomo C A, Oettinger M A. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadofsky M J, Hesse J E, Gellert M. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadofsky M, Hesse J E, van Gent D C, Gellert M. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 12.Kirch S A, Sudarsanam P, Oettinger M A. Eur J Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- 13.Silver D P, Spanopolou E, Mulligan R C, Baltimore D. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steen S B, Han J, Mundy C, Oettinger M A, Roth D B. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekiguchi J A, Whitlow S, Alt F W. Mol Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 16.Peixoto B R, Mikawa Y, Brenner S. Gene. 2000;246:275–283. doi: 10.1016/s0378-1119(00)00091-3. [DOI] [PubMed] [Google Scholar]
- 17.Roman C A, Cherry S R, Baltimore D. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 18.Kirch S A, Rathbun G A, Oettinger M A. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callebaut I, Mornon J P. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind L, Koonin E V. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 21.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 22.Zeremski M, Hill J E, Kwek S S, Grigorian I A, Gurova K V, Garkavtsev I V, Diatchenko L, Koonin E V, Gudkov A V. J Biol Chem. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]
- 23.Bochar D A, Savard J, Wang W, Lafleur D W, Moore P, Cote J, Shiekhattar R. Proc Natl Acad Sci USA. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalkhoven E, Teunissen H, Houweling A, Verrijzer C P, Zantema A. Mol Cell Biol. 2002;22:1961–1970. doi: 10.1128/MCB.22.7.1961-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley Interscience; 1989. [Google Scholar]
- 26.Gartner F, Alt F W, Monroe R, Chu M, Sleckman B P, Davidson L, Swat W. Immunity. 1999;10:537–546. doi: 10.1016/s1074-7613(00)80053-9. [DOI] [PubMed] [Google Scholar]
- 27.Siden E, Alt F, Shinefeld L, Sato V, Baltimore D. Proc Natl Acad Sci USA. 1981;78:1823–1827. doi: 10.1073/pnas.78.3.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strasser A, Rolink A, Melchers F. J Exp Med. 1989;170:1973–1986. doi: 10.1084/jem.170.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlissel M S, Morrow T. J Immunol. 1994;153:1645–1657. [PubMed] [Google Scholar]
- 30.Alt F W, Yancopoulos G D, Blackwell T K, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai E, Bottaro A, Davidson L, Sleckman B P, Alt F W. Proc Natl Acad Sci USA. 1999;96:1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Dordai D I, Lee J, Desiderio S. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 33.Bassing C H, Alt F W, Hughes M M, D'Auteuil M, Wehrly T D, Woodman B B, Gartner F, White J M, Davidson L, Sleckman B P. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 34. Jung, D., Bassing, C. H., Fugmann, S. D., Cheng, H.-L., Schatz, D. G. & Alt, F. W. (2003) Immunity, in press. [DOI] [PubMed]
- 35.Jones J M, Gellert M. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mundy C L, Patenge N, Matthews A G, Oettinger M A. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.