Abstract
The fusion of mammalian cells into syncytia is a developmental process that is tightly restricted to a limited subset of cells. Besides gamete and placental trophoblast fusion, only macrophages and myogenic stem cells fuse into multinucleated syncytia. In contrast to viral cell fusion, which is mediated by fusogenic glycoproteins that actively merge membranes, mammalian cell fusion is poorly understood at the molecular level. A variety of mammalian transmembrane proteins, among them many of the immunoglobulin superfamily, have been implicated in cell-cell fusion, but none has been shown to actively fuse cells in vitro. Here we report that the FGFRL1 receptor, which is up-regulated during the differentiation of myoblasts into myotubes, fuses cultured cells into large, multinucleated syncytia. We used luciferase and GFP-based reporter assays to confirm cytoplasmic mixing and to identify the fusion inducing domain of FGFRL1. These assays revealed that Ig-like domain III and the transmembrane domain are both necessary and sufficient to rapidly fuse CHO cells into multinucleated syncytia comprising several hundred nuclei. Moreover, FGFRL1 also fused HEK293 and HeLa cells with untransfected CHO cells. Our data show that FGFRL1 is the first mammalian protein that is capable of inducing syncytium formation of heterologous cells in vitro.
Keywords: Actin, Cell Surface Protein, Cell Surface Receptor, Cell-Cell Interaction, Cytoskeleton, Glycosaminoglycan, Heparin-binding Protein, Cell Fusion, Cell-Cell Fusion, Fusogen
Introduction
Under non-pathological conditions, the fusion of mammalian cells is a highly regulated process that takes place only in a small number of well defined tissues. The male gamete fuses with the female egg, macrophages fuse into multinucleated osteoclasts, myoblasts differentiate into the syncytial myofibers and placental cells form the trophoblast layer by cell-cell fusion (1). When cells fuse with each other, the lipid bilayer of the plasma membrane has to be actively merged and the energetic barrier that normally prevents membrane merging has to be overcome (2). In contrast to intracellular vesicle fusion and virally induced fusion, the molecular events underlying cell-cell fusion have remained largely elusive (3). Some progress has come from the study of developmental cell-cell fusion events in Caenorhabditis elegans. In worm cells, the membrane proteins EFF and AFF have been shown to mediate homotypic fusion events of a variety of somatic cell types. As genuine fusogens, both proteins also induce the fusion of heterologous cells upon ectopic expression (4, 5).
In mammals, the fusion of the gametes, osteoclasts, and myofibers is much less understood despite intensive research efforts. Although several cell surface and intracellular proteins have been shown to be essential for syncytium formation, none of them have been identified as the molecules that directly mediate the fusion process. The syncytins, which are involved in human placental trophoblast fusion, represent an exception, as syncytin-1 and syncytin-2 exhibit direct fusogenic activity in cell culture assays (6, 7). These two proteins, however, are believed to be of retroviral origin and can therefore not be considered as mammalian fusogenic proteins. To date, no mammalian protein has been shown to induce syncytium formation of heterologous cells in culture (1, 2).
In this context, we investigated a putative role of the fibroblast growth factor receptor-like 1 (FGFRL1)2 in mammalian cell fusion. FGFRL1 is the most recently discovered member of the fibroblast growth factor receptor (FGFR) family. In contrast to the four conventional FGFRs, it lacks the cytoplasmic tyrosine kinase domain that is needed for intracellular signal transduction (8). During mouse embryogenesis it is mainly expressed in cartilage, cartilaginous primordia of bones, and in the developing muscles of the diaphragm and the tongue (9). FGFRL1-deficient mice die immediately after birth because of severe defects in the diaphragm muscle, which is not strong enough to properly inflate the lungs (10, 11). In addition, they completely lack functional kidneys due to a failure to initiate nephrogenesis (12). In agreement with a possible role in myogenesis, FGFRL1 is expressed in the myogenic somites from E10.5 on (13), and its expression is sharply up-regulated during the differentiation of C2C12 myogenic cells into multinucleated myotubes (10, 14). With its short intracellular domain of only 100 amino acids and its extracellular domain that is homologous to the FGFRs, it can function as a negative regulator of FGF signaling (14). However, FGFRL1 also displays characteristics that point to an additional cellular function, which is reminiscent of many cell adhesion proteins. Like the nectins and other cell adhesion proteins it is a member of the Ig-domain superfamily (IgSF) and, as demonstrated by FRET experiments, it forms constitutive dimers that promote cell adhesion (15). Moreover, when FGFRL1 is overexpressed in cultured cells it is often enriched at sites of cell-cell contact, indicating a role in cell-cell signaling or adhesion (15).
In the present study, we demonstrate that FGFRL1 rapidly induces the formation of large syncytia when it is overexpressed in heterologous cells. Ig-domain III and the transmembrane domain of FGFRL1 are sufficient to fuse CHO cells with a diverse set of other cell lines. FGFRL1 is therefore the first mammalian protein that has been shown to fuse heterologous cells in culture, indicative of a possible role in myoblast fusion.
EXPERIMENTAL PROCEDURES
Cell Culture
Regular CHO-K1 cells, glycosaminoglycan-deficient CHO-PgsA-745 cells (ATCC CRL-2242), heparan sulfate-deficient CHO-PgsD-677 cells (ATCC CRL-2244), and HEK293 cells were obtained from the ATCC and kept under standard conditions in DMEM supplemented with 10% FBS. HEK-TetOn advanced cells were obtained from Clontech and cultured under standard conditions.
Stable, Tetracycline-inducible Cell Line
HEK-TetOn cells were co-transfected with FGFRL1 wild type and FGFRL1ΔC in pTRE (Tet Responsive Element plasmid from Clontech) and a linear hygromycin selection marker (Clontech) and selected with 150 μg/ml hygromycin for 2 weeks. Healthy colonies were picked with cloning cylinders and screened for inducible FGFRL1 expression by Northern blotting. To induce expression, 1000 ng/ml doxycycline (Clontech) was added to the culture medium.
Cell-Cell Fusion Assay
CHO cells were grown to 50% confluence in 100 mm dishes and transfected with 2 μg luciferase (pTRE-Luc) in a tetracycline-inducible expression vector (pTRE, Clontech) using the Metafectene transfection reagent (Biontex). HEK-TetOn cells were grown to 50% confluence in 60 mm dishes and transfected with 1 μg of FGFRL1 expression constructs in pcDNA3.1 (Invitrogen). After 36 h, the two cell populations were trypsinized and seeded together into 12-well plates at a slightly overconfluent density. The cells were then left to fuse overnight in the presence of 1000 ng/ml doxycycline (Clontech). Luciferase activity was assayed after cell lysis with the Promega luciferase assay system. The luciferase activity measured in wells with only CHO cells served as a reference and was subtracted as background. As a control, the CHO cells were seeded together with HEK-TetOn wild-type cells. To block the fusion, 2 μg/ml of a monoclonal, humanized Fab-fragment (16) against the human FGFRL1 ectodomain was added to the cell culture medium upon seeding of the cells. 2 μg/ml of an antibody against the cytoplasmic domain of FGFRL1 (Santa Cruz Biotechnology) was used as a control antibody.
To visualize fusion, eGFP (from Clontech living colors expression plasmid) in pTRE was transfected instead of the luciferase construct. For the fusion experiments with the FGFRL1 wild-type construct (Figs. 1 and 2), 10 μg/ml heparin sodium (Sigma) was added to the culture medium upon seeding of the cells.
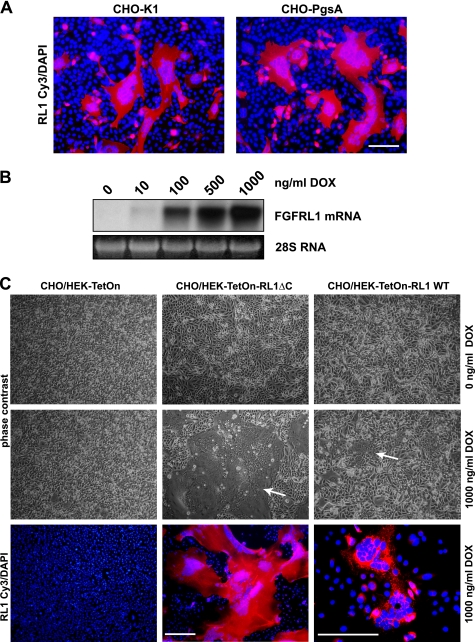
FIGURE 1.
FGFRL1 expression induces changes in cellular morphology that indicate cell-cell fusion and syncytium formation. A, CHO-K1 (left) and glycosaminoglycan deficient CHO-PgsA (right) cells were transfected with C-terminally truncated FGFRL1 (FGFRL1ΔC in pcDNA3.1). One day after transfection, cells were fixed and stained with a monoclonal antibody against FGFRL1 (Cy3, red) and DAPI. Note that most FGFRL1 positive CHO cells appear to be large, syncytial cell clusters with a continuous cytoplasm. Bar, 100 μm. B, Northern blot shows the induction of FGFRL1ΔC expression in a tetracycline inducible HEK-TetOn-RL1ΔC cell line with increasing concentrations of doxycycline. The cell line was derived from a single clone selected for tightly inducible expression. C, CHO-PgsA cells were seeded together with inducible, FGFRL1 wild type (right) and FGFRL1ΔC-expressing cell line (middle), or with regular HEK-TetOn cells (left) as a control. Upon addition of 1000 ng/ml doxycycline to the co-cultures, large “fusion plaques” (indicated by white arrows) appeared in the dishes with the FGFRL1-expressing HEK cell lines. No fusion was observed in the control dishes and in dishes without doxycycline. Antibody staining of FGFRL1 (Cy3, red) shows that the large cell aggregates uniformly expressed FGFRL1. Note that the image showing the fused FGFRL1 wild-type-expressing cells was taken at a higher magnification. Bar, 200 μm.
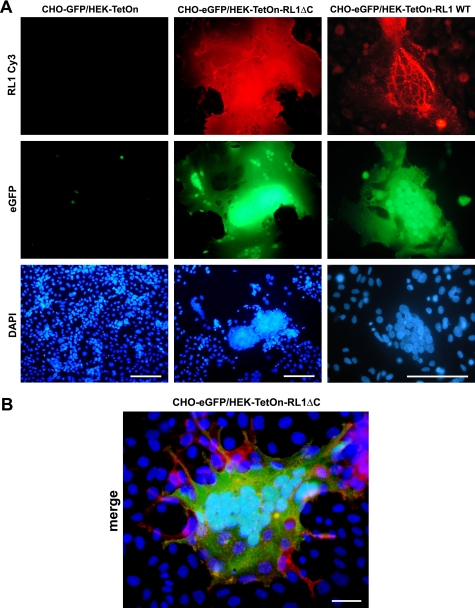
FIGURE 2.
Fusion and cytoplasmic mixing of HEK and CHO cells upon expression of FGFRL1. A, CHO-PgsA cells were transfected with a tetracycline inducible eGFP expression construct, which is only active in the presence of the TetOn transactivator protein and doxycycline. The cells were trypsinized and seeded together with either HEK-TetOn-RL1ΔC, HEK-TetOn-RL1WT cells (express FGFRL1ΔC and FGFRL1 wild type, respectively, in the presence of doxycycline) or with regular HEK-TetOn cells. Doxycycline at 1000 ng/ml was added to all dishes. Note that both HEK-TetOn-RL1 cell lines fused with the CHO cells, leading to the diffusion of the TetOn transactivator from the HEK-TetON-RL1 cells into the syncytial cytoplasm. This activated the expression of eGFP in the large, syncytial cells. No fusion and no activation of eGFP expression was observed with the regular HEK-TetOn cells that served as controls. The image showing the fused FGFRL1 wild-type-expressing cells was taken at a higher magnification. Bar, 100 μm. B, higher powered magnification of a large, syncytial cell consisting of eGFP (green)-transfected CHO and FGFRL1ΔC (red)-expressing HEK-TetOn cells. Note that some FGFRL1-expressing HEK-TetOn cells (red) appear to be attracted by the syncytial cell. Bar, 20 μm.
Latrunculin-B, cytochalasin-D, and nocodazole were purchased from Axxora (Switzerland), diluted in DMSO and added to the culture medium after seeding of the cells. Heparanase I was purchased from Sigma. It was added when cells started to fuse at 5 units/ml in serum-free DMEM. For the complete list of FGFRL1 and FGFR constructs that were tested for their fusogenic activity please see below.
Exogenous Glycosaminoglycans
To test the effect of exogenous GAGs on the cell-cell fusion, each was added at 0.01–10 μg/ml to the culture medium upon seeding. From Sigma were: Cell culture grade heparin sodium salt from porcine mucosa (H3149), dermatan sulfate (C3788), chondroitin sulfate A (chondroitin-4-sulfate) from bovine trachea (C9819), dextran sulfate (D7140), chondroitin-6-sulfate from shark cartilage (C4384), heparan sulfate sodium salt from bovine kidney (H7640). Hyaluronic acid sodium salt from human umbilical chord was from Fluka (53740).
Fusion Time Course Experiment
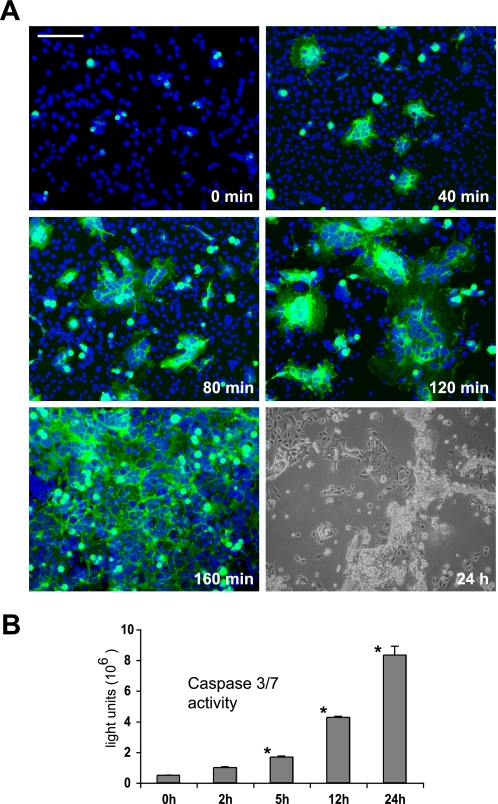
HEK293 cells were transfected with FGFRL1ΔCΔ12-eGFP, trypsinized and seeded together with untransfected, trypsinized CHO-PgsA cells onto coverslips in 30 mm dishes at a confluent density (ratio of CHO to HEK cells 5:1). The time point when the cells started to attach (∼6 h after seeding) was taken as the starting point. From then on, one dish at a time was fixed every 40 min and stained with DAPI. A representative picture from every time point (up to 160 min) was taken to illustrate the time course of the fusion process.
GST Pull-down and Mass Spectrometry
Fusion constructs of FGFRL1ΔC and FGFRL1ΔCΔ12 with glutathione S-transferase (from pGEX, GE-Healthcare) were generated by overlap PCR and cloned into pcDNA3.1. HEK293 cells were transfected with the constructs and stably expressing cells were selected with 800 μg/ml G418 over 3 weeks. The cells were grown to confluence in 150 mm dishes, lysed with 1% Triton-X100 in PBS, supplemented with Roche Complete Mini Protease Inhibitor, and centrifuged at 25,000 × g for 30 min at 4 °C. The supernatant was incubated with glutathione beads for 4 h at 4 °C, followed by repeated washing of the beads with PBS. Bound proteins were eluted with saturated urea (Fluka) in PBS. To evaluate pull-down efficiency, 50% of the eluate was boiled in SDS sample buffer, separated by SDS-PAGE and visualized by silver staining of the gel (silver staining kit from Pierce). To identify the proteins that co-purified with FGFRL1-GST, proteins in the remaining eluate were digested with trypsin, separated by HPLC (Waters Alliance HT2795) and identified with a Bruker Esquire3000plus Ion Trap Mass spectrometer. As control samples, HEK293 wild-type cells or cells expressing only GST were subjected to the same treatment. The proteins detected in four independent control pull-down experiments (two wild type and two stably GST transfected) were subtracted from the proteins detected in four FGFRL1ΔC-GST and FGFRL1ΔCΔ12-GST pull-downs. In addition, contaminant proteins that commonly bind to Sepharose beads, published by Trinkle-Mulcahy et al. (17), were subtracted.
FGFRL1 and FGFR Expression Constructs
The C-terminally truncated human FGFRL1 constructs were generated as described by Rieckmann et al. (16). The final expression plasmid coded for the following amino acids: RL1ΔC-(1–416), RL1ΔHisΔTyr-(1–468), RL1ΔHis-(1–478), and RL1 full-(1–504). The FGFRL1 constructs with deletions in the ectodomain were based upon the C-terminally truncated construct (1–416) and coded for the following amino acids: FGFRL1ΔCΔ1-(1-26 + 113–416), FGFRL1ΔCΔ23-(1–144 + 361–416), FGFRL1ΔCΔ12-(1–29 + 238–416), FGFRL1ΔCΔ3-(1–240 + 357–416), FGFRL1ΔCΔ2-(1–144 + 240–416). The soluble ectodomain (RL1exSol) covered the nucleotide sequence for amino acids 1–367. The FGFRL1ΔC and FGFRL1ΔCΔ12 fusion constructs with GST and eGFP were generated by overlap PCR and encoded the proteins described above with a C-terminal GST (from pGEX, Invitrogen) or eGFP (from Clontech Living Colors expression plasmid) moiety. The C-terminally truncated FGFR constructs corresponded to the following amino acids: FGFR1ΔC-(1–415), FGFR2ΔC-(1–415), FGFR3ΔC-(1–415), FGFR4ΔC-(1–410).
Immunocytochemistry
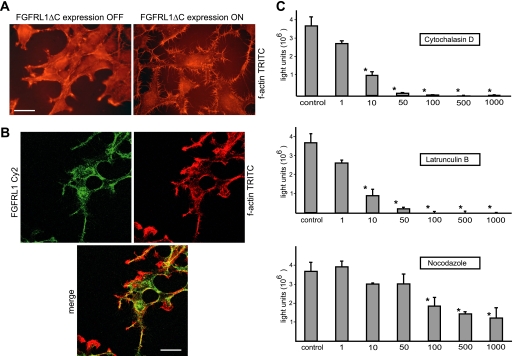
Cells were fixed with 4% paraformaldehyde in PBS, permeabilized and blocked with 0.2% Triton-X100 and 1% BSA in PBS followed by staining with a monoclonal, humanized Fab-fragment antibody against the FGFRL1 ectodomain (1 μg/ml, described in Rieckmann et al. (16)) and secondary anti-human Fab Cy3- or Cy2-coupled antibodies (Jackson Laboratories). The actin cytoskeleton was visualized by staining with TRITC-coupled phalloidin (Sigma). Nuclei were stained with 4′,6-diamidino-2-phenylindol (DAPI, Invitrogen). After mounting with Mowiol, the cells were inspected with a Nikon Eclipse E1000M microscope. The confocal images of the C-terminally truncated FGFRL1 proteins and those of FGFRL1 and the actin cytoskeleton were taken on a Carl Zeiss LSM510 confocal microscope.
Surface Biotinylation of FGFRL1
The different FGFRL1 variants were stably transfected into HEK293 cells via puromycin selection. Whole cell extracts were prepared by boiling of the cells in SDS sample buffer, followed by Western blot analysis of FGFRL1 expression with a polyclonal antibody against the ectodomain of human FGFRL1 (R&D Systems). All cell lines were grown to 80% confluence in 100 mm cell culture dishes followed by surface biotinylation with the Pierce cell surface isolation kit according to manufacturer's instructions. The isolated surface proteins were separated by SDS-PAGE and biotinylated FGFRL1 was detected by Western blotting with the antibody described above.
Apoptosis Detection
HEK293 cells were transfected with FGFRL1ΔCΔ12-eGFP and co-cultured with CHO-PgsA cells. The time point of cell attachment was taken as the starting point. For the caspase 3/7 activity assay, the cells were lysed in the culture dishes by adding 0.2% Triton-X100 to the culture medium at the indicated time points. 50 μl of the lysate was given to 50 μl of luminometric caspase 3/7 detection solution (Promega), incubated for 15 min at room temperature followed by measurement of caspase activity in a luminometer. For the TUNEL staining, the fused cells were fixed after 16 h and stained with the Roche In Situ Cell Death Detection kit according to manufacturer's instructions.
RESULTS
FGFRL1 Induces Syncytium Formation of CHO Cells
For our studies of FGFRL1 function in cultured cells we overexpressed different FGFRL1 constructs in several cell lines. When we transfected a C-terminally truncated form of FGFRL1 (FGFRL1ΔC) into regular CHO wild-type cells (CHO-K1), we observed that the cells underwent peculiar morphological changes. Specifically, we observed many cellular aggregates that appeared to be syncytial cells containing a large number of nuclei. To further investigate this phenomenon, we transfected human FGFRL1ΔC into CHO-K1 cells and stained the cells for FGFRL1 with DAPI as a counterstain. As shown in Fig. 1A, large FGFRL1ΔC expressing cell clusters with up to 100 aggregated nuclei formed 24h post-transfection. The aggregates appeared to be large, syncytial cells with a continuous cytoplasm. No cell clusters and no nuclear aggregation were observed in control transfected cells (not shown).
Because FGFRL1 is a heparin-binding receptor (8), we asked whether heparan sulfate chains are required for the observed morphological changes. We therefore transfected FGFRL1ΔC into glycosaminoglycan-deficient CHO-PgsA-745 (CHO-PgsA) cells. The CHO-PgsA cells also aggregated and the putative fusion process took place to an even greater extent than in the CHO-K1 cells. Because of this, most of the following experiments were performed with both the CHO-PgsA and the regular CHO-K1 cells. If not otherwise indicated, the figures show images obtained with the CHO-PgsA cell line.
We next asked whether other cell lines overexpressing FGFRL1 could “fuse” with the CHO cells. To this end, we generated tetracycline-inducible, clonal HEK cell lines (HEK-TetOn-RL1) that express high levels of human FGFRL1 wild type and FGFRL1ΔC upon induction with doxycycline. The representative Northern blot in Fig. 1B demonstrates that FGFRL1ΔC expression is tightly suppressed in the absence of the inducer but strongly activated in the presence of 1000 ng/ml doxycycline. When the CHO cells were co-cultured with the HEK-TetOn-RL1 cell lines in the absence of doxycycline, no “fusion-plaques” formed (Fig. 1C). However, when FGFRL1 expression was induced with 1000 ng/ml doxycycline, large, syncytia-like cell clusters with aggregated nuclei developed overnight. Notably, the C-terminally truncated form of FGFRL1 induced much larger syncytial cells than the wild type receptor. As expected, doxycycline had no effect on a co-culture of control HEK-TetOn and CHO cells (Fig. 1C). The fluorescent staining of FGFRL1 in the doxycycline-treated co-cultures (Fig. 1C, bottom, right) confirmed that the large cell aggregates uniformly expressed FGFRL1. The DAPI counterstain showed that the nuclei aggregated within these large structures whereas no nuclear aggregation was observed in the control culture with HEK TetOn cells (Fig. 1C, bottom, left). When the dosage of doxycycline was lowered to induce moderate (100 ng/ml) to low (10 ng/ml) expression levels of FGFRL1ΔC, cell fusion still occurred, albeit to a lesser extent (supplemental Fig. S1). Because we did not have to use any transfection reagent for these experiments, we could exclude that the observed phenomenon was in some way caused by a cationic-lipid-mediated membrane disruption.
We next investigated whether the cellular aggregates were in fact of a syncytial nature or merely densely clustered groups of cells. In syncytial cells, the merging of the cell membranes and the resulting fusion pores lead to cytoplasmic mixing followed by the formation of a continuous cytoplasm (1, 2). To confirm cytoplasmic mixing, we developed a reporter system based upon the TetOn transactivator protein from the clonal, FGFRL1 expressing HEK-TetOn-RL1 cell lines. Before co-culturing, the CHO cells were transfected with a tetracycline inducible eGFP construct that is only expressed when the activated TetOn-transactivator protein is present. When cytoplasmic mixing between HEK-TetOn-RL1 and CHO cells occurs in the presence of doxycycline, the TetOn transactivator should diffuse from the HEK cells into the common cytoplasm and activate the expression of the eGFP construct. As a control, we co-cultured eGFP-transfected CHO cells with non-transfected HEK-TetOn cells that did not express FGFRL1. Fig. 2A shows that the HEK-TetON-RL1 cells indeed fused with the CHO cells. The large, syncytial cells expressed FGFRL1 (red) and eGFP (green), indicating that cytoplasmic mixing had occurred. The cells comprised up to several hundred nuclei that clustered together. Again, the C-terminally truncated FGFRL1 induced much larger syncytia than the wild-type receptor. In the control cultures no nuclear clustering and only very little background eGFP expression was observed. The higher powered magnification in Fig. 2B shows a syncytial-, FGFRL1ΔC-, and eGFP-expressing cell, which is surrounded by HEK cells that express only FGFRL1ΔC. The HEK cells appear to be attracted by the large cell, to which they attach before their fusion.
In addition to the HEK cells, we tested whether a diverse selection of cell lines could fuse with the CHO cells upon transfection of FGFRL1 expression plasmids. C2C12 myogenic cells, HeLa, MG63, and COS7 cells all readily fused with the CHO cells. Moreover, FGFRL1-expressing CHO cells also fused with all cell lines tested so far (data not shown). However, FGFRL1-mediated fusion occurred only when at least one of the cell lines was a CHO cell line.
Identification of the Fusogenic Domain of FGFRL1
Now that we had shown that FGFRL1 induces cell fusion of CHO and other cells, we set out to identify the domain(s) of FGFRL1 that mediates the fusogenic activity. For this purpose, we modified the eGFP-based fusion assay described above with a tetracycline inducible luciferase construct (pTRE-Luc) instead of the eGFP. The CHO cells were transfected with pTRE-Luc and, separately, Hek-TetOn cells were transfected with the wild type and mutated FGFRL1 expression constructs. After transfection, the two cell lines were trypsinized and co-cultured overnight to allow cell fusion to occur. The luciferase activity in the fused cells correlated well with the visible amount of cell fusion between the HEK-TetOn and the CHO cells. This assay could thus be utilized to quantify the fusogenic activity of the mutated FGFRL1 constructs.
We first aimed to address the role of the cytoplasmic domain. The short intracellular part of FGFRL1 comprises two conserved motifs, one being a histidine-rich domain at the C terminus, the other a double tyrosine motif further N-terminal (16). To investigate the influence of these motifs and of the entire cytoplasmic domain on the fusion process, we employed the fusion assay described above. The wild-type receptor, the C-terminally truncated receptor (ΔC) and constructs with a deletion of the histidine motif (ΔHis) and an additional deletion of the double tyrosine motif (ΔHisΔTyr) were transfected to test their fusogenic activity. In good agreement with our previous observations (Figs. 1C and 2A), truncations of the C terminus enhanced the fusogenic activity when compared with the wild-type FGFRL1 (Fig. 3A). Although the wild-type receptor did induce considerable cell-cell fusion and luciferase activity, the effects were dramatically enhanced with the mutated constructs. The most pronounced fusion was observed with the C-terminally truncated FGFRL1 (RL1ΔC), indicating that the intracellular domain is not necessary for the cell fusion process.
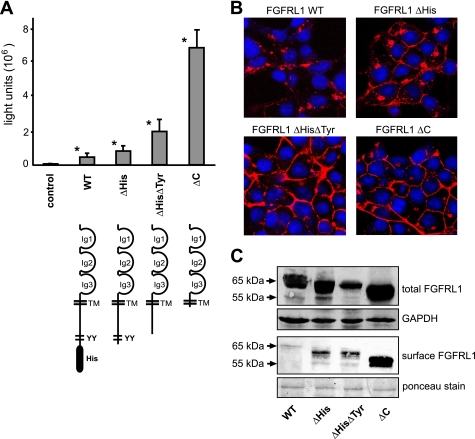
FIGURE 3.
Role of the cytoplasmic domain on the fusion process. A, CHO-K1 cells were transfected with a tetracycline-inducible luciferase expression construct (pTRE-Luc) and seeded together with HEK-TetOn cells that had been transfected with different C-terminally truncated FGFRL1 expression constructs (the corresponding proteins are schematically shown below the respective bars). The luciferase activity, measured 14 h postseeding, correlated well with the visible extent of cell-cell fusion induced by the mutated FGFRL1 constructs. The bars represent the average of three experiments. Successive truncation of cytoplasmic internalization motifs led to increased fusion, with strongest fusion observed for the completely truncated construct FGFRL1ΔC. Note that the wild-type, full-length protein also induces considerable fusion when compared with nontransfected control cells. Asterisks next to bars indicate statistically significant results (p < 0.05) compared with the control group. B, confocal images of immunofluorescent stainings of the C-terminally truncated FGFRL1 proteins. Note that the wild-type protein resided primarily in intracellular compartments, while the truncated forms accumulated in the plasma membrane. C, surface biotinylation of mutated FGFRL1 proteins. HEK293 cells stably expressing the indicated FGFRL1 proteins were subjected to surface biotinylation, followed by isolation with neutravidin agarose and detection of biotinylated FGFRL1 by Western blotting. The upper panel shows total FGFRL1 in the respective cells, with GAPDH as a loading control. The lower panel shows biotinylated FGFRL1 from the cell surface. Ponceau staining of a major membrane protein of ∼85 kDa served as a loading control for the neutravidin purification.
We have previously shown that the histidine-rich region and the double tyrosine motifs of the cytoplasmic domain mediate the rapid internalization of FGFRL1 from the cell surface (16). Therefore, we speculated that truncation of the cytoplasmic domain would lead to higher levels of FGFRL1 at the cell surface, which could in turn result in enhanced cell fusion. To address this, HEK293 cells were stably transfected with the wild type and the truncated FGFRL1 constructs. Confocal images of antibody stained FGFRL1 in these cell lines showed that the wild-type receptor resided primarily in intracellular pools, while the C-terminally truncated receptors accumulated in the plasma membrane (Fig. 3B). To quantify different levels of FGFRL1 at the cell surface, we subjected the stably transfected cells to surface biotinylation, followed by neutravidin-based isolation of biotinylated proteins. Western blot analysis of the amount of biotinylated FGFRL1 revealed that only a very small amount of wild-type FGFRL1 was present at the surface of the HEK293 cells (Fig. 3C). The amount of FGFRL1 at the cell surface was strongly increased for all of the truncated versions, with the highest levels being observed for the fully truncated FGFRL1ΔC protein. From these experiments we concluded that it is the extracellular domain of FGFRL1 that mediates the fusogenic activity. Whereas the cytoplasmic domain is not necessary for the fusion process, it does control the amount of FGFRL1 at the cell surface, thereby regulating the extent of cell-cell fusion in our fusion assays.
In agreement with the role of the extracellular part of FGFRL1 being responsible for the fusion, a monoclonal antibody directed against its ectodomain almost completely blocked the fusogenic activity (Fig. 4A), whereas a control antibody had no effect. To show that the fusogenic activity is not a general feature of all FGFRs, C-terminally deleted versions of all four conventional FGFRs were tested in the same fusion assay. As shown in Fig. 4A, none of the four FGFRs caused any cell-cell fusion. We concluded from these experiments that the fusogenic activity is a unique feature of FGFRL1 and is mediated by its extracellular domain.
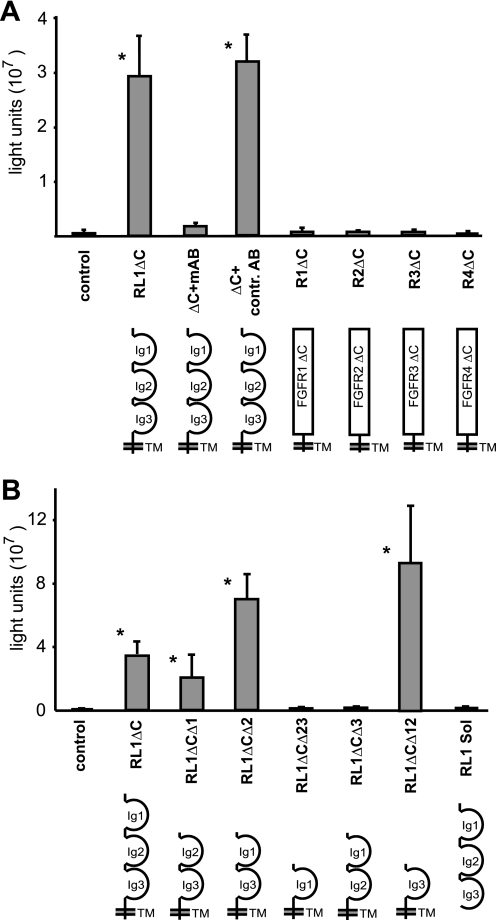
FIGURE 4.
Fusogenic activity is specific to FGFRL1 and mediated by Ig-domain III. FGFRL1ΔC-transfected HEK-TetOn cells were co-cultured with pTRE-Luc-transfected CHO-K1 cells and allowed to fuse overnight, followed by measurement of luciferase activity. A, addition of a monoclonal antibody against the FGFRL1 ectodomain into the culture medium blocked fusion at 2 μg/ml. A control antibody against the intracellular domain of FGFRL1 (not present anymore in FGFRL1ΔC) had no effect. C-terminally truncated FGFR1-FGFR4 did not induce any cell-cell fusion in this cellular system. B, deletion of extracellular Ig-domains I and II leaves fusogenic activity intact. Deletion of Ig-domain III completely disrupts fusion. Note that both the RL1ΔCΔ2 and the RL1ΔCΔ12 proteins show increased activity when compared with the RL1ΔC construct. The soluble FGFRL1 ectodomain does not induce any cell-cell fusion. The asterisks indicate statistically significant results (p < 0.05).
Next, we investigated which part of the FGFRL1 ectodomain is necessary for the fusion process. To this end we started with the C-terminally truncated RL1ΔC protein and systematically deleted each of the three Ig-like domains (RL1ΔCΔ1, RL1ΔCΔ2, RL1ΔCΔ3) and constructed compound deletions of Ig I and Ig II (RL1ΔCΔ12) and of Ig II and Ig III (RL1ΔCΔ23). In addition, we generated a soluble ectodomain to investigate whether the transmembrane domain is necessary for the cell fusion. The fusogenic activity of the resulting contructs was tested in the luciferase fusion assay described above. As shown in Fig. 4B, Ig-domain I and II are dispensable for the fusion process, as the RL1ΔCΔ1, RL1ΔCΔ2, and especially the RL1ΔCΔ12 construct showed pronounced fusogenic activity. Because the RL1ΔCΔ12 protein comprises only Ig-domain III and the transmembrane domain, it is clearly Ig-domain III that mediates the cell fusion. In agreement with this finding, deletion of Ig-domain III in the RL1ΔCΔ3 and the RL1ΔCΔ23 construct completely abrogated the fusogenic activity. The soluble ectodomain did not induce any cell fusion, indicating that Ig III needs to be anchored in the membrane to exert its fusogenic activity.
We were also interested in the kinetics of the fusion process and thus needed a way to better visualize the fusing cells. For that purpose, we generated a fusion construct of Ig-domain III, the transmembrane domain and an intracellular eGFP moiety (RL1ΔCΔ12-eGFP). This protein was highly fusogenic in the CHO cells and readily fused transiently transfected HEK293 cells with untransfected CHO cells. For a time course experiment, the HEK cells were transfected with the RL1ΔCΔ12-eGFP construct and seeded together with the CHO cells. The point of initial attachment of the cells was taken as the starting point. Fig. 5A shows representative snapshots taken every 40 min after the initial attachment. Within 40 min, the first multinucleated, RL1ΔCΔ12-eGFP expressing cells had formed. The syncytia grew rapidly in size within the next 80 min. After 160 min, the entire field of vision and in fact almost the entire culture dish was one continuous syncytial cell. Overnight, the nuclei of the fused cells had clustered together (see Fig. 2) and after 24 h most multinucleated cells had detached from the dish and seemed to have died. To verify this, we measured the activity of effector caspases 3 and 7 in the fusing cells over 24 h. Caspase activity was increased 2 h after fusion had set in and exponentially increased from that time point on (Fig. 5B). A co-culture of HEK293 wild-type cells with the CHO cells did not lead to any increase of caspase activity (not shown). This pronounced increase in effector caspase activity suggested that the cells underwent apoptotic cell death after fusion had begun. TUNEL staining of the fused cells 16 h after seeding furthermore revealed that the cells fragmented their DNA when they detached from the cell culture dish (supplemental Fig. S2). Whereas the fusion took only a few hours, the cells survived between 12 and 24 h before they detached and died.
FIGURE 5.
FGFRL1ΔCΔ12 rapidly fuses HEK293 and CHO-PgsA cells and induces apoptotic cell death. A, kinetics of the FGFRL1-induced cell-cell fusion. HEK293 cells were transfected with FGFRL1ΔCΔ12-eGFP and seeded together with CHO-PgsA cells. The time of initial attachment was taken as the starting point. The figure displays snapshots taken every 40 min after the initial attachment of the cells. The first syncytial, multinucleated cells appeared after 40 min and rapidly expanded until, after 160 min, the entire field of vision was one continuous syncytium. 24 h after the onset of fusion, cells detached and appeared to be dying. Bar, 100 μm. B, activity of caspase 3/7 in the fusing HEK293 and CHO-PgsA cells. A luminometric caspase 3/7 activity kit was used to determine the activity of these effector caspases in the fusing cells. The asterisks indicate significant (p < 0.05) increases in caspase activity relative to unfused cells.
The Actin Cytoskeleton and Surface Glycosaminoglycans Are Involved in FGFRL1-mediated Fusion
Because most mammalian cell-cell fusion processes previously described are dependent on the activity of the actin cytoskeleton (18, 19), we next addressed a potential involvement of actin polymerization in the FGFRL1-mediated fusion of cells. Immunofluorescent staining of FGFRL1ΔC and phalloidin-TRITC staining of polymerized actin provided first evidence for an influence of FGFRL1ΔC on the actin cytoskeleton. Upon doxycycline-induced expression of the C-terminally truncated FGFRL1 construct in the clonal HEK-TetOn-RL1ΔC cells, the cells displayed a pronounced redistribution of polymerized actin to the cell periphery and formed actin-positive protrusions resembling filopodia (Fig. 6A). Moreover FGFRL1ΔC substantially co-localized with cortical f-actin (Fig. 6B). To investigate a potential role of actin polymerization in HEK and CHO cell fusion we employed our luciferase-based cell fusion assay and several cytoskeleton disrupting agents. HEK-TetOn cells were transfected with FGFRL1ΔC and seeded together with pTRE-Luc-transfected CHO cells. Upon seeding, the fusing cells were treated with increasing concentrations of the actin depolymerizing agents cytochalasin-D and latrunculin-B as well as with the microtubule disrupting drug nocodazole. Fourteen hours after seeding, luciferase activity was assessed in the lysed cells. As shown in Fig. 6C, both latrunculin and cytochalasin strongly inhibited syncytium formation at low concentrations and completely eliminated cell-cell fusion at higher concentrations. It should be noted that we observed a pronounced decrease of fusion before overt signs of toxicity could be detected. In contrast, disruption of microtubules with nocodazole barely affected cell-cell fusion before we observed overall cell toxicity (rounded cells, detachment from culture dish). Even at very high, clearly toxic concentrations of nocodazole, cells still fused to a considerable extent. Although off-target effects of the inhibitors cannot be ruled out, we concluded that actin, but not tubulin polymerization was required for FGFRL1-induced fusion.
FIGURE 6.
The actin skeleton is involved in FGFRL1-mediated cell-cell fusion. A, stable HEK-TetOn cell line with inducible expression of FGFRL1ΔC was cultured with (right) and without (left) doxycyline in the medium. After fixing of the cells, the actin cytoskeleton was stained with phalloidin-TRITC. FGFRL1 expression induces the formation of actin rich protrusions resembling filopodia. We also observed a redistribution of f-actin to the cell periphery. Bar, 8 μm. B, confocal image of antibody stained FGFRL1ΔC (green) and phalloidin-TRITC stained f-actin (red). The overlay shows that there is substantial overlap between FGFRL1ΔC and f-actin. Bar, 20 μm. C, luciferase-based cell-cell fusion assay was utilized to test the effect of the f-actin destabilizing compounds latrunculin-B and cytochalasin-D and of the microtubule disrupting agent nocodazole (all at 1–1000 ng/ml) on FGFRL1ΔC-induced cell-cell fusion. Either of the actin affecting drugs was strongly inhibitory on the fusogenic activity of FGFRL1ΔC and completely blocked fusion at higher concentrations. Nocodazole did not reduce the fusion at lower concentrations and moderately affected fusion at highly toxic concentrations. The bars represent the average luciferase activity measured in three wells. Asterisks indicate statistically significant (p < 0.05) differences relative to untreated control.
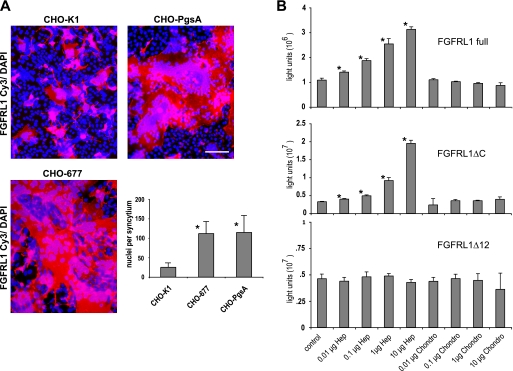
Because the CHO-PgsA cells fused better than the CHO-K1 cells, we had already obtained evidence that cellular surface glycosaminoglycans (GAG) might have an inhibitory effect on the FGFRL1 mediated fusion. A recent study also suggested that GAGs have a general role in many cell-cell fusion events (20). We therefore aimed to address the potential role of cell surface GAGs, particularly heparan sulfates, in the FGFRL1-mediated cell fusion process in more detail. To this end FGFRL1ΔC-transfected HEK-TetOn cells were cultured together with either CHO-K1 cells, CHO-PgsA cells (which lack all surface glycosaminoglycans) or CHO-PgsD-677 (CHO-677) cells (which are deficient in heparan sulfates). The cells were allowed to fuse overnight, FGFRL1 and nuclei were stained and the number of nuclei per syncytium was determined to establish a “fusion index” for each of these cell lines. As shown in Fig. 7A, both CHO-PgsA and CHO-677 cells fused more efficiently than the CHO-K1 cells, resulting in a higher number of nuclei per syncytium. The extent of fusion of the CHO-677 cells was nearly identical to that of the CHO-PgsA cells, indicating that mainly cell surface heparan sulfates inhibited FGFRL1-induced fusion. Furthermore, treatment of FGFRL1ΔC-transfected CHO-K1 cells with heparanase I increased the size of FGFRL1ΔC-induced syncytia (supplemental Fig. S4A). These findings are in agreement with our previous binding studies that showed binding of FGFRL1 to heparin but not to chondroitin sulfates (8, 15).
FIGURE 7.
Cell surface heparan sulfates have an inhibitory effect on FGFRL1-mediated cell fusion. CHO-K1 cells, CHO-677 cells (lack heparan sulfates) and CHO-PgsA cells (lack all glycosaminoglycans) were seeded together with FGFRL1ΔC-transfected HEK-TetOn cells and left to fuse overnight. A, representative pictures of the resulting HEK/CHO syncytia are shown for each CHO cell line. Bar, 100 μm. The diagram gives the average number of nuclei per syncytium of the fusing CHO-K1, CHO-677, and CHO-PgsA cells with FGFRL1-transfected HEK cells. Both CHO-677 and CHO-PgsA cells displayed dramatically increased cell fusion efficiency when compared with the wild-type CHO-K1 cells, indicating that heparan sulfates have an inhibitory effect on FGFRL1-induced cell-cell fusion. B, luciferase (pTRE-Luc)-transfected CHO-K1 and FGFRL1 wild-type, FGFRL1ΔC-, and FGFRL1ΔCΔ12-transfected HEK-TetOn cells were co-cultured overnight in the absence and presence of exogenous heparin or chondroitin-4-sulfate in the culture medium, followed by luciferase measurement.
In further experiments, we tested the effect of exogenous GAGs on the fusion of HEK and CHO cells. For this purpose, wild-type FGFRL1, FGFRL1ΔC, and FGFRL1ΔCΔ12 were transfected into the HEK cells, which were subsequently co-cultured with luciferase-transfected CHO cells in the presence of increasing concentrations of various GAGs. Interestingly, exogenous heparin strongly increased the fusogenic activity of the wild-type FGFRL1 and FGFRL1ΔC in a dose-dependent manner, while the FGFRL1ΔCΔ12 mediated fusion was not affected. Chondroitin-4-sulfate had no effect in any of these assays (Fig. 7B). Heparan sulfate and dermatan sulfate exerted an effect similar to heparin, while hyaluronan, dextran sulfate, and chondroitin-6-sulfate did not affect FGFRL1-mediated fusion (supplemental Fig. S3A). Heparin, heparan sulfate, and dermatan sulfate all increased the fusogenic activity of FGFRL1 WT and FGFRL1ΔC but not that of FGFRL1ΔCΔ12 (supplemental Fig. S3B). This effect could also be observed when the fusion assay was performed in CHO cells only (supplemental Fig. S4). Addition of exogenous heparin increased the fusion of FGFRL1ΔC transfected CHO-K1 cells, but had no effect on CHO-K1 cells transfected with FGFRL1ΔCΔ12. Obviously, direct binding of FGFRL1 to cellular heparan sulfates via its heparin binding site (located on Ig-domain II) is inhibitory for the fusion process. Given this is the case, exogenous heparin but not chondroitin sulfates, would competitively displace FGFRL1 from cellular heparan sulfates and result in increased fusogenic activity. In agreement with this notion, heparin, heparan sulfate and dermatan sulfate had no effect on the fusogenic activity of the FGFRL1ΔCΔ12 receptor, which does not possess the heparin binding site.
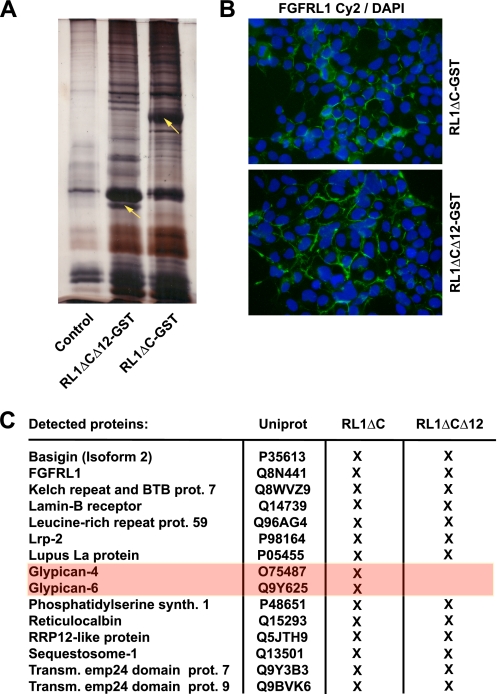
To investigate this further, we generated an FGFRL1ΔC and an FGFRL1ΔCΔ12 (only IgIII and transmembrane domain) construct with a C-terminal glutathione S-transferase tag (GST-tag). These fusion constructs were stably transfected into HEK293 cells and the resulting proteins were precipitated from Triton X-100-solubilized whole cell extracts with glutathione-Sepharose beads. Proteins bound to FGFRL1ΔC and FGFRL1ΔCΔ12 were eluted with saturated urea, digested in solution with trypsin and identified with liquid chromatography followed by mass spectrometry (LC-MS). The proteins detected in GST-only transfected or untransfected HEK293 cells, as well as the published “Sepharose bead proteome” (17) served as background reference lists and were therefore subtracted from the FGFRL1 pull-down to eliminate unspecific hits. Fig. 8A shows a silver-stained gel of control and FGFRL1 pull-down eluates, indicating that a number of proteins were complexed with the overexpressed FGFRL1 proteins. As shown in Fig. 8B, the addition of a GST moiety did not alter the distribution and membrane localization of the FGFRL1-GST constructs, suggesting that the receptor was purified from a membrane environment. The list in Fig. 8C and the supplemental Table S1 contain the identified proteins for FGFRL1ΔC and FGFRL1ΔCΔ12 after the filtering criteria described above had been applied. Among a number of proteins, the heparan sulfate proteoglycans glypican-4 and glypican-6 co-purified with FGFRL1ΔC but not with FGFRL1Δ12ΔC. This indicated that the major heparin-binding site of FGFRL1 on Ig-domain II (15) indeed binds to heparan sulfate chains of cell surface proteoglycans.
FIGURE 8.
Mass spectrometric identification of proteins that co-purified with GST-tagged FGFRL1. A, GST-tagged FGFRL1 proteins were expressed in HEK293 cells (stable expression) and purified from Triton X-100 lysates with glutathione beads. Bound proteins were eluted from the beads with saturated urea. The silver-stained gel shows resolved proteins from a control pull-down (untransfected cells) versus proteins from FGFRL1ΔC-GST and FGFRL1ΔCΔ12-GST pull-downs from the transfected cells. The presence of additional silver-stained bands indicated that a number of proteins co-purified with the GST-tagged FGFRL1 proteins (yellow arrows indicate FGFRL1ΔC-GST and FGFRL1ΔCΔ12-GST). B, immunofluorescent staining of FGFRL1ΔC-GST and FGFRL1ΔCΔ12-GST proteins in the stably transfected HEK293 cells. Both constructs retained a plasma membrane localization. C, eluates were subjected to LC-MS shotgun sequencing to identify the proteins that were purified together with the FGFRL1 proteins. The table lists proteins that associated with FGFRL1ΔC-GST and FGFRL1ΔCΔ12-GST in HEK293 cells.
DISCUSSION
In the present study we showed that ectopic expression of the human FGFRL1 receptor leads to extensive cell-cell fusion and syncytium formation of CHO cells and also induces fusion of diverse other cells with CHO cells. We demonstrated that the cytoplasmic domain of FGFRL1 is dispensable for the fusion process but controls the extent of cell-cell fusion mediated by the ectodomain. Within the ectodomain, we traced the fusogenic activity to the most membrane proximal Ig-domain III, which, together with the transmembrane domain, was sufficient for full activity. Furthermore we showed that actin polymerization is necessary for the fusion process and that the binding of FGFRL1 to cellular heparan sulfates negatively regulates FGFRL1-mediated fusion.
IgSF Proteins and Cell-Cell Fusion
FGFRL1 is certainly not the first member of the immunoglobulin superfamily (IgSF) of transmembrane proteins that has been implicated in cell-cell fusion. In fact, IgSF proteins have been shown to be involved in every mammalian cell fusion process investigated to date. In sperm-egg fusion the Ig-like domain protein Izumo, expressed in sperm, is essential for the fusion of the gametes. Accordingly, Izumo deficient male mice are infertile due to a fusion block of sperm and egg. Izumo, with its single Ig-domain and its short intracellular domain (21) is strikingly similar to the minimal fusogenic part of the FGFRL1 receptor (FGFRL1ΔCΔ12) described in this study.
The fusion of macrophages into osteoclasts and multinucleated giant cells is another, relatively well studied process, which involves several members of the IgSF. The interaction of SIRPα, initially termed the macrophage fusion receptor, with CD47 has been shown to be required for syncytium formation. Accordingly, monoclonal antibodies directed against either SIRPα or against CD47 block fusion in culture (22, 23). CD44, another IgSF protein, is transiently expressed prior to multinucleation and is thought to be involved in the fusion process, as soluble, recombinant CD44 blocks macrophage fusion in culture (24). The domain structures of SIRPα and FGFRL1 are very similar since both proteins have three extracellular Ig-like domains and a short cytoplasmic tail with poorly defined function (25).
The fusion of myoblasts into multinucleated myofibers has been an area of intensive research efforts over the past decade. Although a very large number of surface and intracellular proteins has been implicated in the differentiation and fusion process, it is again IgSF proteins that constitute core components of the essential fusion machinery in insects and in vertebrates (18). In Drosophila myoblast fusion, the Ig-domain containing membrane proteins Sticks and Stones (Sns) and Hybris (Hbs), expressed by fusion competent myoblasts, as well as Rst and Duf/Kirre in the founder myotube mediate cell-cell attachment and are required for the fusion of myoblasts with founder myotubes (26–28). Accordingly, compound deletion of both Rst and Duf/Kirre leads to a complete fusion block of Drosophila body wall musculature (29). Sns and Hbs also function redundantly on the surface of fusion competent myoblasts by binding to Duf/Kirre and Rst on the founder cells to direct muscle fiber fusion (26).
In mammals the situation is more complex and the adhesion/fusion machinery is less understood than in Drosophila. A recent study, however, has shown that nephrin, the mammalian orthologue of Sns and Hybris, is involved in vertebrate myogenesis, as nephrin-deficient zebrafish and mice displayed fusion defects in skeletal muscles and in cultured myoblasts, respectively (30).
FGFRL1 and Myogenesis
In mouse embryos FGFRL1 is expressed in the myogenic somites from E10.5 on (11, 13) and it is also sharply up-regulated in differentiating C2C12 myogenic cells exactly when the cells start to fuse (10, 14). Similar to nephrin, FGFRL1 is also expressed in some developing skeletal muscles and in cultured primary myoblasts during myotube formation (9, 10, 14, 30). Moreover, FGFRL1 is often enriched at sites of cell-cell contacts, for example in cultured A204 myosarcoma cells (15), supporting a role in cell-cell adhesion and potentially in cell-cell fusion. For these reasons, we feel that the observed fusion phenomenon is of physiological importance.
To date, however, we have not been able to obtain direct evidence for a fusion phenotype in our FGFRL1-deficient mice: Although the homozygous null embryos have a fully penetrant and lethal diaphragm phenotype (10, 11), we could not detect obvious fusion defects in the affected diaphragm muscle. The fibers were normally developed and the reduced thickness of the diaphragm was rather due to a smaller number of fibers than to a reduction of nuclei per fiber. Furthermore, RNAi knockdown of FGFRL1 in C2C12 cells did not result in any obvious fusion defects and overexpression of the receptor did not increase the number of nuclei per differentiated myotube.3 Nonetheless, we speculate that it is possible that FGFRL1, like Duf/Kirre and Rst in Drosophila, contributes redundantly to myoblast fusion during murine development. In that case other surface adhesion factors could substitute for its function in the FGFRL1-deficient mice. It is also conceivable that only a small subset of fibers needs FGFRL1 for fusion, making a potential fusion phenotype very hard to detect.
Mechanism of FGFRL1-mediated Fusion
Despite years of intensive research, the precise mechanisms underlying the fusion processes described above have remained largely elusive. Whereas intracellular vesicle fusion relies on the fusogenic, force generating SNARE proteins (3) and virus-cell fusion is mediated by fusogenic glycoproteins that actively merge membranes (31), no comparable mechanism has been described in mammalian cell-cell fusion. A common theme, however, is an active role of the actin cytoskeleton in the merging of the two opposing cell membranes (18, 19). Several transmembrane proteins, among them Sns, Hbs, Rst, and Duf/Kirre as well as GTP exchange factors, small GTPases and actin associated proteins have been shown to be required for the fusion of myoblasts into myotubes (32–35). These proteins direct intensive actin filament reorganization at the site of cell-cell attachment, leading to an electron dense, actin-rich structure, termed “actin focus,” where eventually fusion pores open and cytoplasmic mixing occurs (33, 36, 37).
Although FGFRL1 expression rapidly fused cultured cells similar to viral fusogens, we think that FGFRL1 itself does not actively merge membranes. Unlike the SNARE proteins and viral glycoproteins, the Ig-domain III of FGFRL1 does not have obvious helices that could generate force to pull membranes together (3, 38). It also lacks hydrophobic loop structures that would serve as membrane inserting fusion peptides, which are commonly found in viral fusogenic glycoproteins (31). We therefore propose that FGFRL1 induces cell-cell fusion by a more complex, indirect mechanism, which involves binding to other cell surface proteins and probably requires active force generation by the actin cytoskeleton. Accordingly, we demonstrated in the present study that the cell fusion induced by FGFRL1 expression is dependent on actin polymerization, placing it in line with other mammalian cell fusion processes. Because intracellularly truncated FGFRL1 was most active in the cell fusion assay, it is clearly the ectodomain, more specifically the IgIII domain, which is responsible for the fusogenic activity. The fusion and the observed actin reorganization upon FGFRL1ΔC expression can therefore only be mediated by binding to other cell surface proteins, either in cis or in trans. These binding partners remain to be investigated.
Although our proteomics approach in HEK293 cells did not reveal any FGFRL1-associated proteins that could explain its fusogenic activity, the screen identified two members of the glypican family as binding partners of FGFRL1. This clearly helped to explain why cell surface heparan sulfates had an inhibitory effect on the FGFRL1-induced cell-cell fusion. The heparin binding site of FGFRL1, located at the beginning of Ig-domain II, most likely binds to heparan sulfate chains of glypicans (and possibly other proteoglycans), which seems to reduce the fusogenic activity of the receptor. The addition of exogenous heparin displaced FGFRL1 from this binding, which increased its fusogenic activity. In further agreement with this notion, deletion of Ig-domain II as well as compound deletion of Ig-domains I and II dramatically increased the fusogenic activity of FGFRL1. In contrast, the deletion of Ig-domain I had only minimal effect on the fusion efficiency. Finally, addition of exogenous heparin had no effect on the fusogenic activity of the FGFRL1ΔCΔ12 protein, indicating that heparin was only effective when it could antagonize the binding of FGFRL1 to surface heparan sulfates. A possible explanation for the inhibitory effect of heparan sulfate binding could be steric hindrance of binding to other cell surface proteins or a partial “neutralization” of FGFRL1 activity, when the receptor is bound to heparan sulfate chains.
We do not know why FGFRL1 exerted its fusogenic activity only in CHO cells and in cell lines co-cultured with CHO cells. Most plausibly, CHO cells express a “receptor” for FGFRL1, which, together with FGFRL1, mediates the merging of the membranes. Interestingly, this fusion machinery works in cis and in trans: Every cell that was transfected with FGFRL1 could fuse with CHO cells while FGFRL1-transfected CHO cells also fused with all cell lines tested. Notably, we did not observe background fusion of HEK cells with wild-type CHO cells, suggesting that CHO cells do not generally fuse with other cells. This is supported by the fact that CHO cells are routinely used as components in cell-cell fusion assays to assess the fusogenic activity of viral glycoproteins (39, 40). If CHO cells had a pronounced propensity to fuse with each other or with other cells, they would not be suitable for quantitative fusion assays. We therefore suggest that the FGFRL1-induced fusion reflects a physiological role of this receptor in cell-cell fusion or a specialized function in cell-cell adhesion. How this function of FGFRL1 can be reconciled with a putative role in the FGF signaling system remains to be investigated. Regardless of the specific role of FGFRL1 in myoblast fusion, we think that the rapid fusion of CHO cells with other cell lines represents an exciting model for mammalian cell-cell fusion that will be of help to understand this developmentally important process.
This study was supported by grants from the Swiss National Science Foundation (31003A-127046) and the Swiss Foundation for Research on Muscular Diseases as well as by the Olga Mayenfisch Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
B. Trueb, unpublished data.
- FGFRL1
- fibroblast growth factor receptor-like 1
- LC-MS
- liquid chromatography-mass spectrometry
- FGF
- fibroblast growth factor
- FGFR
- fibroblast growth factor receptor
- GAG
- glycosaminoglycan.
REFERENCES
- 1.Chen E. H., Grote E., Mohler W., Vignery A. (2007) FEBS Lett. 581, 2181–2193 [DOI] [PubMed] [Google Scholar]
- 2.Oren-Suissa M., Podbilewicz B. (2007) Trends Cell Biol. 17, 537–546 [DOI] [PubMed] [Google Scholar]
- 3.Martens S., McMahon H. T. (2008) Nat. Rev. Mol. Cell Biol. 9, 543–556 [DOI] [PubMed] [Google Scholar]
- 4.Sapir A., Choi J., Leikina E., Avinoam O., Valansi C., Chernomordik L. V., Newman A. P., Podbilewicz B. (2007) Dev. Cell 12, 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podbilewicz B., Leikina E., Sapir A., Valansi C., Suissa M., Shemer G., Chernomordik L. V. (2006) Dev. Cell 11, 471–481 [DOI] [PubMed] [Google Scholar]
- 6.Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., Heidmann T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esnault C., Priet S., Ribet D., Vernochet C., Bruls T., Lavialle C., Weissenbach J., Heidmann T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17532–17537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trueb B., Zhuang L., Taeschler S., Wiedemann M. (2003) J. Biol. Chem. 278, 33857–33865 [DOI] [PubMed] [Google Scholar]
- 9.Trueb B., Taeschler S. (2006) Int. J. Mol. Med. 17, 617–620 [PubMed] [Google Scholar]
- 10.Baertschi S., Zhuang L., Trueb B. (2007) FEBS J. 274, 6241–6253 [DOI] [PubMed] [Google Scholar]
- 11.Catela C., Bilbao-Cortes D., Slonimsky E., Kratsios P., Rosenthal N., Te Welscher P. (2009) Dis. Model Mech. 2, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber S. D., Steinberg F., Beyeler M., Villiger P. M., Trueb B. (2009) Dev. Biol. 335, 106–119 [DOI] [PubMed] [Google Scholar]
- 13.Bertrand S., Somorjai I., Garcia-Fernandez J., Lamonerie T., Escriva H. (2009) BMC Evol. Biol. 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg F., Zhuang L., Beyeler M., Kälin R. E., Mullis P. E., Brändli A. W., Trueb B. (2010) J. Biol. Chem. 285, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rieckmann T., Kotevic I., Trueb B. (2008) Exp. Cell Res. 314, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 16.Rieckmann T., Zhuang L., Flück C. E., Trueb B. (2009) Biochim. Biophys. Acta 1792, 112–121 [DOI] [PubMed] [Google Scholar]
- 17.Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochlin K., Yu S., Roy S., Baylies M. K. (2010) Dev. Biol. 341, 66–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFife K. M., Jenney C. R., Colton E., Anderson J. M. (1999) FASEB J. 13, 823–832 [DOI] [PubMed] [Google Scholar]
- 20.O'Donnell C. D., Shukla D. (2009) J. Biol. Chem. 284, 29654–29665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue N., Ikawa M., Isotani A., Okabe M. (2005) Nature 434, 234–238 [DOI] [PubMed] [Google Scholar]
- 22.Saginario C., Sterling H., Beckers C., Kobayashi R., Solimena M., Ullu E., Vignery A. (1998) Mol. Cell. Biol. 18, 6213–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X., Sterling H., Chen Y., Saginario C., Brown E. J., Frazier W. A., Lindberg F. P., Vignery A. (2000) J. Biol. Chem. 275, 37984–37992 [DOI] [PubMed] [Google Scholar]
- 24.Sterling H., Saginario C., Vignery A. (1998) J. Cell Biol. 143, 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang L., Falquet L., Trueb B. (2010) Exp. Therap. Med. 1, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelton C., Kocherlakota K. S., Zhuang S., Abmayr S. M. (2009) Development 136, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artero R. D., Castanon I., Baylies M. K. (2001) Development 128, 4251–4264 [DOI] [PubMed] [Google Scholar]
- 28.Dworak H. A., Charles M. A., Pellerano L. B., Sink H. (2001) Development 128, 4265–4276 [DOI] [PubMed] [Google Scholar]
- 29.Strünkelnberg M., Bonengel B., Moda L. M., Hertenstein A., de Couet H. G., Ramos R. G., Fischbach K. F. (2001) Development 128, 4229–4239 [DOI] [PubMed] [Google Scholar]
- 30.Sohn R. L., Huang P., Kawahara G., Mitchell M., Guyon J., Kalluri R., Kunkel L. M., Gussoni E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9274–9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison S. C. (2008) Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gildor B., Massarwa R., Shilo B. Z., Schejter E. D. (2009) EMBO Rep. 10, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson B. E., Beckett K., Nowak S. J., Baylies M. K. (2007) Development 134, 4357–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson B. E., Nowak S. J., Baylies M. K. (2008) Traffic 9, 1050–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger S., Schäfer G., Kesper D. A., Holz A., Eriksson T., Palmer R. H., Beck L., Klämbt C., Renkawitz-Pohl R., Onel S. F. (2008) J. Cell Sci. 121, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 36.Kesper D. A., Stute C., Buttgereit D., Kreisköther N., Vishnu S., Fischbach K. F., Renkawitz-Pohl R. (2007) Dev. Dyn. 236, 404–415 [DOI] [PubMed] [Google Scholar]
- 37.Kim S., Shilagardi K., Zhang S., Hong S. N., Sens K. L., Bo J., Gonzalez G. A., Chen E. H. (2007) Dev. Cell 12, 571–586 [DOI] [PubMed] [Google Scholar]
- 38.Sleeman M., Fraser J., McDonald M., Yuan S., White D., Grandison P., Kumble K., Watson J. D., Murison J. G. (2001) Gene 271, 171–182 [DOI] [PubMed] [Google Scholar]
- 39.Huerta L., Lamoyi E., Báez-Saldaña A., Larralde C. (2002) Cytometry 47, 100–106 [DOI] [PubMed] [Google Scholar]
- 40.McShane M. P., Longnecker R. (2005) Methods Mol. Biol. 292, 187–196 [DOI] [PubMed] [Google Scholar]