Abstract
The p53 DNA-binding domain harbors a conformationally flexible multiprotein binding site that regulates p53 ubiquitination. A novel phosphorylation site exists within this region at Ser269, whose phosphomimetic mutation inactivates p53. The phosphomimetic p53 (S269D) exhibits characteristics of mutant p53: stable binding to Hsp70 in vivo, elevated ubiquitination in vivo, inactivity in DNA binding and transcription, increased thermoinstability using thermal shift assays, and λmax of intrinsic tryptophan fluorescence at 403 nm rather than 346 nm, characteristic of wild type p53. These data indicate that p53 conformational stability is regulated by a phosphoacceptor site within an exposed flexible surface loop and that this can be destabilized by phosphorylation. To test whether other motifs within p53 have similarly evolved, we analyzed the effect of Ser215 mutation on p53 function because Ser215 is another inactivating phosphorylation site in the conformationally flexible PAb240 epitope. The p53S215D protein is inactive like p53S269D, whereas p53S215A is as active as p53S269A. However, the double mutant p53S215A/S269A was transcriptionally inactive and more thermally unstable than either individual Ser-Ala loop mutant. Molecular dynamics simulations suggest that (i) solvation of phospho-Ser215 and phospho-Ser269 by positive charged residues or solvent water leads to local unfolding, which is accompanied by local destabilization of the N-terminal loop and global destabilization of p53, and (ii) the double alanine 215/269 mutation disrupts hydrogen bonding normally stabilized by both Ser215 and Ser269. These data indicate that p53 has evolved two serine phosphoacceptor residues within conformationally flexible epitopes that normally stabilize the p53 DNA-binding domain but whose phosphorylation induces a mutant conformation to wild type p53.
Keywords: Protein Denaturation, Protein Stability, Transcription Factors, Tryptophan, Ubiquitination, DNA Binding, p53, Phosphorylation, Thermal Shift, Transcription
Introduction
p53 protein is a sequence-specific DNA-binding protein and transcription factor that can regulate the cellular response to cellular stresses. Activating signals include virus infection, irradiation, hypoxia, and metabolic stress (1). The molecular basis for p53 activation involves coordinated inhibition of the ubiquitin-proteasome degradation system that is regulated by the E3 ubiquitin ligase MDM2 and induction of sets of activating enzymes including protein kinases, proline isomerases, and acetyltransferases that regulate p53 responsive gene expression. Of these signaling pathways that activate p53 that include enzymes like ATM, PIN, and p300, the most well characterized signals are those mediated by phosphorylation.
The most highly conserved phosphorylation sites of human p53 occur in the N-terminal transactivation domain at Ser15/18/20 and the C-terminal CK2 phosphorylation site at Ser392 (1). Phosphomimetic mutation of these sites can stimulate p53-dependent transcription (2), and mouse transgenes with alanine-substituted mutations have increased cancer incidence in stress or tissue-specific manner (3–5). The biochemical basis for these effects has been reported; transactivation domain phosphorylation at Thr18 can directly inhibit MDM2 binding (6, 7), whereas Ser20 phosphorylation can stabilize p300 binding (8, 9). Ser392 phosphorylation can stimulate p53 sequence-specific DNA binding according to the ensemble model of allostery (10) in part by stabilizing p53 tetramers from the dimeric state (11). Phosphomimetic mutation of codon 392 increases the thermostability of the core DNA-binding domain, demonstrating an allosteric effect to the activation (12). There are other phosphorylation sites on p53 that stimulate its function as defined using transgenics; one such site includes the MAPK kinase site at Ser46 (13), but the molecular basis for these activating effects on p53 structure and function is not as well defined as is the former cluster of phosphoacceptor sites.
Post-translational modifications can also catalyze the inhibition of p53 protein; the most well characterized are those induced by ubiquitination-degradation pathways, but there are also kinase-signaling pathways that can inhibit p53. MDM2-mediated degradation of p53 is the best characterized of the p53-inhibiting pathways (14) that can in turn be stimulated by MDM2 interactions with other proteins, including TAFII250 (15) and MDMX (16). Although there are many phosphoacceptor sites on MDM2 that have been identified, the best characterized site with potential to stimulate MDM2-mediated inhibition of p53 is in the N-terminal MDM2 pseudosubstrate motif, or “lid” (17, 18). In addition to the MDM2-stimulated degradation pathway, there are three distinct kinase pathways that directly target p53 and inhibit the protein function, although the molecular basis for this inhibition is not known. One inactivating phosphorylation site on p53 is at Ser315. Although Ser315 phosphorylation can stimulate the specific DNA-binding function of p53 (19) and DNA damage can activate p53 function via Ser315 phosphorylation in cells (20), this phosphorylation in dividing cells is catalyzed by a GSK3-signaling pathway that can promote the nuclear export and inhibition of p53 under conditions of endoplasmic reticulum stress (21). These data indicate that phosphorylation at Ser315 can inhibit or activate p53 and therefore depends upon the context. A second p53 inactivating kinase pathway is at Ser215, whose phosphorylation can be catalyzed by an Aurora-dependent signaling pathway (22), but the molecular basis for how this inhibits p53 is not known. The final kinase-inactivating pathway is triggered by COP9, which phosphorylates p53 at Thr155, and this triggers p53 degradation (23). As with the other inactivating site reviewed above, the protein-protein interactions that are driven by Thr155 phosphorylation are not defined.
In an accompanying paper (24), we report the identification of a novel p53 phosphorylation site in a multiprotein docking site in the DNA-binding domain of p53 at Ser269. This phosphorylation site is notable in that it occurs within the MDM2-binding site (e.g. “ubiquitination signal”) that triggers p53 ubiquitination and that it also forms docking sites for distinct class of protein kinases (25). Phosphomimetic mutation at Ser269 suggested that this phosphorylation would inactivate rather than activate p53 function in vivo. In this paper, we report on the biochemical basis for p53 inactivation by Ser269 phosphorylation. Using phosphomimetic mutants, we propose that Ser269 phosphorylation inactivates p53 by direct destabilization of the p53 core DNA-binding domain, thus inducing a mutant conformation to wild type p53. Molecular dynamics simulations further suggest specific mechanisms to explain how the phosphate at Ser269 could destabilize p53 protein folding by altering the allosteric effects of Gln100 and Thr102 on p53 conformational stability. These data indicate that p53 has evolved a kinase pathway(s) that can regulate the conversion of WT p53 between folded and unfolded conformational ensembles and suggest the existence of signaling cascades that can induce a mutant conformation to the wild type p53 tetramer.
MATERIALS AND METHODS
Reagents and Plasmids
N-terminally tagged biotinylated peptides were obtained from Mimotopes (Carlton, Australia). Anti-p53 antibodies were DO-1, DO-12, PAb240, PAb1620, CM-1, Ab-1 (anti-p21, Cell Signaling), 2A10 (anti-Mdm2), Hsp70, and Hsp90 (Cell Signaling). pcDNA 3.1 p53 and pCMV-Mdm2 plasmids were described (26); pRSET p53 core domain expression plasmid was obtained from Dr. P. Nikolova (27). Single amino acid mutations were introduced into wild type p53 at Ser269 according to the QuikChange site-directed mutagenesis kit (Stratagene) using pcDNA 3.1 p53, pExpr p53, and pRSET p53 DNA core domain as templates, and the following oligonucleotides were used for mutagenesis (underlined): p53 S215A, 5′-c act ttt cga cat gct gtg gtg gtg ccc-3′ (forward primer) and 5′ ggg cac cac cac agc atg tcg aaa agt g 3′ (reverse phase); p53 S215D, 5′-c act ttt cga cat gat gtg gtg gtg ccc-3′ (forward primer) and 5′-ggg cac cac cac atc atg tcg aaa agt g (reverse primer); p53 S269A, 5′-g gga cgg aac gcc ttt gag gtg cg-3′ (forward primer) and 5′-cg cac ctc aaa ggc gtt ccg tcc c-3′ (reverse primer); p53 S269D, 5′-ctg gga cgg aac gac ttt gag gtg cg-3′ (forward primer) and 5′-cg cac ctc aaa gtc gtt ccg tcc cag-3′ (reverse primer).
Peptide ELISA
Biotinylated unphosphorylated and phosphorylated peptides were captured onto ELISA wells coated with streptavidin and blocked with 3% BSA in PBS-Tween as described previously to measure Mdm2 binding (17) or mAb binding (6).
Cell Culture, Transfection, and Analysis
H1299 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cells were harvested and lysed using urea lysis buffer as described previously (28) unless otherwise stated. For mutant p53 conformation analyses, p53 within these lysates was analyzed by ELISA using PAb1620 and DO-1 as described previously (29) or by immunoprecipitation. For immunoprecipitation, cell lysates (100 ng) were precleared with protein G beads (Sigma) for 1 h before incubation with 1 μg of DO-1, PAb1620, or PAb240 at 4 °C.
DNA Binding Assays
BL21 AI Escherichia coli were transformed with pRSET p53 core wild type or S269A or S269D mutant form of the p53 core domain (residues 94–312) and were expressed and purified from soluble lysates using SP cation exchange and heparin affinity Hi-Trap columns (Amersham Biosciences), as described previously (27, 30). The DNA binding activity of p53 was examined by EMSA. p21 promoter-derived sequences (31) were labeled with [γ-32P]ATP and incubated with purified recombinant wild type or serine 269 mutants of p53 core domain in 30 mm Hepes pH 7.5, 50 mm KCl, 5% glycerol, 0.4 mm DTT, 0.1 mg/ml BSA, and 0.5% Triton X-100 containing 1 μg of poly(dI-dC) DNA (Sigma) and 500 ng of salmon sperm DNA in a final volume of 12 μl for 30 min at room temperature. Reactions were processed by adding 6× DNA sample buffer, loaded onto 5% polyacrylamide Tris borate gels, and separated by electrophoresis at 35 mA for 3 h at 4 °C. Gels were dried, and images were analyzed following exposure to a phosphor storage screen.
Intrinsic Fluorescence
Fluorescence emission spectra of the purified wild type, S269A, and S269D forms of the p53 core domain were measured using a SPEX FLUOROMAX-3 spectrofluorometer as described previously (32). Excitation wavelengths of 280 and 295 nm were used for tyrosine and tryptophan residues, respectively, and tyrosine and tryptophan fluorescence spectra were recorded from 300 to 350 nm and from 320 to 450 nm, respectively, at 10 °C using 0.5-nm steps and an integration time of 1 s. The final spectrum was the average of three emission scans minus the background buffer (50 mm Tris, pH 7.2, 5 mm DTT) fluorescence scan alone.
Thermal Protein Unfolding Assay
p53 protein unfolding was monitored using fluorescent SYPRO Orange dye (Invitrogen). Recombinant p53 core proteins were diluted to a final concentration of 2.5 μm in buffer (50 mm Tris, pH 7.2, and 2 mm DTT) in the presence or absence of increasing concentrations of ligand (0.1–30 mm MgCl2 or 1–8 μm annealed consensus DNA (27)) and incubated on ice for 15 min before SYPRO Orange was added to a final concentration of 5× (from a stock of 5000×). Samples were aliquoted into a 96-well PCR plate and sealed with optical quality sealing film (Bio-Rad). Thermal protein unfolding was carried out using an iCycler iQ real-time PCR system (Bio-Rad) by heating samples from 15 to 55 °C in 0.5 °C increments with a 30-s incubation at each increment. The fluorescent intensity was measured using excitation/emission wavelengths of 485/575 nm in relative fluorescent units (RFU), and the thermal denaturation graphs were plotted as a function of the gradient of protein unfolding against the temperature gradient (d(RFU)/dT).
Molecular Dynamics Methods
For the modeling studies, we used chain A from the crystal structure of the p53 core domain dimer bound to DNA (Protein Data Bank entry 2AHI resolved at 1.85 Å) (33). This chain was chosen because it had the least number of disordered residues. Modeling studies were carried out using the AMBER9 (34) package. The missing atoms were built using standard geometries as implemented in AMBER. Mutants were constructed in silico using SCWRL (35). The parameters for phosphoserine were taken from the AMBER parameter data base at University of Manchester (available on the World Wide Web).
The DNA-binding domain of p53 consists of a zinc ion that is coordinated to three Cys residues and one His residue, and the parameters for this coordination were taken from earlier studies (36, 37). Each system was solvated with a box of TIP3P (38) water molecules such that the boundary of the box was at least 10 Å from any protein atom. The positive charges in the system were balanced by adding chloride ions. The Parm99 force field was used for intermolecular interactions. The particle mesh Ewald method (39) was used for treating the long range electrostatics. All bonds involving hydrogen were constrained by SHAKE (40). A time step of 2 fs was used for propagating the dynamics. Each system was initially minimized for 2000 steps using steepest descent and conjugate gradient minimizers to remove any unfavorable interactions between the protein and the solvent. This was followed by heating each system to 300 K over 75 ps under normal pressure/temperature conditions. Subsequently, each system was simulated for ∼40–48 ns at constant temperature (300 K) and pressure (1 atm) (41), and the structures were stored every 2 ps for analysis. Analysis was carried out using VMD (42), and figures were made using PyMOL (43).
RESULTS
The Phosphomimetic p53S269D Is in an Unfolded or Mutant Conformation
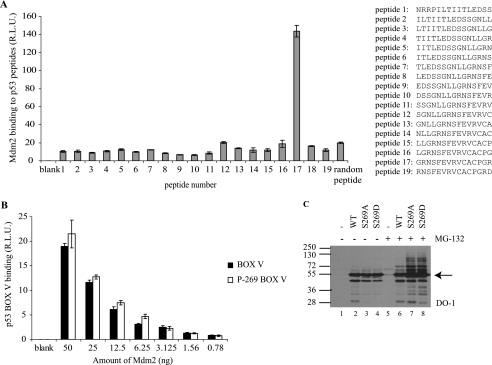
A variety of mechanisms could account for inactivation of p53 following phosphorylation at Ser269 (24). Inactivating p53 mutation of one neighboring residue within this MDM2-binding site in the S9-S10 linker region (p53F270A) has been shown to unfold p53 protein and promote “hyperubiquitination” of p53 in vivo (44). This is attributed to enhanced MDM2 binding affinity to destabilized, unfolded p53 mutants (like p53R175H) due to exposure of the second MDM2 binding site in the DNA-binding domain of p53 (29, 45). Indeed, MDM2 protein preferentially binds to peptides derived from this conformationally sensitive region of p53 (Fig. 1A, peptide 17), and phosphorylation of p53 at Ser269 does not inhibit MDM2 interactions with this motif (Fig. 1B).
FIGURE 1.
Mutation of p53 at codon 269 increases p53 ubiquitination. A, mapping the region of p53 DNA-binding domain bound by MDM2. MDM2 binding to p53 peptides was determined by ELISA. Streptavidin-coated plates were coated with biotinylated peptides and incubated with MDM2, and the amount of MDM2 captured was determined using monoclonal 2A10 followed by chemiluminescence. The data are plotted as MDM2 binding (relative light units) as a function of MDM2 levels. B, effects of serine 269 phosphorylation on MDM2 binding to its p53-DNA-binding domain docking site. MDM2 binding to wild type and Ser269-phosphorylated p53 BOX-V domain peptide (LGRNSFEVR) was examined by ELISA as in A. C, mutation of p53 at codon 269 increases p53 ubiquitination. pcDNA expression vectors encoding p53, p53S269A, or p53S269D were transfected into H1299 cells (without or with 10 μm MG-132 treatment for 4 h prior to harvesting). Lysates (10 μg) were immunoblotted with DO-1 to detect total p53 and total ubiquitin-like modification of p53. Arrow indicates p53. R.L.U., relative light units. Error bars, S.D.
We examined therefore whether p53S269D is phenotypically equivalent to mutant, inactive, and unfolded p53 as defined by sensitivity to ubiquitin-like modification in cells. Immunoblotting lysates from H1299 cells transfected with wild type p53 and with p53S269A and p53S269D mutants reveals a ladder of higher molecular mass bands in lysates from cells expressing p53S269A and p53S269D mutants (Fig. 1C, lanes 7 and 8 versus lane 6), a phenomenon indicative of p53 ubiquitin-like modifications (29). The intensity of the high molecular mass p53 ladder of “ubiquitin-like adducts” was significantly increased when protein degradation was inhibited by treating H1299 cells with the proteasome inhibitor, MG-132 (Fig. 1C, compare lanes 1–4 with lanes 5–8). Thus, p53S269D is phenotypically equivalent to p53R175H in that it is inactive (24) and highly sensitive to ubiquitin-like adducts in cells. By contrast, because transfected p53S269A can be more active than WT p53 at inducing elevated levels of MDM2 protein (24), the observed ubiquitin-like adducts of p53S269A can be attributed to enhanced induction of endogenous MDM2 protein. This phenomenon has been observed previously using the gain-of-function wild type p53 mutants in the S9-S10 loop: p53S261A and p53S264A (44) (highlighted in Fig. 2) (24).
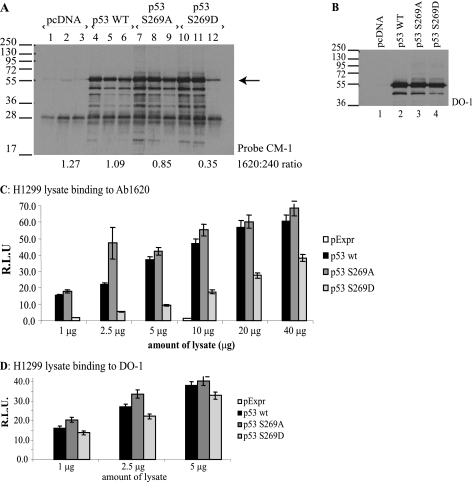
FIGURE 2.
Phosphomimetic substitution at codon 269 induces a mutant conformation to p53. A–D, effects of the S269D mutation on p53 conformation. A and B, H1299 cells were transfected with either wild type or the mutant p53 vectors. p53 forms were immunoprecipitated from lysates using monoclonal antibodies: (i) DO-1 (panspecific; lanes 1, 4, 7, and 10); (ii) PAb240 (detecting misfolded p53; lanes 2, 5, 8, and 11), or (iii) PAb1620 (detecting native p53; lanes 3, 6, 9, and 12). p53 in the immunoprecipitates was detected using CM-1 (panspecific p53 rabbit IgG) (A), whereas total expression of p53 protein isoforms was determined by immunoblotting lysates (B). The intensity of p53-reactive bands was quantified by Scion Image software, and the ratio of PAb1620- to PAb240-reactive p53 is indicated below A. C and D, H1299 cells were transfected with wild type or the indicated mutant p53 expression vectors, and p53 forms were captured on solid phase precoated with PAb1620 (C) or DO-1 (D) by incubations with lysate as indicated. Captured p53 was quantified using chemiluminescence. The data are plotted as p53 bound to the respective monoclonal antibody as a function of lysate concentration in relative light units (R.L.U.). Error bars, S.D.
We next evaluated whether the phosphomimetic p53S269D protein exhibits a mutant conformation in vivo, thus explaining the inability of p53S269D to act as a transcription factor (24). To test this, we used conformation-specific monoclonal antibodies to examine folding of the p53 isoforms. p53 alleles were transfected into H1299 cells, and the proteins were immunoprecipitated from lysates using PAb1620 and PAb240 monoclonal antibodies, which specifically recognize folded/wild type and denatured/mutant conformations of p53, respectively (46). The DO-1, PAb1620, and PAb240 antibodies immunoprecipitated equivalent levels of transfected wild type p53 (Fig. 2A, lanes 4–6, respectively). The equivalent amounts of PAb1620 (native/folded) and PAb240 (mutant/unfolded) reactive wild type p53 are due to the equilibrium that exists between the “folded” and “unfolded” states of p53 protein (47). Transfection of p53S269A into cells also produced p53 protein in an equivalent folded and unfolded equilibrium (Fig. 2A, lanes 7–9). However, the amount of p53S269D immunoprecipitated using PAb1620 was significantly lower than that of wild type p53 (Fig. 2A, compare lane 12 with lanes 6 and 9). This was not due to lowered expression of the p53S269D mutant because blotting of the total cellular p53 pool shows equivalent expression of p53 wild type, p53S269A, and p53S269D forms (Fig. 2B). The ratio of folded to non-folded p53S269D mutant was further examined by quantitative ELISA. Lysates from cells transfected with wild type and p53S269A showed significant and comparable binding to PAb1620 (Fig. 2, C versus D). In contrast, p53S269D showed significantly less binding to PAb1620 compared with wild type p53 (Fig. 2, C versus D), confirming that the phosphomimetic mutant is in a non-native conformation.
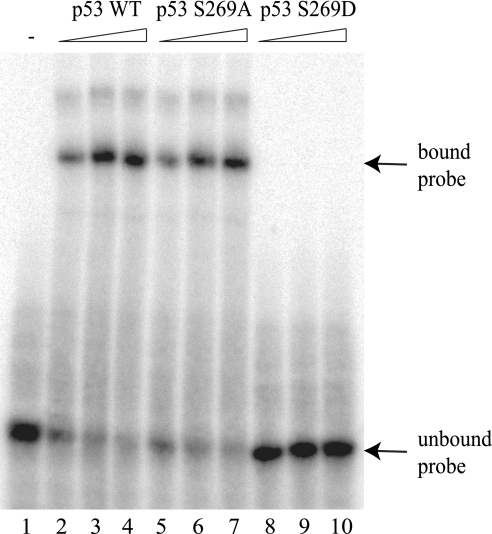
Codon 269 is adjacent to Phe270 and Asn268, and mutation of either residue to Ala270 or Asp268 can either destabilize or stabilize the p53 tetramer, respectively (29, 44, 48). For example, the N268D mutation can form an altered hydrogen bond network that links the S1 and S10 sheets of the β-sandwich in a more energetically stable manner. It is therefore possible that the S269D mutation destabilizes the p53 core domain in a manner similar to the F270A mutation (29). To determine whether loss of transcriptional activity of p53S269D was due to reduced DNA binding, we examined the ability of the p53 core domain variants to bind DNA in a sequence-specific manner at 4 °C. Recombinant wild type p53, p53S269A, and p53S269D core domain mutants were expressed and purified from E. coli. The wild type p53 and p53S269A core domain proteins both demonstrated a concentration-dependent increase in binding to the p21 promoter sequence (Fig. 3). By contrast, p53S269D did not bind to the p21 promoter element (Fig. 3), demonstrating that phosphomimetic mutation of p53 at serine 269 ablates its sequence-specific DNA binding function.
FIGURE 3.
Phosphomimetic mutation of p53 at serine 269 ablates its specific DNA binding function. The DNA binding function of p53 containing codon 269 mutations was measured using a radiolabeled p21 DNA sequence using native gel electrophoresis. The binding activity of p53 wild type, p53S269A, or p53S269D (200, 300, and 400 ng) was determined in reactions containing the p21 promoter sequence. DNA-p53 complexes were resolved using a native polyacrylamide gel, dried, and detected by storage phosphor screen. Bound and free probe are highlighted by arrows.
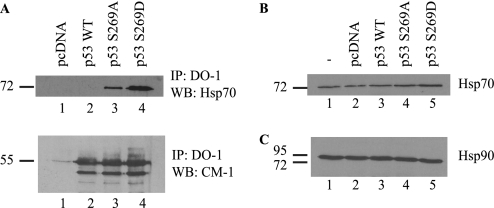
In order to further evaluate whether the phosphomimetic mutant is in a misfolded conformation in cells, we analyzed whether this p53 mutant can interact with Hsp70. This molecular chaperone interacts with unfolded mutant forms of p53 and can target the mutant protein for degradation (49). Analysis of immunoprecipitated p53 shows that Hsp70 associates with the p53S269D mutant (Fig. 4A, lane 4 versus lane 1) yet does not co-immunoprecipitate with wild type p53, supporting the hypothesis that mutation of serine 269 to aspartate leads to p53 unfolding. Surprisingly, p53S269A was also found to co-immunoprecipitate Hsp70, despite its wild type-like activity (24). This suggests that this mutant (S269A) is partially destabilized, despite being fully active. In fact, biophysical studies (see below) confirm and indicate that p53 protein instability in vitro can be uncoupled from loss-of-function effects.
FIGURE 4.
Hsp70 co-immunoprecipitates with p53-S269D. A, H1299 cells were transiently transfected with p53 wild type, S269A, or S269D, and total p53 was immunoprecipitated (IP) using DO-1. Co-immunoprecipitating Hsp70 was detected by immunoblotting (IB). Bottom panel, equal immunoprecipitation of p53 from lysates was determined by blotting with anti-rabbit p53 (CM-1). Total Hsp70 (B) and Hsp90 (C) levels in H1299 cells following transfection with p53 wild type, S269A, or S269D were determined by immunoblotting.
Together, these findings indicate that mutation of serine 269 to the phosphomimetic aspartate form leads to p53 protein unfolding in vivo and suggests that phosphorylation of p53 protein at serine 269 may induce conformational changes within p53 that cause it to adopt a mutant conformation. This may account for the observed loss of DNA binding in vitro and lowered transcriptional activity toward endogenous p21 and Mdm2 promoters. Biophysical analyses were thus initiated to determine whether the phosphomimetic p53S269D was in fact misfolded because these methodologies could reflect fundamental thermodynamic properties of the wild type and mutant p53 core DNA-binding domains.
Aspartate Mutation of Codon 269 Produces a Mutant p53 Conformation; Implications for Control of p53 Folding by Phosphorylation at Serine 269
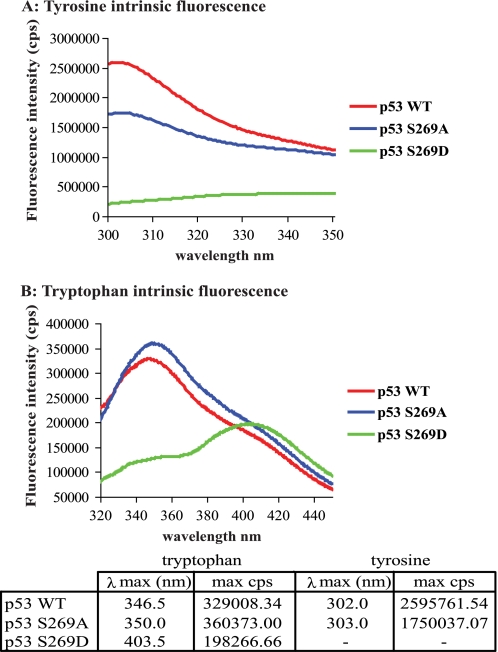
Intrinsic tyrosine or tryptophan fluorescence is highly sensitive to its local environment, and changes in fluorescence can reflect conformational changes, ligand binding, or denaturation. These properties have been useful in defining the conformational flexibility and thermodynamic instability of wild type p53 at physiological temperatures and for defining the enhanced instability of tumor-derived mutations in the p53 core domain (27, 30, 50). The structural integrity of p53 wild type, p53S269A, and p53S269D mutants was examined by studying the intrinsic fluorescent properties within the purified core domain. The p53 core domain contains a single tryptophan residue (Trp146), which is not freely accessible on the surface and faces the interior of the p53 protein (PyMOL, Protein Data Bank entry 2FEJ, in solution). The p53 core domain also contains eight tyrosine residues, some of which are accessible on the surface of the protein and some of which are buried in the interior. Strong tyrosine fluorescence spectra were observed for the wild type p53 and p53S269A core domain proteins, peaking at 302 and 303 nm, respectively (Fig. 5A). The intrinsic fluorescent properties of the tryptophan residue within the wild type p53 and p53S269A core domains was also examined, and both showed similar tryptophan fluorescent spectra (Fig. 5B). The spectra of p53S269A was, however, shifted to a longer wavelength (red shift) relative to the wild type core domain with a λmax peak of tryptophan emission at 350 nm compared with 346.5 nm (Fig. 5B). Previous studies have shown that natively folded p53 exhibits a tyrosine-dominated fluorescent spectrum with a maximum tyrosine emission at 305 nm (27, 50), suggesting that like p53 wild type, p53S269A exists in a native conformation. The red shift in tryptophan and tyrosine fluorescence may, however, suggest that p53S269A differs from that of p53 wild type and possibly exists in a more open or flexible conformation.
FIGURE 5.
Alanine or aspartate substitution of serine 269 generates conformational differences within the p53 core domain. Tyrosine (A) and tryptophan (B) fluorescence spectra of the wild type p53, p53S269A, and p53S269D core domain variants were determined and corrected for the presence of buffer background spectra as described previously (32). The fluorescent maxima for both tryptophan and tyrosine emission and the corresponding wavelength are detailed in the table below. cps, counts per second.
By contrast, the tryptophan and tyrosine fluorescent spectra obtained for the p53S269D core domain mutant were very different from that of the p53 wild type (Fig. 5, A and B). We were unable to detect distinct peaks of tyrosine fluorescence characteristic of wild type p53 (Fig. 5A), and the tyrosine spectrum obtained for p53S269D was only marginally above that of the background buffer. By contrast, a strong peak of tryptophan fluorescence was detected; however, the fluorescence spectra obtained did not exhibit the characteristic peak (∼346 nm) observed in the presence of the wild type p53 and instead displayed a significant red shift to longer wavelengths peaking with a λmax of 403.5 nm (Fig. 5B). Because the fluorescent spectrum of p53 core domain changes from a tyrosine-dominated to a tryptophan-dominated spectrum upon denaturation (27, 50), these findings indicate that p53S269D exists in a denatured or aggregated form and suggest that phosphomimetic mutation of p53 at serine 269 leads to denaturation of the core domain.
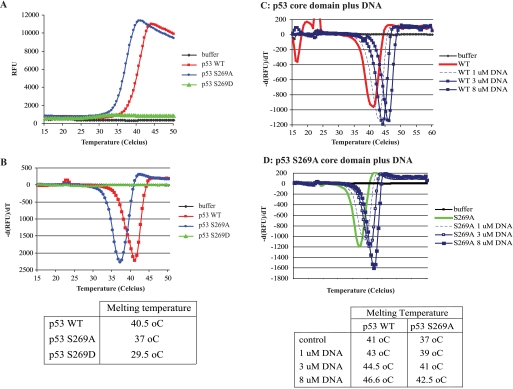
The DNA-binding domain of p53 is highly structured; however, the thermodynamic stability of this region is relatively low, and mutations can reduce the thermodynamic stability of p53 (51, 52). To examine whether phosphomimetic mutation of serine 269 alters the thermodynamic stability of p53, we monitored the thermal unfolding transition of the p53 core domain using SYPRO Orange fluorescent dye, an environmentally sensitive dye whose fluorescence is low in aqueous solutions yet increases in a hydrophobic environment. This dye is a useful tool to monitor the transition in protein folding during thermal denaturation because exposure of the hydrophobic interior of a protein upon heating enhances the fluorescent intensity of the dye (53, 54). SYPRO Orange fluorescence was measured at temperatures ranging from 15 to 50 °C. The fluorescent emission in the presence of p53 wild type and p53S269D began to increase around ∼32–34 °C and increased to a maximum around ∼41–44 °C (Fig. 6A). Thereafter, the fluorescent intensity decreased, presumably due to aggregation of unfolded p53 protein-dye complexes quenching emission. By plotting the gradient of fluorescent emission against the temperature (Fig. 6B), the midpoint temperature (Tm) of p53 core domain folding-unfolding transition can be obtained. Wild type p53 core domain displayed an unfolding transition peak with midpoint temperature of 40.5 °C (Fig. 6B), whereas p53S269A displayed a midpoint transition peak at 37 °C (Fig. 6B), suggesting that wild type and p53S269A core domains exist in two thermodynamically distinct states. Because the intrinsic fluorescence data suggests p53S269A may exist in a more open conformation (Fig. 5, A and B), the lowered thermodynamic stability of p53S269A may be due to greater conformational flexibility within the p53S269A protein.
FIGURE 6.
Thermal stability of p53 serine 269 substitution mutants. A, thermal melting profile of p53 mutants. Unfolding of p53 mutants was monitored between 15 and 50 °C using SYPRO Orange fluorescent dye, and melt curves were generated by plotting SYPRO Orange fluorescence as a function of temperature in relative fluorescent units (RFU). B, phase transitions in the thermal melting profile. The gradient of p53 unfolding was plotted as a function of temperature to obtain the midpoint temperature transition (Tm) for each variant. C and D, effect of ligands on p53 unfolding. Unfolding of p53 wild type (C) and p53S269A (D) was measured between 15 and 60 °C in the presence of increasing concentrations of p21 consensus DNA. The midpoint unfolding transition temperatures are indicated below. Thermal analyses were performed in triplicate; however, a single trace representative of each mutant is shown.
Very little change in SYPRO Orange fluorescent emission was observed in the presence of p53S269D (Fig. 6A), although a small but noticeable shoulder was obtained when the gradient of fluorescent emission in the presence of p53S269D was plotted against temperature (Fig. 6B). This gave p53S269D a calculated midpoint temperature of transition of 29.5 °C. This is significantly lower than Tm calculated for wild type p53 and p53S269A, suggesting that phosphomimetic mutation of serine 269 increases the thermodynamic instability of the p53 core domain. Little if any unfolding of p53S269D was observed at the temperatures required to unfold the wild type p53 and p53S269A proteins, suggesting that only a small proportion of the purified p53S269D core domain pool has a thermostability similar to that of wild type p53. The intrinsic fluorescent data show that p53S269D exists in an unfolded, aggregated form (Figs. 5A and 6B). This would account for the lack of SYPRO Orange binding because greater melting temperatures may be required to unfold higher order aggregates.
To further examine the differences in thermodynamic stability of p53 and p53S269A, we investigated the impact ligands would have on thermal unfolding transitions. p53 consensus DNA had a significant effect on the thermal stability of both wild type p53 and p53S269A. The unfolding transition peak of p53 wild type was increased to 43, 44.5, and 46.5 °C in the presence of 1, 3, and 8 μm consensus DNA, respectively (Fig. 6C). Because the consensus DNA did not display any change in fluorescence above background at any of the temperatures tested (data not shown), these data indicate that binding to consensus DNA induces changes that significantly enhance the thermodynamic stability of the p53 core domain.
Consensus DNA also altered the fluorescent profile of the p53S269A mutant (Fig. 6D). The unfolding kinetics of p53S269A was shifted in the presence of 1, 3, and 8 μm consensus DNA, and the unfolding transition midpoint temperatures were increased from 37 to ∼42.5 °C (Fig. 6D). These data show that, like that of wild type p53, the stability of p53S269A is significantly enhanced upon binding to DNA. However, DNA binding does not overcome the marginal thermal instability incurred by alanine mutation of serine 269 because even in complex with DNA, the p53S269A core domain remains slightly less thermodynamically stable than the wild type p53 core domain complexed to DNA (Tm of ∼42.5 °C versus Tm of ∼46.5 °C), and the p53S269A-DNA complex appears structurally distinct from the wild type p53 core domain-DNA complex. As a control, the addition of magnesium ions did not qualitatively alter the unfolding kinetics of wild type p53 or p53S269A (data not shown).
Two Conformationally Flexible Loops Containing Phosphoacceptor Sites Regulate p53 Conformation and Activity
Characterization of the codon 269 mutants has shown that the alanine mutant can induce a more open conformation as defined by intrinsic fluorescence and that this mutant does not reduce the specific activity of p53 in cells. However, the Ala269 mutant does increase thermoinstability in vitro and increases Hsp70 interactions in vivo. By contrast, the phosphomimetic mutant p53S269D is highly destabilized, inactive, and characteristic of unfolded mutant p53. These data suggest first that this conformationally flexible loop in p53 has evolved a serine residue to maintain a degree of intrinsic inflexibility (i.e. flexibility or a more open conformation can be improved upon by alanine mutation of codon 269). Second, the serine has evolved to function as a phosphoacceptor site whose phosphorylation drives the unfolding and destabilization of wild type p53. This would create a rapid and flexible mechanism to convert the wild type p53 tetramer to a mutant conformation. Although the physiological functions of this switch are not yet evident and will require additional research, the existence of this phosphorylation site provides a novel mechanism to unfold and inactivate wild type p53 in vivo.
In order to further evaluate the evolutionary significance of this post-translational mode of p53 inactivation, we also examined a previously identified phosphorylation site of p53 at Ser215. Phosphorylation at the Ser215 site of p53 was previously shown to occur in cells, and phosphomimetic mutation at Ser215 created a transcriptionally inactive p53 (22). The mechanism accounting for p53 inactivation following Ser215 phosphorylation has not been described. In analyzing this Ser215 phosphorylation site, we noticed that it occurs at another so-called conformationally flexible epitope, bound by the monoclonal antibody PAb240 (55). This mAb was the original probe used to demonstrate that mutant p53 protein is unfolded in human cancer cells (56). We evaluated the alanine-substituted mutants of codon 215 mutants to determine whether this flexible motif behaved like the codon 269 mutants and secondarily whether these two conformationally flexible motifs interact allosterically and alter the folding of the p53 DNA-binding domain. For example, we could predict that Ala215 mutation could overcome the Asp269 mutation or vice versa. Alternatively, because Ala269 and Ala215 are individually fully active but with enhanced conformational flexibility, it could be predicted that the double mutant S215A/S269A would be more active than WT p53. However, neither of these two outcomes was observed (see below). The structural integrity of wild type p53, p53S215A, p53S269A, and the double mutant p53S215A/269A was tested to evaluate these possible outcomes.
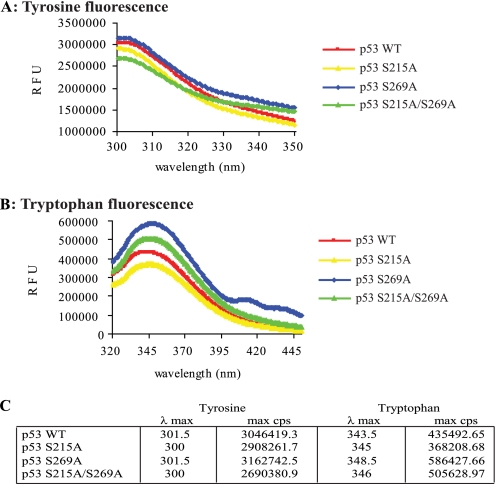
Tyrosine fluorescence spectra observed for the wild type p53, p53S215A, p53S269A, and the double mutant p53S215A/S269A core domain proteins peaked from ∼300 to 301.5 nm (Fig. 7A), indicating that all alanine-substituted mutants were similar in their folding properties to wild type p53 and that the double alanine-substituted mutant did not have characteristics of unfolded mutant p53 (Fig. 5). However, the intrinsic tryptophan fluorescence within the p53S215A and the double mutant p53S215A/S269A core domains also revealed differences similar to those seen with the p53S269A mutant. The spectrum of p53S215A was shifted to a longer wavelength (red shift) relative to the wild type core domain although not to the degree observed using p53S269A (Fig. 7B). The double mutant p53S215A/S269A also exhibited a red shift in intrinsic tryptophan fluorescence that is also indicative of a more open or flexible conformation. There was no apparent synergy in red shift using the double alanine-substituted p53 mutant. Together, these studies suggest that, like p53S269A, p53S215A also exists in a native and more “opened” conformation.
FIGURE 7.
Alanine substitution of serine 215 also generates conformational differences within the p53 core domain. Tyrosine (A) and tryptophan (B) fluorescence spectra of the wild type p53, p53S215A, p53S269A, and p53S215A/S269A core domain variants were determined and corrected for the presence of buffer background spectra as described previously (32). The fluorescent maxim for either tryptophan or tyrosine emission and the corresponding wavelength are detailed in C. RFU, relative fluorescent units.
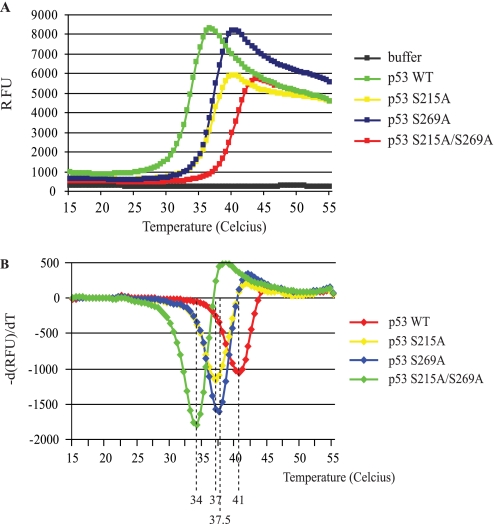
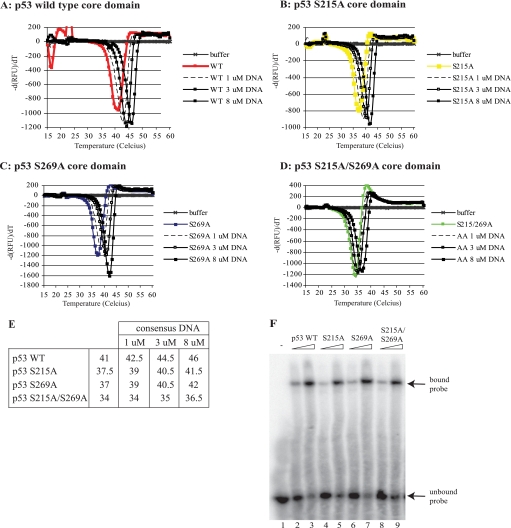
The stability of wild type p53, p53S215A, p53S269A, and the double mutant p53S215A/S269A proteins was also examined in thermal shift assays in order to compare the rate of p53S215A unfolding with that of p53S269A. Relative to wild type p53, which exhibits a thermal transition at 41 °C, both Ala215 and Ala269 mutations showed an equivalent increase in the thermal instability of p53 by approximately the same temperature of 4 °C (Fig. 8, A and B). However, the double mutant displayed a further enhanced thermoinstability 7 °C below wild type p53 (Fig. 8). Together, these data indicate that each of these conformationally flexible loops maintains p53 thermostability to equivalent degrees, and removing both serine residues precludes further the acquisition of a wild type conformation. The thermal shift assays were also performed in the presence of increasing DNA concentrations, where enhanced thermostability of ∼4–5 °C was observed using wild type p53, p53S215A, and p53S269A proteins (Fig. 9, A–C). The double mutant p53S215A/S269A exhibited an increased thermal shift of 2.5 °C (Fig. 9D), indicating that the protein can still interact with the DNA ligand (as seen in Fig. 9F) but not to the extent of wild type p53 or the single alanine-substituted mutants (see Fig. 9E).
FIGURE 8.
Thermal stability of p53 serine 215/269 substitution mutants. A, thermal melting profile of p53 mutants. Unfolding of p53 mutants was monitored between 15 and 55 °C using SYPRO Orange fluorescent dye, and melt curves were generated by plotting SYPRO Orange fluorescence as a function of temperature. B, phase transitions in the thermal melting profile. The gradient of p53 unfolding was plotted as a function of temperature to obtain the midpoint temperature transition (Tm) for each variant. Midpoint transition temperatures for each mutant are indicated. RFU, relative fluorescent units.
FIGURE 9.
The effect of consensus DNA on the stability of p53 core domains S215A and S269A mutants. Unfolding of p53 wild type (A), p53S215A (B), p53S269A (C), and p53S215/269A (D) was measured between 15 and 60 °C in the presence of increasing concentrations of consensus DNA. The midpoint unfolding transition temperatures are indicated in E. Thermal analyses were performed in triplicate; however, a single trace representative of each mutant is shown. F, the binding activity of p53 wild type, p53S215A, p53S269A, or p53S215A/S269A (300 or 600 ng) was determined in reactions containing the p21 promoter sequence. DNA-p53 complexes were resolved using a native polyacrylamide gel, dried, and detected by storage phosphor screen. Bound and free probe are highlighted by arrows.
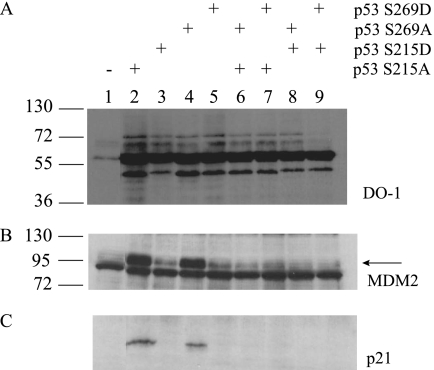
We finally evaluated the activity of the p53S215A, p53S269A, and p53S215A/S269A and mixed alanine/aspartate mutants in cells to determine whether single and double loop mutants affected p53 function in vivo. As observed using the single S269A or S269D mutant p53s (Fig. 10, lanes 4 and 5), alanine or aspartate mutations at codon 215 produced a p53 that is either fully active or inactive, respectively (Fig. 10, D and E, lane 2 versus lane 3). The double mutants provided additional evidence that p53 conformational integrity can be controlled by the surface loop mutants. The double alanine mutant (S215A/S269A), although in a wild type conformation as defined by (i) enhanced flexibility (Fig. 7), (ii) significant destabilization in thermal shift (Fig. 8), and (iii) ability to be stabilized by DNA in vitro at 4 °C (Fig. 9), was completely inactive as a transcription factor as defined by loss of p21 and MDM2 protein production (Fig. 10, lane 6). The mixed aspartate/alanine mutant was also inactive (Fig. 10, lanes 7–9). Together, these data suggest that p53 has evolved two serine residues that maintain a degree of thermodynamic stability but whose individual phosphorylation can unfold and produce a p53 with a mutant-like conformation. The negative dominance of the double alanine-substituted mutant further suggests that at least one of the two serine residues is required to maintain thermostability of p53.
FIGURE 10.
The effects of Ser215 and Ser269 mutation on p53 function; phosphomimetic substitution or double alanine substitution of p53 at Ser215 or Ser269 inactivates p53. The effect of serine 215 and serine 269 mutation on expression of the downstream target genes of p53 was examined by immunoblotting lysates derived from H1299 cells transfected with pcDNA vector containing p53 S215A, S215D, S269A, S269D, S215A/S269A, S215A/S269D, S215D/S269A, or S215D/S269D. Lysates were blotted for reactivity to DO-1 (A), MDM2 (B) (2A10), and p21 (C).
The Effects of Phosphoserine on p53 Conformation Using Molecular Dynamics Simulations
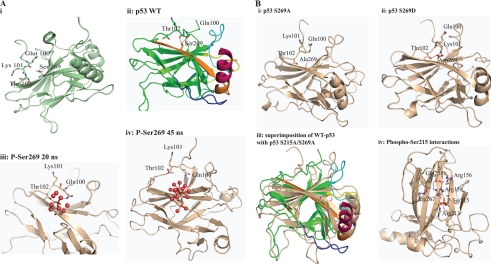
Molecular dynamics simulations were carried out on the core domain of wild type p53 and its phosphorylated states (phosphorylated at Ser215 and separately at Ser269). We also carried out simulations on S269D, S269A, and S215A/S269A double mutants. In the wild type state, molecular dynamics simulations show (Fig. 11A, ii) that the side chain of Ser269 makes hydrogen bonds with Gln100 and Thr102 and also with solvent water molecules (the crystal structure by itself does not show any hydrogen bonds between Ser and the other two residues). The N-terminal loop is tethered to the surface of the protein by a few hydrogen bonds, and these are made largely with the S10 β-strand. Upon phosphorylation at Ser269, the highly negatively charged phosphate moiety involves Gln100 and Thr102 in hydrogen bonds for ∼20 ns (Fig. 11A, iii). However, with no cationic residues in the vicinity of this phosphate to stabilize it through salt bridges, the negatively charged phosphate is energetically more stable if well solvated by water molecules. As this site fills with water, the N-terminal loop moves away from it by up to 8 Å after 45 s (Fig. 11A, iv). These events are also accompanied by destabilization of the DNA binding regions and the dimerization interface.
FIGURE 11.
Model summarizing the effect of phosphorylation at the conformationally flexible motif on p53 activity. A, i, diagram of the structure of the wild type p53 core domain (Protein Data Bank entry 2FEJ) highlighting the Ser269 region and its proximity to the flexible motif (residues 100–103). ii, the epitope is in orange color, whereas the loop and helix that interact with the DNA are shown in yellow and brown, respectively. ii–iv, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are phospho-Ser269 effects on p53. ii, wild type p53 highlighting interactions of Ser269 with Thr102 and Gln100; iii, the phospho-Ser269 intermediate; iv, phospho-Ser269. Because no stabilizing cationic side chains exist near the phosphate, the negatively charged phosphate is energetically more stable when solvated by water. This is accompanied by the movement away of the N-terminal loop and destabilization of the DNA binding regions plus the dimerization interface. B, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are Asp269 or Ala269 effects on p53. i, S269A final molecular dynamics snapshot; ii, S269D final molecular dynamics snapshot; iii, superposition of wild type and double alanine final molecular dynamics snapshot; iv, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are phospho-Ser215 effects on p53 and the phospho-Ser215 intermediate showing the network of interactions. Phosphorylation at Ser215 leads to the formation of an extended network that involves Arg213-Ser(P)215-Arg158-Asp258-Arg156.
When Ser269 is mutated to alanine, the hydrogen bonds that Ser269 made with Gln100 and Thr102 are replaced by transient hydrogen bonds to solvent and to various surrounding atoms (Fig. 11B, i). The side chain of Ala is stabilized by the hydrophobic side chains of Leu252 and Ile254 located on β-strand S9. Overall, this mutation does not appear to cause as much perturbation as the phosphorylation at Ser269. In contrast, the S269D mutation initially maintains the hydrogen bond with Gln100 and Thr102 for ∼40 ns, after which a major conformational change in the N-terminal loop occurs, which is accompanied by a flip of Lys101 (its terminal amine moves by ∼11 Å). This results in the formation of a salt bridge with the side chain of Asp269 (the hydrogen bond with Gln100 is lost; Fig. 11B, ii). This is also accompanied by the formation of a new hydrogen bond between Asp269 and Gln130 that in turn perturbs the spatially contiguous DNA binding region. Together, these data highlight the importance of the interaction of the S10-β strand and the motif preceding the first β-strand in the DNA-binding domain of p53.
Ser215 is initially hydrogen-bonded to the backbone of Leu206. Within 2 ns, Arg209, which is located on the loop connecting S6 and S7 and is exposed to solvent in the crystal structure, undergoes a local conformational change and moves ∼11 Å and makes a salt bridge with Asp258 (located on S4), which is also hydrogen-bonded to Arg158. In this process, Ser215 gets buried (Fig. 11B, iv). Phosphorylation at Ser215 initially leads to the formation of a salt bridge between the phosphate and Arg158. The cavity near the phosphate initially widens as the phosphate is also hydrated. Accompanying this is a conformational change in the S6-S7 loop and an ∼11 Å movement of Arg213 by ∼10 ns as it moves to make a salt bridge with the phosphate (Fig. 11B, iv). It appears that this takes place because the space between the phosphate and Arg213 is separated by only a few water molecules when the phosphate is temporarily hydrated. This gives the space between the negatively charged phosphate and the positively charged Arg213 a low dielectric character, which results in the Arg213 moving toward the phosphate. The resulting salt bridge extends an existing network that involves Arg213-Ser(P)215-Arg158-Asp258-Arg156, and this remains quite robust. During the transition of Arg213, a hydrophobic interaction between Pro98, Met160, Ile162, and Thr211 that appears to weakly tether the N-terminal loop to the surface of p53 is broken by the migrating Arg213. This triggers the peeling away of the N-terminal loop and also results in destabilization of regions that are at the dimerization and DNA binding interface.
In the S215A/S269A double mutant, the lack of hydrogen bonds between the side chain at 215 or 269 with Leu206 and Gln100, respectively, leads to local destabilization, which propagates in such a manner that the loop containing Leu206 loses its interactions with the Pro98 region, undergoes a conformational change, and is accompanied by the moving away of the N-terminal loop (Fig. 11B, iii).
DISCUSSION
Unfolding of the p53 Core DNA-binding Domain by Phosphorylation at Ser269
The transcriptional activity of p53, its stability, and its turnover are regulated by multiple post-translational modifications, including acetylation, ubiquitination-like modifications, and phosphorylation (57). Ubiquitination targets p53 for degradation by the proteasome, p53 acetylation facilitates recruitment of co-activators, and phosphorylation facilitates changes in its specific activity as a transcription factor by altering protein-protein interactions (58). The binding of MDM2 to the N-terminal LXXLL motif of p53 induces conformational changes within MDM2 that enhance its binding to the ubiquitination peptide signal (SXXLXGXXXF) within the flexible linker in the DNA-binding domain of p53 (26, 59). Phosphorylation of p53 also requires allosteric interactions between kinases that bind within the flexible linker in the DNA-binding domain or at sites in the tetramerization domain. Thus, this central flexible motif (Fig. 2) (24) forms a common docking site that allosterically modulates the activity of several enzymes that regulate p53 function. This underscores the importance of the flexible linker in the DNA-binding domain of p53 in coordinating incoming stress signals and p53 activity.
In the accompanying study (24), the p53 multiprotein docking domain containing the MDM2 ubiquitination signal is shown to harbor a phosphorylation site at Ser269 (Fig. 11). The inactivity of p53S269D in cells suggests that this phosphorylation would be inactivating.
In this paper, we have characterized biochemically this phosphomimetic p53 mutant to determine how phosphorylation could inactive p53. The phosphomimetic mutant destabilized the p53 core DNA-binding domain and reduced sequence-specific DNA binding, thus explaining its reduced transactivation potential. Molecular dynamics simulations also suggest that the introduction of negatively charged phosphate groups at either Ser215 or Ser269 requires either solvent water or positive charges to stabilize it. There are no positively charged residues near Ser269, so solvent waters preferentially hydrate this site, leading to the N-terminal loop in its vicinity being pushed away, accompanied by the exposure of the second MDM2 binding site in the S10 β-strand. In contrast, two positively charged residues, Arg158 and Arg213 are coordinated by Ser(P)215; the latter results from a large conformational change, which also disrupts the interaction of the N-terminal loop with the p53 surface. In the case of S269D, the planar anionic environment of the carboxyl group leads to the formation of a novel salt bridge with Lys101 that is accompanied by local structural rearrangements. Both events lead eventually to destabilization of the p53 core DNA-binding domain (Fig. 11).
In light of our current studies, the interaction between the S10-β-strand (containing the MDM2 binding site and the Ser269 phosphorylation site) and the motif (residues 100–103) preceding the S1-β-strand requires further evaluation. This flexible motif between positions 100 and 102 is not as well studied, given that a significant number of p53 DNA-binding domain analyses begin with residue 102/103, which is just prior to the start of the structured S1-β-strand. However, there are previous reports suggesting an important role for this motif. First, although residues 100–102 do not generally represent hot spots in a large range of human cancers, there is a report indicating that it is a hot spot for mutations in adrenal cancers (60). These mutations in this motif can include Q100R, K101N, and T102I, any of which could destabilize the interaction of this motif with the S10-β-strand (Fig. 11A). Additionally, NMR studies have shown that conformational changes can be detected in this motif of p53 (residues 101–103) after the interaction with peptides from the central domain of MDM2 (61). This might lead to “unfolding” of p53 as a prerequisite for substrate ubiquitination catalyzed by MDM2. A recent structural analysis of p53 tetramers also suggested an important role for Gln100 and Lys101 at the dimer interface (62), which we have not evaluated in our current study. The Glu224 of one monomer forms a pair of hydrogen bonds with the main-chain amide of Gln100 and Lys101 of an adjacent monomer. Also, Lys100 from one monomer donates a hydrogen bond to the carbonyl of Val225 of an adjacent monomer. These clinical and biophysical data suggest an important functional interaction between the S10-β-strand and the flexible motif (residues 100–103) and suggest a mechanism underlying the ability of Ser269 phosphorylation to destabilize p53 conformation.
The overproduction of the kinases and phosphatases that target this serine 269 site on p53 has implications for how cells might produce two distinct forms of wild type p53 in response to DNA damage; (i) one form of wild type p53 is transcriptionally active coordinated in part by phosphorylation at sites including Ser20 and Ser15, and (ii) the other form of wild type p53 with a radiation-induced phosphorylation at Ser269 (24) is inactive and would have function independent of the classic wild type p53 transcription program. Because mutant forms of p53 have distinct subcellular localizations and functions (63), the phospho-Ser269 form of wild type p53 protein might have similar physiological functions. The identification of the kinase and phosphatases that regulate this equilibrium in wild type p53, between hypophosphorylation and hyperphosphorylation at Ser269, has implications for wild type p53 inactivation in human cancers. The kinases and/or kinase-signaling pathways that target the Ser269 in vivo remain to be defined.
The Role of the Two Conformationally Flexible Phosphorylation Sites in Regulating the Folding of the p53 DNA-binding Domain
The core DNA binding drives specific DNA binding and forms a highly structured domain (52). The β sandwich loop-sheet-helix motif in p53 is stabilized by many hydrophobic and electrostatic interactions; however, the thermodynamic stability of this region is low, and missense mutations can easily destabilize the core domain (52). There are three major classes of p53 mutants: (i) DNA contact mutations (R273H) remaining in a folded conformation, (ii) weakly destabilized and partially unfolded mutants (G245S) that are unable to bind DNA, and (iii) mutants (R175H) that are highly destabilized and globally unfolded and aggregate at physiological temperatures (50). Phosphorylation of p53 (or S269D phosphomimetic mutation) should lead to global unfolding of the core domain and create a mutant with characteristics like p53R175H. Not all p53 mutations inactivate transcriptional activity; some mutants show a gain of function on a subset of genes that may promote tumorogenic growth (reviewed in Refs. 64 and 65). Many gain-of-function mutants show altered selectively for subsets of p53 target genes. For example, p53R213Q is able to potently induce Mdm2 yet is unable to induce expression of apoptosis-inducing genes PIG3 and PIG11 (66). Other mutant forms of p53 display differential gene expression through binding to DNA at sites lacking p53-responsive elements or as a result of altered interaction with transcription factors such as Ets1 and Sp1 and act in a manner opposite that of wild type p53 (67). The increase in specific activity and thermoinstability of p53S269A appears different mechanistically from the incremental increase in p53 thermostability and activity previously induced via mutation of up to four amino acids in the DNA-binding domain (48, 68). By contrast to the inactive S269D mutant p53, p53S269A forms a control for specificity because it was (i) more active than the wild type p53 at inducing endogenous p21 and Mdm2, (ii) more flexible as defined by red shift in tryptophan fluorescence, and (iii) more thermally unstable as defined by thermal shift assays. Simulations revealed that although the S269A mutation looses the hydrogen bonding with Gln100 and Thr102, providing a rationale for reductions in thermostability (Fig. 7), Ala269 does not form a destabilizing bridge with this chain (Fig. 11) because p53 activity in cells can be partially improved upon by the serine to alanine substitution at codon 269 and produce a structurally modified form of p53 with a reduced thermostability but with an elevated specific activity in cells. The latter finding suggests that increasing the flexibility and thermoinstability of p53 can perhaps increase its ability to adopt different conformations with transcriptional components in cells.
A phosphorylation site was also previously reported at Ser215, but a mechanism to account for p53 inactivity was not demonstrated (22). This site is notable in that it contains the conformationally flexible PAb240 monoclonal antibody epitope that can be used to define p53 mutant unfolding in human cancers (56). Similar to the p53S269A protein, p53S215A is as active as wild type p53, but the phosphomimetic mutant p53S215D is thermally unstable (Figs. 7–9). These data suggest that both Ser215 and Ser269 phosphorylation convert wild type p53 to the mutant misfolded conformation. Surprisingly, the double Ala mutant protein, p53S215A/S269A, is completely inactive in cells and is significantly more thermally unstable than the individual, active alanine mutants (Figs. 7–9). Molecular dynamics simulations also suggest that the double alanine mutant is unable to maintain the intradomain interactions required to maintain p53 conformation (Fig. 11B, iv) and suggest an important dual role for Ser215 and Ser269 in stabilizing WT p53.
A previous report also highlighted a novel role of phosphorylation in driving destabilization of a target protein. The splicing regulatory protein KSRP has a phosphoacceptor site at Ser193 whose phosphorylation leads to unfolding of the protein as defined by NMR (70). This unfolding allows access of the phosphomotif to 14-3-3 that in cells regulates compartmentalization of the protein. Similar to our studies on p53, biophysical studies demonstrated that an alanine mutation at codon 193 has little effect on the stability of the protein, but asparate mutation increases significantly the thermoinstability of KSRP. The hydration of phosphate observed here from the molecular dynamics simulations (Fig. 11) may be a feature that is more ubiquitous. A recent report examining the binding of the POLO box binding domain to phosphopeptides (71) using molecular dynamics simulations found similarly that negatively charged phosphate groups can contribute significantly to binding by stabilizing several waters of hydration; indeed, the authors found that these very same water molecules in the absence of phosphorylation, are energetically unfavorable. Further work on other systems will clearly reveal more information on how water molecules of hydration modulate interactions in biology.
In summary, biophysical studies have shown that the p53 core domain is thermodynamically unstable and that phosphorylation in the DNA-binding domain could inactivate p53 by enhancing intrinsic thermoinstability. This reversible thermoinstability of p53 is the feature by which there is promise in reactivating mutant, inactive p53 (72). p53 stability can be enhanced by interacting proteins, and evidence for this dynamic regulation through docking interactions has emerged in recent years as an additional layer of regulation for the complex signaling network (69, 73). In this study, we show that phosphomimetic mutation within the MDM2 ubiquitination signal also destabilizes p53 structure and function. This provides a model to describe how phosphorylation at Ser269 would inactivate p53 in cells. Because the Ser215 phosphorylation site in another conformationally flexible loop also has a phosphorylation site, these data indicate that the two surface loop serine residues can function not only as phosphorylation sites but that they together are required to maintain the thermostability of p53. This also identifies an intriguing paradigm in p53 protein evolution; it has two kinase phosphoacceptor sites that regulate the thermostability of p53 by virtue of controlling the equilibrium between folded and unfolded states.
Acknowledgment
We acknowledge the use of the Edinburgh Biophysical Characterization Facility (supported by the Scottish University Life Sciences Alliance and the Biotechnology and Biological Sciences Research Council).
Footnotes
This work was supported by CRUK Programme Grant C483/A6354 (to T. R. H.).
REFERENCES
- 1.Maclaine N. J., Hupp T. R. (2009) Aging 1, 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jabbur J. R., Zhang W. (2002) Cancer Biol. Ther. 1, 277–283 [DOI] [PubMed] [Google Scholar]
- 3.Bruins W., Bruning O., Jonker M. J., Zwart E., van der Hoeven T. V., Pennings J. L., Rauwerda H., de Vries A., Breit T. M. (2008) Mol. Cell. Biol. 28, 1974–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruins W., Zwart E., Attardi L. D., Iwakuma T., Hoogervorst E. M., Beems R. B., Miranda B., van Oostrom C. T., van den Berg J., van den Aardweg G. J., Lozano G., van Steeg H., Jacks T., de Vries A. (2004) Mol. Cell. Biol. 24, 8884–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacPherson D., Kim J., Kim T., Rhee B. K., Van Oostrom C. T., DiTullio R. A., Venere M., Halazonetis T. D., Bronson R., De Vries A., Fleming M., Jacks T. (2004) EMBO J. 23, 3689–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig A. L., Burch L., Vojtesek B., Mikutowska J., Thompson A., Hupp T. R. (1999) Biochem. J. 342, 133–141 [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C. J., Srinivasan D., Jun L. H., Coomber D., Verma C. S., Lane D. P. (2008) Cell Cycle 7, 608–610 [DOI] [PubMed] [Google Scholar]
- 8.Teufel D. P., Bycroft M., Fersht A. R. (2009) Oncogene 28, 2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornan D., Shimizu H., Perkins N. D., Hupp T. R. (2003) J. Biol. Chem. 278, 13431–13441 [DOI] [PubMed] [Google Scholar]
- 10.Hupp T. R., Lane D. P. (1995) J. Biol. Chem. 270, 18165–18174 [DOI] [PubMed] [Google Scholar]
- 11.Sakaguchi K., Sakamoto H., Lewis M. S., Anderson C. W., Erickson J. W., Appella E., Xie D. (1997) Biochemistry 36, 10117–10124 [DOI] [PubMed] [Google Scholar]
- 12.Nichols N. M., Matthews K. S. (2002) Biochemistry 41, 170–178 [DOI] [PubMed] [Google Scholar]
- 13.Feng L., Hollstein M., Xu Y. (2006) Cell Cycle 5, 2812–2819 [DOI] [PubMed] [Google Scholar]
- 14.Horn H. F., Vousden K. H. (2007) Oncogene 26, 1306–1316 [DOI] [PubMed] [Google Scholar]
- 15.Allende-Vega N., Saville M. K., Meek D. W. (2007) Oncogene 26, 4234–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 17.Worrall E. G., Wawrzynow B., Worrall L., Walkinshaw M., Ball K. L., Hupp T. R. (2009) J. Chem. Biol. 2, 113–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worrall E. G., Worrall L., Blackburn E., Walkinshaw M., Hupp T. R. (2010) J. Mol. Biol. 398, 414–428 [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Farmer G., Soussi T., Prives C. (1995) Oncogene 10, 779–784 [PubMed] [Google Scholar]
- 20.Blaydes J. P., Luciani M. G., Pospisilova S., Ball H. M., Vojtesek B., Hupp T. R. (2001) J. Biol. Chem. 276, 4699–4708 [DOI] [PubMed] [Google Scholar]
- 21.Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A. E. (2004) Genes Dev. 18, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q., Kaneko S., Yang L., Feldman R. I., Nicosia S. V., Chen J., Cheng J. Q. (2004) J. Biol. Chem. 279, 52175–52182 [DOI] [PubMed] [Google Scholar]
- 23.Bech-Otschir D., Kraft R., Huang X., Henklein P., Kapelari B., Pollmann C., Dubiel W. (2001) EMBO J. 20, 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser J., Vojtesek B., Hupp T. R. (2010) J. Biol. Chem. 285, 37762–37772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig A. L., Chrystal J. A., Fraser J. A., Sphyris N., Lin Y., Harrison B. J., Scott M. T., Dornreiter I., Hupp T. R. (2007) Mol. Cell. Biol. 27, 3542–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 27.Bullock A. N., Henckel J., DeDecker B. S., Johnson C. M., Nikolova P. V., Proctor M. R., Lane D. P., Fersht A. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14338–14342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser J. A., Hupp T. R. (2007) Biochemistry 46, 2655–2673 [DOI] [PubMed] [Google Scholar]
- 29.Shimizu H., Saliba D., Wallace M., Finlan L., Langridge-Smith P. R., Hupp T. R. (2006) Biochem. J. 397, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bullock A. N., Henckel J., Fersht A. R. (2000) Oncogene 19, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 31.Walerych D., Kudla G., Gutkowska M., Wawrzynow B., Muller L., King F. W., Helwak A., Boros J., Zylicz A., Zylicz M. (2004) J. Biol. Chem. 279, 48836–48845 [DOI] [PubMed] [Google Scholar]
- 32.Wawrzynow B., Pettersson S., Zylicz A., Bramham J., Worrall E., Hupp T. R., Ball K. L. (2009) J. Biol. Chem. 284, 11517–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T. E., Shakked Z. (2006) Mol. Cell 22, 741–753 [DOI] [PubMed] [Google Scholar]
- 34.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bower M. J., Cohen F. E., Dunbrack R. L., Jr. (1997) J. Mol. Biol. 267, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 36.Walker R. C., Mercer I. P., Gould I. R., Klug D. R. (2007) J. Comput. Chem. 28, 478–490 [DOI] [PubMed] [Google Scholar]
- 37.Walker R. C. (2003) The Development of a QM/MM Based Linear Response Method and Its Application to Proteins, Ph.D. thesis, Imperial College London, London [Google Scholar]
- 38.Jorgensen W. L., Chandrasekhar J., Buckner J. K., Madura J. D. (1986) Ann. N.Y. Acad. Sci. 482, 198–209 [DOI] [PubMed] [Google Scholar]
- 39.Darden T., York D. M., Pedersen L. J. (1993) J. Chem. Phys. 98, 10089–10099 [Google Scholar]
- 40.Van Gunsteren W. F., Berendsen H. J. (1977) Mol. Phys. 34, 1311–1327 [Google Scholar]
- 41.Berendsen H. J., Postma J. P., Van Gunsteren W. F., Dinola A., Haak J. R. (1984) J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 27–28 [DOI] [PubMed] [Google Scholar]
- 43.Delano W. L. (2002) The Pymol Molecular Graphics System, DeLano Scientific, LLC, San Carlos, CA [Google Scholar]
- 44.Shimizu H., Burch L. R., Smith A. J., Dornan D., Wallace M., Ball K. L., Hupp T. R. (2002) J. Biol. Chem. 277, 28446–28458 [DOI] [PubMed] [Google Scholar]
- 45.Terzian T., Suh Y. A., Iwakuma T., Post S. M., Neumann M., Lang G. A., Van Pelt C. S., Lozano G. (2008) Genes Dev. 22, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vojtesek B., Dolezalova H., Lauerova L., Svitakova M., Havlis P., Kovarik J., Midgley C. A., Lane D. P. (1995) Oncogene 10, 389–393 [PubMed] [Google Scholar]
- 47.Vojtĕsek B., Lane D. P. (1993) J. Cell Sci. 105, 607–612 [DOI] [PubMed] [Google Scholar]
- 48.Joerger A. C., Allen M. D., Fersht A. R. (2004) J. Biol. Chem. 279, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 49.Muller P., Hrstka R., Coomber D., Lane D. P., Vojtesek B. (2008) Oncogene 27, 3371–3383 [DOI] [PubMed] [Google Scholar]
- 50.Friedler A., Veprintsev D. B., Hansson L. O., Fersht A. R. (2003) J. Biol. Chem. 278, 24108–24112 [DOI] [PubMed] [Google Scholar]
- 51.Joerger A. C., Fersht A. R. (2007) Oncogene 26, 2226–2242 [DOI] [PubMed] [Google Scholar]
- 52.Joerger A. C., Fersht A. R. (2008) Annu. Rev. Biochem. 77, 557–582 [DOI] [PubMed] [Google Scholar]
- 53.Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 54.Lo M. C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. (2004) Anal. Biochem. 332, 153–159 [DOI] [PubMed] [Google Scholar]
- 55.Stephen C. W., Helminen P., Lane D. P. (1995) J. Mol. Biol. 248, 58–78 [DOI] [PubMed] [Google Scholar]
- 56.Gannon J. V., Greaves R., Iggo R., Lane D. P. (1990) EMBO J. 9, 1595–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dornan D., Shimizu H., Burch L., Smith A. J., Hupp T. R. (2003) Mol. Cell. Biol. 23, 8846–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersson S., Kelleher M., Pion E., Wallace M., Ball K. L. (2009) Biochem. J. 418, 575–585 [DOI] [PubMed] [Google Scholar]
- 60.Lin S. R., Yang Y. C., Jung J. H., Tsai J. H. (1996) DNA Cell Biol. 15, 793–803 [DOI] [PubMed] [Google Scholar]
- 61.Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y., Dey R., Chen L. (2010) Structure 18, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klotzsche O., Etzrodt D., Hohenberg H., Bohn W., Deppert W. (1998) Oncogene 16, 3423–3434 [DOI] [PubMed] [Google Scholar]
- 64.Brosh R., Rotter V. (2009) Nat. Rev. Ca. 9, 701–713 [DOI] [PubMed] [Google Scholar]
- 65.Kim E., Deppert W. (2004) J. Cell. Biochem. 93, 878–886 [DOI] [PubMed] [Google Scholar]
- 66.Pan Y., Haines D. S. (2000) Oncogene 19, 3095–3100 [DOI] [PubMed] [Google Scholar]
- 67.Sampath J., Sun D., Kidd V. J., Grenet J., Gandhi A., Shapiro L. H., Wang Q., Zambetti G. P., Schuetz J. D. (2001) J. Biol. Chem. 276, 39359–39367 [DOI] [PubMed] [Google Scholar]
- 68.Nikolova P. V., Henckel J., Lane D. P., Fersht A. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14675–14680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zúñiga A., Torres J., Ubeda J., Pulido R. (1999) J. Biol. Chem. 274, 21900–21907 [DOI] [PubMed] [Google Scholar]
- 70.Díaz-Moreno I., Hollingworth D., Frenkiel T. A., Kelly G., Martin S., Howell S., García-Mayoral M., Gherzi R., Briata P., Ramos A. (2009) Nat. Struct. Mol. Biol. 16, 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huggins D. J., McKenzie G. J., Robinson D. D., Narváez A. J., Hardwick B., Roberts-Thomson M., Venkitaraman A. R., Grant G. H., Payne M. C. (2010) PLoS Comput. Biol. Vol. 6, pii:el000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown C. J., Lain S., Verma C. S., Fersht A. R., Lane D. P. (2009) Nat. Rev. Cancer 9, 862–873 [DOI] [PubMed] [Google Scholar]
- 73.Saxena M., Williams S., Taskén K., Mustelin T. (1999) Nat. Cell Biol. 1, 305–311 [DOI] [PubMed] [Google Scholar]