Abstract
Hepatitis B virus (HBV) infects humans and certain nonhuman primates. Viral clearance and acute disease are associated with a strong, polyclonal, multispecific cytotoxic T lymphocyte response. Infiltrating T cells, as well as other activated inflammatory cells, produce cytokines that can regulate hepatocellular gene expression. Using an HBV transgenic mouse model, our laboratory has previously demonstrated that adoptive transfer of HBV-specific cytotoxic T lymphocytes or injection of IL-2 can noncytopathically inhibit HBV gene expression by a posttranscriptional IFN-γ- and/or tumor necrosis factor α-dependent mechanism. Here, we report that HBV gene expression can also be controlled at the posttranscriptional level during persistent lymphocytic choriomeningitis virus infection. In contrast, it is controlled at the transcriptional level during acute murine cytomegalovirus infection or after repetitive polyinosinic-polycytidylic acid injection. Finally, we show that transcriptional inhibition of HBV is associated with changes in liver-specific gene expression. These results elucidate pathways that regulate the viral life cycle and suggest additional approaches for the treatment of chronic HBV infection.
Hepatitis B virus (HBV) is a hepatotropic DNA virus that infects humans and certain nonhuman primates. Although the host range of HBV is thought to be controlled at the stage of viral entry, the hepatic tropism of HBV is also due to tissue-specific viral gene expression (1–4). Upon entry into a cell, the 3.2-kb viral genome is delivered to the nucleus and converted into a covalently closed circular DNA molecule, which the cellular RNA polymerase II transcriptional machinery uses as a template for the synthesis of four 3′ coterminal viral mRNAs (3.5, 2.4, 2.1, and 0.7 kb). Transcription of each viral RNA is driven by a unique promoter in conjunction with shared enhancer elements. All of these regulatory elements are composed of binding sites for ubiquitous as well as liver-enriched transcription factors including CCAAT/enhancer-binding protein (C/EBP), hepatocyte nuclear factor (HNF)-1, -3, and -4, and peroxisomal proliferation-activated receptor (PPAR)/retinoid X receptor (RXR) heterodimers (5–26).
HBV causes acute and chronic necroinflammatory liver disease and hepatocellular carcinoma. In patients that resolve the infection, acute disease and viral clearance are associated with a strong, polyclonal, multispecific cytotoxic T lymphocyte (CTL) response (27). Activated CTLs not only kill target cells, but like other activated inflammatory cells they also produce cytokines that can act directly on hepatocytes to alter intracellular gene expression. Using an HBV transgenic mouse model, we have demonstrated that the induction of cytokines in the liver can noncytopathically inhibit HBV DNA replication and gene expression (28–34). Additionally, direct cytokine-mediated antiviral effects on HBV replication and gene expression have been confirmed in vitro in immortalized HBV transgenic mouse hepatocytes (35, 36).
Importantly, inhibition of HBV DNA replication and HBV gene expression occurs through independent mechanisms that exhibit distinct kinetics and cytokine dependence (33). HBV gene expression appears to be more resistant to inhibition because reduction of HBV RNA occurs later than the reduction of HBV DNA, and only a subset of the stimuli that inhibit HBV DNA replication also affect HBV gene expression. For example, whereas adenovirus infection or activation of NKT cells by α-galactosylceramide efficiently reduce intrahepatic levels of HBV DNA replicative intermediates, they do not affect viral mRNA levels (31, 37).
In contrast, both HBV DNA replication and HBV gene expression are inhibited in transgenic mice after adoptive transfer of HBV-specific CTLs (32), during acute and persistent lymphocytic choriomeningitis virus (LCMV) infection (34), during acute murine cytomegalovirus (MCMV) infection (31), and after treatment with IL-2, tumor necrosis factor α (TNF-α), or IFN-α/β (28, 29, 38). Nuclear run-on assays have demonstrated that after adoptive transfer of HBV-specific CTLs, HBV RNA is eliminated at the posttranscriptional level by an IFN-γ- and TNF-α-mediated mechanism (32, 33, 39). Likewise, IL-2 treatment reduced viral RNA by a TNF-α-dependent posttranscriptional mechanism (29, 38). Although it was shown that the IFN-α/β-induced effects on HBV gene expression were independent of TNF-α (38), the mechanisms by which LCMV, MCMV, and IFN-α/β reduce HBV RNA have remained unknown.
Thus, in this study we investigated whether LCMV, MCMV, and IFN-α/β inhibit HBV gene expression transcriptionally or posttranscriptionally. We found that HBV gene expression was controlled at the transcriptional or posttranscriptional level depending on the antiviral stimulus applied. Whereas LCMV infection decreased HBV RNA posttranscriptionally, HBV gene expression was controlled transcriptionally during acute MCMV infection and after repeated injections of the IFN-α/β inducer polyinosinic-polycytidylic acid (polyI/C). Furthermore, transcriptional, but not posttranscriptional, regulation of HBV gene expression coincided with the modulation of liver-associated genes, including liver-enriched transcription factors that have been shown to control HBV gene expression in other systems.
Materials and Methods
Mice.
The 1.3.32 HBV transgenic mouse lineage [Tg(HBV 1.3 genome) Chi32] has been described (3). These mice express and replicate HBV in the liver from a greater than genome length integrated transgene. Inbred C57BL/6 HBV transgenic mice were bred against B10D2 or BALB/c mice to produce the b/d F1 experimental mice used. Animals were housed in pathogen-free rooms under strict barrier conditions. Mice were matched by age, sex, and serum levels of hepatitis B e-antigen (HBeAg) (EBK 125I RIA Kit, DiaSorin, Stillwater, MN).
LCMV Infection.
The WE clone 54 of LCMV was provided by J. C. del la Torre and M. B. A. Oldstone (The Scripps Research Institute). LCMV was titered by plaque assay on Vero cells (40). To establish persistent LCMV infection, 1.3.32 B6D2 mice were infected by an intracranial inoculation of 103 plaque-forming units of virus within 24 h after birth.
MCMV Infection.
The MCMV Smith strain was provided by A. Campbell (Eastern Virginia Medical School, Norfolk). The virus was grown and titered on NIH 3T3 cells (American Type Culture Collection), and then passaged in vivo to produce the virulent MCMV stock used for these studies, as described (31). For experiments, 1.3.32 B6 BALB/c mice were given i.p. injections of 0.9% NaCl solution (saline) or 2.5 × 104 plaque-forming units of MCMV in a volume of 200 μl.
PolyI/C Injection.
Intravenous injections of 200 μg of polyI/C (Sigma) were delivered in 200 μl of saline. PolyI/C or control saline injections were administered three times at 24-h intervals.
Probes.
The ORFs of mouse GAPDH (GenBank accession no. M32599), albumin (AJ011413), PPAR-α (X57638), and RXR-α (X66223) were cloned from a 1.3.32B6D2 mouse by RT-PCR (One-Step RT-PCR, Qiagen, Chatsworth, CA). RT-PCR primers were designed based on the GenBank entries noted. PCR products of the expected size were generated and ligated into the pTEasy vector (Promega). Clones were subsequently verified by sequence analysis (The Scripps Research Institute, MEM DNA core facility). The rat HNF-4α plasmid (41) and the C/EBP-α plasmid (42) were provided by M. Tripodi (Fondazione Istituto Pasteur-Cenci Bolognetti, Università La Sapienza, Rome). The mouse HNF-1α plasmid was provided by G. Crabtree (Stanford University, Stanford, CA). The mouse major urinary protein (MUP) plasmid (43), was provided by W. A. Held (Roswell Park Memorial Institute, Buffalo, NY). The mouse metallothionine (MT) plasmid was provided by R. D. Palmiter (University of Washington, Seattle). The mouse chemokine Crg2 plasmid (44) was provided by J. Farber (National Institutes of Health, Bethesda). The mouse 2′5′-oligoadenylate synthetase (OAS) cloned by Zhou et al. (45) was provided by R. H. Silverman (The Cleveland Clinic, Cleveland).
Probes were generated from plasmid DNAs either by direct excision and purification of gene fragments or by PCR amplification. HBV RNA was detected with a complete genome-length fragment capable of detecting all viral transcripts.
Northern Blot.
Total cellular RNA was isolated by the guanidine thiocyanate method using standard protocols (46). Twenty micrograms of RNA was resolved in formaldehyde agarose gels and transferred to Nytran nylon membranes (Schleicher & Schuell). Transcripts were detected by hybridization with 32P-labeled cDNA probes, followed by analysis using a storage phosphor system (Cyclone, Packard).
RNase Protection Assay.
A mouse liver-specific transcription factor RNase protection assay (RPA) template set was created and used for these studies. Using the GenBank entries noted below, PCR primers were designed to amplify a fragment of specified length from each of the following genes: HNF-1α (M57966), HNF-1β (BC025189), HNF-4α (XM123982), C/EBP-α (NM007678), PPAR-α (X57638), RXR-α (M84817), and HNF-3β (L10409). Each fragment was subjected to blast analysis to ensure that no unwanted protected species would be generated. A HindIII restriction site was added to all 5′ primers to allow for the linearization of transcriptional templates for antisense RNA synthesis. Gene fragments were cloned from a 1.3.32B6D2 mouse as indicated above. Colonies were screened for correct insert size and direction (T7 synthesis) and subsequently verified by sequence analysis (The Scripps Research Institute, MEM DNA core facility). An RPA template for the ribosomal L32 gene was included as a loading standard (32, 47). Using RNA samples with known levels of each transcript, RPA probes were tested individually to confirm correct probe sizes and accurate representation of the specific gene. The probe set was assembled by mixing equal amounts of each linearized template to a final concentration of 50 ng/μl. For experiments, 10 μg of each RNA sample was subjected to RPA, as described (32, 47).
Isolation of Nuclei.
Nuclei were isolated by dounce homogenization and sucrose gradient centrifugation. Liver tissues were suspended in 5 ml of ice cold buffer A (10 mM Hepes, pH 7.9/25 mM KCl/1 mM EGTA/1 mM EDTA/0.32 M sucrose/0.15 mM spermine/0.5 mM spermidine/1 mM DTT/0.5 mM PMSF/0.5 μg/ml leupeptin/1 μg/ml aprotinin/1 μg/ml pepstatin) and homogenized in a 15-ml Potter-Elvehjem (Wheaton Science Products, Millville, NJ) tissue grinder. Samples were diluted with 2 vol of buffer B (buffer A with 2 M sucrose) and layered onto buffer B sucrose cushions in polyallomer centrifuge tubes. The gradient was centrifuged for 45 min at 24,000 rpm by using an SW40-Ti rotor (Beckman Coulter). The nuclear pellet was then resuspended in 2.5 ml of buffer A, diluted with 5 ml of buffer B, and layered onto a second 3.5-ml cushion of buffer B for centrifugation. Nuclei were resuspended in storage buffer (50 mM Tris, pH 8/0.1 mM EDTA/2 mM MgCl2/30% glycerol), counted, and frozen in liquid nitrogen.
Run-On Assay.
Approximately 1 × 107 nuclei were thawed on ice and mixed with an equal volume of 2× transcription reaction mix for a final composition of 25 mM Tris (pH 8), 100 mM KCl, 7 mM MgCl2, 12.5% glycerol, 0.5 mM ATP, GTP, and CTP, 10 mM creatine phosphate, 1 mM DTT, 10 μg/ml PMSF, and 250 μCi [32P]UTP (New England Nuclear). Transcription reactions were incubated 20 min at 26°C. RNase-free DNase was added and samples were incubated for 10 min at 30°C, followed by proteinase K digestion for 30 min at 45°C. After adding 1/10th volume of 2 M NaOAc, pH 4, RNA was extracted with 500 μl of H2O-saturated phenol and 200 μl of chloroform, and precipitated with 3 vol of ethanol at −80°C. Labeled RNA was pelleted, resuspended in STE (10 mM Tris, pH 8/1 mM EDTA/140 mM NaCl), and run through a G-50 column to remove free nucleotides (Amersham Pharmacia).
Slot Blot Hybridization.
Two micrograms of each linearized gene fragment or pUC19 negative control DNA was alkaline-denatured with 1/6th vol of 1 M NaOH and neutralized by 10 vol of 6× SSC. Probes were vacuum-transferred onto Nytran membranes by using a slot blot apparatus (Schleicher & Schuell) and immobilized by UV-crosslinking. Membranes were prehybridized at 60°C overnight [5× SSPE (3M NaCl, 200 mM NaH2PO4, 20 mM EDTA), 10× Denhardt's, 0.2% SDS, 200 μg/ml single-stranded DNA, 100 ng/ml Escherichia coli RNA]. The next day, ≈1 × 107 cpm of run-on RNA was added to 2 ml of fresh prewarmed hybridization buffer, and samples were incubated at 68°C for 60 h. Membranes were washed (2× SSC, 0.1% SDS) once for 20 min at room temperature, twice for 20 min at 65°C, and then treated with 5 μg/ml RNaseA in 2× SSC for 20 min at 37°C. After a final 20-min wash at room temperature, membranes were exposed to PhosphorImage screens.
Results
Posttranscriptional Inhibition of HBV Gene Expression During Persistent LCMV Infection.
The level of viral transcripts in the liver of HBV transgenic mice is decreased during persistent LCMV infection (34). To determine whether this inhibition occurs transcriptionally or posttranscriptionally, we performed Northern blot, RPA, and nuclear run-on experiments to monitor changes in the steady-state and transcription level of HBV RNA in persistently LCMV-infected versus noninfected HBV transgenic mice. At 3 months of age, LCMV-infected and noninfected littermates were killed. Total RNA and transcriptionally competent nuclei were isolated from the liver.
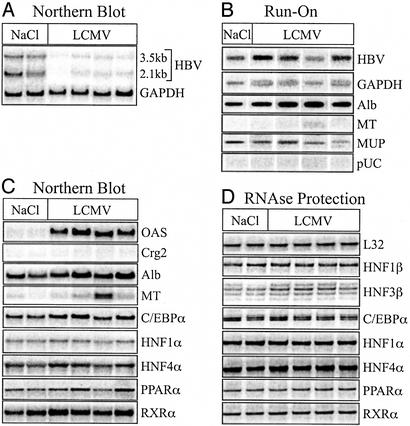
Northern blot analysis of liver RNA confirmed that steady-state levels of the HBV 3.5- and 2.1-kb transcripts were reduced in persistently LCMV-infected animals (Fig. 1A). When normalized to GAPDH levels to control for loading differences, HBV RNA was calculated to be reduced 9-fold in persistently LCMV-infected mice (Fig. 1A).
Figure 1.
Posttranscriptional inhibition of HBV gene expression during persistent LCMV infection. Total RNA and transcriptionally competent nuclei were isolated from the liver of uninfected (NaCl) and persistently LCMV-infected mice. Results from two uninfected and four LCMV-infected mice are shown. (A) Northern blot analysis of HBV and GAPDH steady-state RNA levels. (B) Nuclear run-on transcription analysis in liver nuclei. Nuclei of the two uninfected mice were pooled. (C) Steady-state RNA levels of the indicated genes analyzed by reprobing the Northern blot from A with cDNA probes. (D) The steady-state RNA levels of the indicated genes analyzed by RPA.
Nuclear run-on assays demonstrated that transcription of HBV was not significantly different in persistently LCMV-infected animals compared to uninfected animals (Fig. 1B). When normalized to GAPDH to control for the transcriptional competence of each nuclei preparation, the transcription of HBV RNA in persistently LCMV-infected mice was ≈74% of that observed in uninfected animals (Fig. 1B). Hence, changes in HBV transcription could not account for the large reduction in steady-state HBV RNA. Thus, similar to what has been observed after adoptive transfer of CTLs (39) and IL-2 injection (29), persistent LCMV infection inhibited HBV gene expression posttranscriptionally.
The steady-state RNA level of other hepatocellular genes in these mice was analyzed by Northern blot and/or RPA (Fig. 1 C and D, respectively). As expected, IFN-α/β was induced in LCMV-infected mice as indicated by the induction of OAS, a well-known IFN-α/β-induced gene (14-fold over background) (Fig. 1C). In contrast, expression of the IFN-γ-responsive chemokine Crg2 was not observed (Fig. 1C), nor was IFN-γ mRNA detected by RPA (data not shown). RNA levels of the positive acute-phase gene MT varied between mice in both groups irrespective of LCMV infection (Fig. 1C), and this was reflected at the transcriptional level (Fig. 1B). Steady-state expression and transcription of the negative acute-phase gene albumin were comparable in infected versus uninfected animals, although on average albumin expression in LCMV-infected mice was 30% higher (Fig. 1 B and C). Likewise, transcription of the negative acute-phase gene MUP was not significantly altered (Fig. 1B). Consistent with a lack of global changes in liver-specific gene expression, we did not detect changes in the expression of the transcription factors, C/EBP-α, HNF-1α, HNF-4α, PPAR-α, RXR-α, or HNF-1β, between persistently LCMV-infected and noninfected mice by either Northern blot (Fig. 1C) or RPA (Fig. 1D), although a slight 1.5-fold increase in HNF-3β mRNA was detectable by RPA (Fig. 1D). Hence, persistent LCMV-infection of HBV transgenic mice induced posttranscriptional elimination of HBV transcripts without significantly changing expression of liver-specific transcription factors that are known to regulate HBV gene expression in vitro.
Transcriptional Inhibition of HBV Gene Expression During Acute MCMV Infection.
To further investigate the mechanisms that regulate HBV gene expression in vivo, we determined whether HBV RNA was reduced transcriptionally or posttranscriptionally during acute MCMV infection. Groups of three age-, sex-, and HBeAg-matched HBV transgenic mice were injected with saline or 2.5 × 104 plaque-forming units of MCMV. Four days postinfection, total RNA and transcriptionally competent nuclei were isolated from the liver.
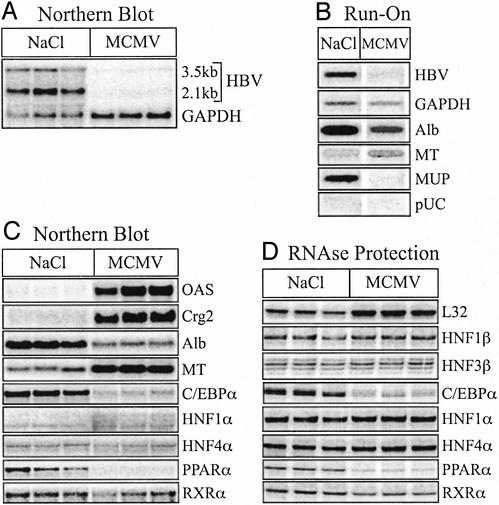
As reported (31), Northern blot analysis revealed that HBV RNA was reduced in the livers of MCMV-infected mice (Fig. 2A). Compared with the housekeeping gene GAPDH, HBV RNA was decreased 34-fold (Fig. 2A). Nuclear run-on analysis indicated that HBV transcription was inhibited 6-fold in MCMV-infected mice, relative to the transcriptional activity of GAPDH (Fig. 2B). Although these results do not preclude the possibility that MCMV infection also induced posttranscriptional control of HBV gene expression, they clearly demonstrate that HBV gene expression was transcriptionally inhibited during acute MCMV infection.
Figure 2.
Transcriptional inhibition of HBV gene expression during MCMV infection. Groups of sex-, age- and HBeAg-matched mice were injected i.p. with saline (NaCl) or MCMV. At 4 days postinfection, total RNA and transcriptionally competent nuclei were isolated from each liver. Results for three saline-injected and three MCMV-infected animals are shown. (A) Northern blot of HBV and GAPDH steady-state RNA levels. (B) Nuclear run-on transcriptional analysis in nuclei isolated from the same mice. Nuclei of the three mice in each group were pooled. (C) Steady-state RNA levels of the indicated genes analyzed by reprobing the Northern blot from A with the cDNA probes. (D) Steady-state RNA levels of the indicated genes analyzed by RPA.
In contrast to what was observed during persistent LCMV infection, transcriptional inhibition of HBV during acute MCMV infection was associated with multiple changes in liver-specific gene expression (Fig. 2 C and D). We detected an induction of OAS, as well as the IFN-γ-responsive chemokine, Crg2 (12- and 22-fold over background, respectively) (Fig. 2C). MCMV-infected mice also exhibited a 3-fold increase in MT, as well as a 5-fold reduction in albumin mRNA (Fig. 2C). Nuclear run-on analysis indicated that both of these changes, as well as the shut-off of MUP expression, occurred at the transcriptional level (Fig. 2B). Notably, the respective changes in expression observed for these three genes are characteristic of an acute-phase response in the liver (48, 49).
Consistent with transcriptional changes in multiple liver genes, we observed changes in liver-specific transcription factor expression. MCMV infection not only resulted in an 8-fold decrease in C/EBP-α expression, but there was also the appearance of additional HNF-1α transcripts (Fig. 2C), which was reflected as a 2-fold increase in HNF-1α expression by Northern blot (Fig. 2C). Of the three transcription factors that have been shown to be sufficient for HBV transcription in nonhepatic cells in vitro (4), PPAR-α mRNA was reduced the most as measured by Northern blot (17-fold) and RPA (13-fold), whereas RXR-α and HNF-4α remained relatively unchanged (Fig. 1 C and D). Likewise, there was no change in HNF-1β or HNF-3β RNA levels in MCMV-infected mice (Fig. 2D). Thus, transcriptional inhibition of HBV during MCMV infection coincided with specific changes in the transcriptional environment in the liver, most notably additional HNF-1α transcripts, and a significant reduction in C/EBP-α and PPAR-α, which all potentially regulate HBV transcription. It should be noted, however, that because expression of the housekeeping genes, GAPDH (Fig. 2A) and ribosomal L32 (Fig. 2D), appeared to be increased 2-fold in MCMV-infected mice in multiple assays, quantification of relative gene expression levels adjusted to these common loading controls may be off by up to a factor of 2 (increases underestimated; decreased overestimated).
Repeated Injection of PolyI/C Inhibits HBV Gene Expression at the Transcriptional Level.
A single injection of 200 μg of polyI/C into HBV transgenic mice transiently inhibits HBV DNA replication by an IFN-α/β-dependent mechanism without affecting the steady-state levels of HBV mRNA (50, 51). To determine whether repeated, and thus prolonged, exposure to polyI/C could influence HBV gene expression, groups of three age-, sex-, and HBeAg-matched HBV transgenic mice were injected daily for 3 days with saline or 200 μg of polyI/C. Twenty-four hours after the last injection, mice were killed and HBV gene expression was analyzed.
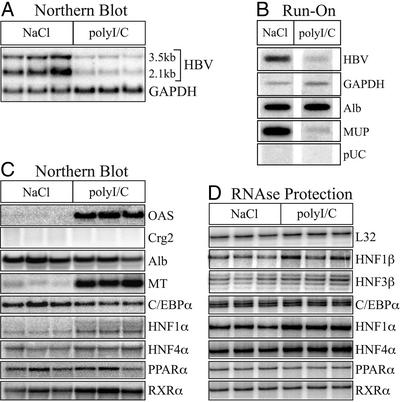
Northern blot analysis of total liver RNA indicated that repetitive polyI/C injection decreased the steady-state levels of HBV RNA in the liver (Fig. 3A). When normalized to GAPDH to control for loading differences, HBV transcripts in polyI/C-treated mice were reduced 7-fold (Fig. 3A). Nuclear run-on analysis on nuclei pooled from the mice in each group revealed that this reduction in HBV RNA was associated with a 14-fold inhibition of HBV transcription relative to GAPDH (Fig. 3B).
Figure 3.
Transcriptional inhibition of HBV gene expression after repetitive injection of polyI/C. Groups of sex-, age-, and HBeAg-matched mice were injected daily with saline (NaCl) or polyI/C for 3 days. At 24 h after the last injection, total RNA and transcriptionally competent nuclei were isolated from each liver. Results from three saline- and three polyI/C-injected mice are shown. (A) Northern blot of HBV and GAPDH steady-state RNA levels. (B) Nuclear run-on transcriptional analysis in nuclei isolated from the same mice. Nuclei of the three mice in each group were pooled. (C) Steady-state RNA levels of the indicated genes analyzed by reprobing the Northern blot from A with cDNA probes. (D) Steady-state RNA levels of the indicated genes analyzed by RPA.
Transcriptional inhibition of HBV after multiple injections of polyI/C was associated with changes in liver gene expression, similar to acute MCMV infection. OAS and MT mRNA were induced in polyI/C-treated mice, 17- and 9-fold, respectively (Fig. 3C). Likewise, polyI/C also induced higher molecular weight HNF-1α transcripts (Fig. 3C), which was reflected as a 2-fold and 3-fold increase in HNF-1α expression by Northern blot (Fig. 2C) and RPA (Fig. 2D), respectively. Another similarity between polyI/C injection and MCMV infection was the 30-fold decrease in MUP transcription (Fig. 3B). Although polyI/C treatment was also calculated to have caused a 40% reduction in steady-state levels of albumin mRNA (Fig. 3C), this apparent reduction may be an artifact of the normalization to GAPDH, which appeared to be increased in polyI/C-injected mice (Fig. 3A). Again, there was also no significant change in the RNA levels of HNF-4α, RXR-α, HNF-1β, or HNF-3β measured by Northern blot and/or RPA (Fig. 3 C and D). Unlike MCMV infection, however, there was no evidence of IFN-γ induction, as indicated by a lack of Crg2 expression (Fig. 3C) and the absence of IFN-γ detection by RPA (data not shown), nor was there a decrease in C/EBP-α or PPAR-α mRNA (Fig. 3 C and D).
Discussion
The data presented here demonstrate that HBV gene expression can be inhibited transcriptionally or posttranscriptionally depending on the nature of the antiviral stimulus applied. Consistent with previous findings attained after in vivo CTL, IL-2, and TNF-α injection (32, 33, 39), we found that HBV RNA was controlled posttranscriptionally in HBV transgenic mice during persistent LCMV infection. In contrast, we now also document transcriptional inhibition of HBV gene expression in the liver of HBV transgenic mice under at least two conditions: acute MCMV infection and repeated polyI/C injection.
To more fully characterize the intracellular events associated with these changes in HBV gene expression, we simultaneously examined the expression of representative endogenous liver transcripts. OAS and Crg2 were used as indicators of IFN-α/β and IFN-γ signaling, respectively. MT, albumin, and MUP were used as indicators of changes in liver-specific gene expression. Additionally, we directly monitored the expression of the liver-enriched transcription factors, C/EBP-α, HNF-1α, HNF-4α, PPAR-α, RXR-α, HNF-1β, and HNF-3β, because HBV transcription in vitro can be regulated by many of these factors (4–6, 8, 10–12, 15–19, 22–24, 52–54), and they are thought to mediate the tissue-restricted expression of HBV in vivo (1–3).
Consistent with the hypothesis that expression of liver-enriched transcription factors is directly related to the transcription of HBV in vivo, we observed changes in hepatocyte-specific gene expression coincident with transcriptional inhibition of HBV, but not under conditions in which HBV RNA was eliminated posttranscriptionally. Specifically, when HBV RNA was reduced posttranscriptionally during persistent LCMV infection, expression of MT, albumin, MUP, C/EBP-α, HNF-1α, HNF-4α, PPAR-α, RXR-α, HNF-1β, and HNF-3β remained relatively unchanged. Furthermore, analysis of the steady-state and transcriptional levels of the same genes in HBV transgenic mice 10 days after adoptive transfer of HBV-specific CTLs gave identical results, again indicating posttranscriptional elimination of HBV RNA in the absence of significant change in liver-specific gene expression (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org).
In contrast, when HBV gene expression was inhibited transcriptionally during acute MCMV infection or after repeated injections of polyI/C, liver-specific gene expression changed, and some of these changes were similar under both conditions. First, after both stimuli, we detected additional HNF-1α transcripts, and an overall increase in HNF-1α RNA. This finding raises the possibility that alterations in HNF-1α could be related to the transcriptional changes in HBV. Based on previous in vitro studies (15–17), one might expect HNF-1α to positively affect HBV transcription; however, HBV transgenic mice that lack HNF-1α have increased levels of HBV replication (55), which is thought to be associated with small increases in viral RNA. Hence, an increase in HNF-1α coincident with decreased HBV transcription is consistent with this in vivo observation. One could postulate various explanations for this type of unexpected effect in vivo. Increased concentrations of HNF-1α could shift the equilibrium such that HNF-1α now out competes a stronger transcriptional activator or the additional isoforms of HNF-1α induced could be less active. Alternatively, an increase in HNF-1α transcription from the preS1 promoter (5, 15, 17) could sterically interfere with transcription from the other viral promoters, as discussed below.
Second, both acute MCMV infection and repetitive polyI/C injection induced expression of MT and transcriptional repression of MUP. Again this finding suggests that similar gene expression changes occurred under both conditions, in particular these changes could be indicators of an acute-phase response. Notably, the transcriptional changes in HBV mirrored that of MUP. Also like MUP, HBV gene expression is higher in male mice and can be induced by testosterone (56–58). Thus, under some conditions hormone-related regulatory effects may exert dominant effects on HBV transcription. Importantly, this finding underscores the idea that multiple regulatory networks most likely intersect and together determine the level of HBV transcription in vivo. In this regard, it is noteworthy that HBV is also subject to hormonal regulation by glucocorticoids (58–60) and contains an intact IFN-stimulated response element and IFN-regulatory element (ref. 61 and references therein). The ability of all of these various regulatory pathways to potentially effect HBV gene expression adds enormous complexity to the control of HBV transcription in vivo. Indeed, the capacity of HBV promoters and enhancers to bind and be activated by numerous transcriptional regulators (5–23, 25, 26) may prohibit the identification of a single transcription factor whose activity parallels that of HBV in all cases of transcriptional regulation (i.e., a “master switch”).
Taken together our data support a link between the state of hepatocyte-specific gene regulation and the synthesis of HBV RNA in vivo. Consistent with in vitro data (4–6, 8, 10–12, 15–19, 22–24, 52–54), expression changes in liver-specific transcription factors were detected coincident with changes in HBV promoter activity. Importantly however, this in vivo analysis also implicated additional liver-specific regulatory pathways (e.g., hormonal regulation/acute-phase liver homeostasis) as participants in the control of HBV gene expression. Further experiments are now needed to determine whether changes in the activity of one or more regulatory factors is directly responsible for the transcriptional inhibition of HBV gene expression in our system. Answering this question requires analysis of protein levels, as well as posttranslational modifications known to affect the function of these proteins. In the case of HNF-4α, PPAR-α, and RXR-α, which are all members of the nuclear hormone receptor family, assessing the availability of their ligands may also be informative. Finally, information about the engagement of transcription factors on the HBV promoters and enhancers in vivo could be obtained by DNA footprint analysis.
Further experiments should also address the upstream signal(s) that initiate the antiviral inhibition of HBV gene expression. During persistent LCMV infection, the absence of detectable IFN-γ or TNF-α (data not shown) suggests that posttranscriptional inhibition of HBV in this case may result from IFN-α/β signaling. This observation extends our previous findings that posttranscriptional inhibition of HBV gene expression in HBV transgenic mice can be mediated by IFN-γ and TNF-α (29, 38, 39).
Curiously, transcriptional inhibition of HBV after MCMV infection and repetitive polyI/C injection also occurred in the presence of a strong IFN-α/β response, suggesting that IFN-α/β may also be involved in the transcriptional inhibition of HBV. The hypothesis that IFN-α/β may mediate posttranscriptional inhibition of HBV RNA under some conditions and transcriptional regulation of HBV RNA under others is not necessarily contradictory because IFN-α/β signaling may be necessary, but not sufficient to inhibit HBV transcription. It is also possible that sustained IFN-α/β signaling as it occurs during persistent LCMV infection differs from the transient induction of IFN-α/β that occurs during acute MCMV infection or polyI/C injection, in that some aspect(s) of IFN-α/β signaling may become exhausted over time or the host may become tolerant to a given level of IFN-α/β at the cellular and/or systemic level.
It is also intriguing that expression of the viral 3.5- and 2.1-kb RNAs are regulated in concert, despite the fact that they contain distinct combinations of regulatory elements. Although HNF-3 binding sites are present in all HBV promoters (6, 10, 14, 18, 21, 24), we did not detect changes in HNF-3β expression coincident with transcriptional changes in HBV. Yet, because HNF-3 activity is thought to be modulated by the ratio between its multiple isoforms (α, β, and γ), as well as by collaborative interactions with other unrelated factors such as STAT3, STAT5b, peptide hormones, and glucocorticoids (62–64), it is possible that there were HNF-3 effects that were not identified by our gene expression analysis. Alternatively, the viral enhancers might be responsible for the global regulation of the separate viral promoters, as they are able to exert regulatory effects on multiple HBV promoters in vitro (25, 26, 65). It is also possible that rather than being independently regulated by a common transcription factor(s) or regulatory element(s), coordinate regulation of the various viral transcripts occurs because of the close proximity of their promoters. In fact, positional regulation has already been demonstrated to occur between the preS1 and preS2 promoters (66) and between the nucleocapsid promoter and the downstream envelope promoters (67).
Finally, we also observed kinetic differences between transcriptional and posttranscriptional regulation of HBV; specifically, transcriptional inhibition occurred earlier than posttranscriptional inhibition. After multiple polyI/C injections and acute MCMV infection, HBV RNA was reduced transcriptionally by day 1 or 3, respectively. In contrast, posttranscriptional clearance of HBV RNA was not detected until 5 or more days after adoptive transfer of CTLs (ref. 39 and Fig. 4). Likewise, if HBV reporter constructs are delivered to mice persistently infected with LCMV, it takes 6 days before posttranscriptional inhibition of HBV occurs (S.L.U. and F.V.C., unpublished data).
Although the gene expression and kinetic differences we have observed between transcriptional and posttranscriptional inhibition of HBV may provide insight into possible mechanisms responsible for these antiviral activities, the molecular basis of these different pathways has yet to be determined. Elucidating the means by which HBV gene expression can be controlled is important not only in terms of understanding the viral life cycle, but also in practical terms as it may provide additional avenues of research into the treatment of chronically infected patients. Noncytopathic inhibition of viral gene expression is an attractive therapeutic means of controlling infections, especially in the case of HBV where the 3.5-kb RNA serves as the template for reverse transcription of the encapsidated viral DNA genome. The presented data demonstrate that hepatocellular mechanisms exist that can inhibit HBV gene expression either transcriptionally or posttranscriptionally. Further understanding the nature of these antiviral mechanisms should provide valuable insights into novel strategies for controlling HBV infection.
Supplementary Material
Acknowledgments
We thank Bryan Boyd for technical assistance, Heike Mendez and Amber Morris for maintenance and screening of rodent colonies, Luca G. Guidotti for helpful discussions and technical assistance, and Alan McLachlan for critical reading of the manuscript. We also thank all of the colleagues who provided plasmids, viruses, and other necessary reagents. This work was supported by National Institutes of Health Grant CA40489 (to F.V.C.). S.L.U. was supported by National Research Service Award Individual Postdoctoral Fellowship AI49670 from the National Institutes of Health. This is manuscript 15467-MEM from The Scripps Research Institute.
Abbreviations
- HBV
hepatitis B virus
- C/EBP
CCAAT/enhancer-binding protein
- HNF
hepatocyte nuclear factor
- PPAR
peroxisomal proliferation-activated receptor
- RXR
retinoid X receptor
- CTL
cytotoxic T lymphocyte
- LCMV
lymphocytic choriomeningitis virus
- MCMV
murine cytomegalovirus
- TNF-α
tumor necrosis factor α
- polyI/C
polyinosinic-polycytidylic acid
- HBeAg
hepatitis B e-antigen
- MUP
major urinary protein
- MT
metallothionine
- OAS
2′5′-oligoadenylate synthetase
- RPA
RNase protection assay
References
- 1.Araki K, Miyazaki J, Hino O, Tomita N, Chisaka O, Matsubara K, Yamamura K. Proc Natl Acad Sci USA. 1989;86:207–211. doi: 10.1073/pnas.86.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farza H, Hadchouel M, Scotto J, Tiollais P, Babinet C, Pourcel C. J Virol. 1988;62:4144–4152. doi: 10.1128/jvi.62.11.4144-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidotti L G, Matzke B, Schaller H, Chisari F V. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang H, McLachlan A. Proc Natl Acad Sci USA. 2001;98:1841–1846. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang H-K, Ting L. Virology. 1989;170:176–183. doi: 10.1016/0042-6822(89)90364-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen M, Hieng S, Qian X, Costa R, Ou J. Virology. 1994;205:127–132. doi: 10.1006/viro.1994.1627. [DOI] [PubMed] [Google Scholar]
- 7.Dikstein R, Faktor O, Shaul Y. Mol Cell Biol. 1990;10:4427–4430. doi: 10.1128/mcb.10.8.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia A, Ostapchuk P, Hearing P. J Virol. 1993;67:3940–3950. doi: 10.1128/jvi.67.7.3940-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Chen M, Yen T, Ou J. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson J L, Raney A K, McLachlan A. Virology. 1995;208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Cabrera M, Letovskyk J, Hu K Q, Siddiqui A. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Cabrera M, Letovsky J, Hu K, Siddiqui A. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 13.Trujillo M A, Letovsky J, Maguire H F, Lopez-Cabrera M, Siddiqui A. Proc Natl Acad Sci USA. 1991;88:3797–3801. doi: 10.1073/pnas.88.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ori A, Shaul Y. Virology. 1995;207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 15.Raney A K, Milich D R, Easton A J, McLachlan A. J Virol. 1990;64:2360–2368. doi: 10.1128/jvi.64.5.2360-2368.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raney A K, Easton A J, Milich D R, McLachlan A. J Virol. 1991;65:5774–5781. doi: 10.1128/jvi.65.11.5774-5781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raney A, Easton A, McLachlan A. J Gen Virol. 1994;75:2671–2679. doi: 10.1099/0022-1317-75-10-2671. [DOI] [PubMed] [Google Scholar]
- 18.Raney A K, Zhang P, McLachlan A. J Virol. 1995;69:3265–3272. doi: 10.1128/jvi.69.6.3265-3272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuh C-H, Ting L-P. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou D-X, Yen T S B. Mol Cell Biol. 1991;11:1353–1359. doi: 10.1128/mcb.11.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raney A, McLachlan A. J Gen Virol. 1997;78:3029–3038. doi: 10.1099/0022-1317-78-11-3029. [DOI] [PubMed] [Google Scholar]
- 22.Raney A, Johnson J, Palmer C, McLachlan A. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Mertz J. J Virol. 2001;75:11354–11364. doi: 10.1128/JVI.75.23.11354-11364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosovsky M, Huan B, Siddiqui A. J Biol Chem. 1996;271:21859–21869. doi: 10.1074/jbc.271.36.21859. [DOI] [PubMed] [Google Scholar]
- 25.Antonucci T K, Rutter W J. J Virol. 1989;63:579–583. doi: 10.1128/jvi.63.2.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu K-Q, Siddiqui A. Virology. 1991;181:721–726. doi: 10.1016/0042-6822(91)90906-r. [DOI] [PubMed] [Google Scholar]
- 27.Chisari F V, Ferrari C. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 28.Gilles P N, Fey G, Chisari F V. J Virol. 1992;66:3955–3960. doi: 10.1128/jvi.66.6.3955-3960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilhot S, Guidotti L G, Chisari F V. J Virol. 1993;67:7444–7449. doi: 10.1128/jvi.67.12.7444-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanaugh V J, Guidotti L G, Chisari F V. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavanaugh V J, Guidotti L G, Chisari F V. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel R D, Schreiber R D, Chisari F V. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 34.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B A, Chisari F V. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasquetto V, Wieland S, Uprichard S, Tripodi M, Chisari F. J Virol. 2001;76:5646–5653. doi: 10.1128/JVI.76.11.5646-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland S, Vega R, Muller R, Evans C, Hilbush B, Guidotti L, Sutcliffe J, Schultz P, Chisari F. J Virol. 2003;77:1227–1236. doi: 10.1128/JVI.77.2.1227-1236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakimi K, Guidotti L, Koezuka Y, Chisari F. J Exp Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guidotti L G, Guilhot S, Chisari F V. J Virol. 1994;68:1265–1270. doi: 10.1128/jvi.68.3.1265-1270.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsui L V, Guidotti L G, Ishikawa T, Chisari F V. Proc Natl Acad Sci USA. 1995;92:12398–12402. doi: 10.1073/pnas.92.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutko F J, Oldstone M B A. J Gen Virol. 1983;64:1689–1698. doi: 10.1099/0022-1317-64-8-1689. [DOI] [PubMed] [Google Scholar]
- 41.Sladek F, Zhong W, Lai E, Darnell J J. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 42.Friedman A, Landschulz W, McKnight S. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 43.Kuhn N, Woodworth-Gutai M, Gross K, Held W. Nucleic Acids Res. 1984;12:6073–6090. doi: 10.1093/nar/12.15.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanguri P, Farber J. J Biol Chem. 1990;265:15049–15057. [PubMed] [Google Scholar]
- 45.Zhou A, Hassel B, Silverman R. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 46.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 47.Hobbs M V, Weigle W O, Noonan D J, Torbett B E, McEvilly R J, Koch R J, Cardenas G J, Ernst D N. J Immunol. 1993;150:3602–3614. [PubMed] [Google Scholar]
- 48.Ramadori G, Christ B. Semin Liver Dis. 1999;19:141–155. doi: 10.1055/s-2007-1007106. [DOI] [PubMed] [Google Scholar]
- 49.Moshage H. J Pathol. 1997;181:257–266. doi: 10.1002/(SICI)1096-9896(199703)181:3<257::AID-PATH756>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 50.Wieland S, Guidotti L, Chisari F. J Virol. 2000;74:4165–4173. doi: 10.1128/jvi.74.9.4165-4173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClary H, Koch R, Chisari F, Guidotti L. J Virol. 2000;74:2255–2264. doi: 10.1128/jvi.74.5.2255-2264.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou D-X, Yen T S B. J Biol Chem. 1991;266:23416–23421. [PubMed] [Google Scholar]
- 53.Raney A, Kline E, Tang H, McLachlan A. Virology. 2001;289:239–251. doi: 10.1006/viro.2001.1169. [DOI] [PubMed] [Google Scholar]
- 54.Tang H, Raney A, McLachlan A. J Virol. 2001;75:8937–8948. doi: 10.1128/JVI.75.19.8937-8948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raney A, Eggers C, Kline E, Guidotti L, Pontoglio M, Yaniv M, McLachlan A. J Virol. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knopf J, Gallagher J, Held W. Mol Cell Biol. 1983;2:2232–2240. doi: 10.1128/mcb.3.12.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derman E. Proc Natl Acad Sci USA. 1981;78:5425–5429. doi: 10.1073/pnas.78.9.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farza H, Salmon A M, Hadchouel M, Moreau J L, Babinet C, Tiollais P, Pourcel C. Proc Natl Acad Sci USA. 1987;84:1187–1191. doi: 10.1073/pnas.84.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tur-Kaspa R, Burk R D, Shaul Y, Shafritz D A. Proc Natl Acad Sci USA. 1986;83:1627–1631. doi: 10.1073/pnas.83.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chou C-K, Wang L-H, Lin H-M, Chi C-W. Hepatology. 1992;16:13–18. doi: 10.1002/hep.1840160104. [DOI] [PubMed] [Google Scholar]
- 61.Alcantara F, Tang H, McLachlan A. Nucleic Acids Res. 2002;30:2068–2075. doi: 10.1093/nar/30.9.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa R. In: Liver Gene Expression. Tronche F, Yaniv M, editors. Austin, TX: Landes; 1994. [Google Scholar]
- 63.Waris G, Siddiqui A. J Virol. 2002;76:2721–2729. doi: 10.1128/JVI.76.6.2721-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park S, Waxman D. J Biol Chem. 2001;276:43031–43039. doi: 10.1074/jbc.M107597200. [DOI] [PubMed] [Google Scholar]
- 65.Ganem D. In: Fields Virology. Fields B N, Knipe D M, Howely P M, editors. Vol. 2. Philadelphia: Lippincott–Raven; 1996. pp. 2703–2730. [Google Scholar]
- 66.Bulla G, Siddiqui A. Virology. 1989;170:251–260. doi: 10.1016/0042-6822(89)90373-5. [DOI] [PubMed] [Google Scholar]
- 67.Tang H, McLachlan A. J Virol. 2002;76:8572–8581. doi: 10.1128/JVI.76.17.8572-8581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.