Abstract
The filoviruses, Ebola virus (EBOV) and Marburg virus (MARV), cause frequently lethal viral hemorrhagic fever. These infections induce potent cytokine production, yet these host responses fail to prevent systemic virus replication. Consistent with this, filoviruses have been found to encode proteins VP35 and VP24 that block host interferon (IFN)-α/β production and inhibit signaling downstream of the IFN-α/β and the IFN-γ receptors, respectively. VP35, which is a component of the viral nucleocapsid complex and plays an essential role in viral RNA synthesis, acts as a pseudosubstrate for the cellular kinases IKK-ɛ and TBK-1, which phosphorylate and activate interferon regulatory factor 3 (IRF-3) and interferon regulatory factor 7 (IRF-7). VP35 also promotes SUMOylation of IRF-7, repressing IFN gene transcription. In addition, VP35 is a dsRNA-binding protein, and mutations that disrupt dsRNA binding impair VP35 IFN-antagonist activity while leaving its RNA replication functions intact. The phenotypes of recombinant EBOV bearing mutant VP35s unable to inhibit IFN-α/β demonstrate that VP35 IFN-antagonist activity is critical for full virulence of these lethal pathogens. The structure of the VP35 dsRNA-binding domain, which has recently become available, is expected to provide insight into how VP35 IFN-antagonist and dsRNA-binding functions are related. The EBOV VP24 protein inhibits IFN signaling through an interaction with select host cell karyopherin-α proteins, preventing the nuclear import of otherwise activated STAT1. It remains to be determined to what extent VP24 may also modulate the nuclear import of other host cell factors and to what extent this may influence the outcome of infection. Notably, the Marburg virus VP24 protein does not detectably block STAT1 nuclear import, and, unlike EBOV, MARV infection inhibits STAT1 and STAT2 phosphorylation. Thus, despite their similarities, there are fundamental differences by which these deadly viruses counteract the IFN system. It will be of interest to determine how these differences influence pathogenesis.
Introduction
Filoviruses, marburg virus (MARV) and Ebola viruses (EBOV), are enveloped negative-sense RNA viruses associated with zoonotic infections in humans. Several species of EBOV have been identified, including Zaire, Sudan, Ivory Coast, Bundibugyo, and Reston. Only 1 species of MARV has been identified. These pathogens are notable because of their propensity to cause lethal disease in humans and non-human primates. Some outbreaks in humans of Zaire EBOV have been associated with fatality rates near 90%, while Sudan EBOV epidemics have exhibited fatality rates around 50%. Notably, the few documented cases of human infection with Reston EBOV have not been associated with illness or death, suggesting that this virus may be attenuated in humans. Filovirus illness in humans typically has an abrupt onset characterized by fever, myalgias, headache, and gastrointestinal symptoms including nausea, vomiting, and diarrhea (reviewed in Geisbert and Hensley (2004)). Many patients also develop a maculopapular rash, and changes in coagulation are common, although bleeding is not seen in all patients and is not thought, in most cases, to cause patient deaths. Fatal outcome correlates with increasing viremia over time as the infection leads to shock, convulsions, and disseminated intravascular coagulation (Geisbert and Hensley 2004).
Fatal filovirus disease is characterized by a failure of the infected host to clear the infection, apparently dysregulated host inflammation, activation of coagulation cascades, and lymphocyte apoptosis (reviewed in Geisbert and Hensley (2004); Bray and Geisbert (2005)). Therefore, it is critical to develop a greater understanding as to how these viruses trigger host innate immune responses without triggering effective antiviral responses. This review will focus upon cellular responses to filovirus infection focusing, in particular, on how these viruses modulate production of and cellular responses to interferons (IFNs). Suppression of the innate antiviral responses, in particular the IFN-α/β response, is likely to promote the sustained systemic replication of these viruses, thereby promoting viral disease. While the mechanisms by which these viruses evade IFN responses has come into focus, the viral and host factors that trigger the inflammatory and coagulation responses associated with lethal infection remain to be defined.
Cellular Responses to Filovirus Infection
A number of studies have addressed the impact of filovirus infection upon cellular responses to infection. For example, a micorarray study examined gene expression changes in Zaire EBOV, Reston EBOV, and MARV-infected Huh-7 cells (Kash and others 2006). These cells were chosen because hepatocytes are an important site of replication in vivo (Geisbert and Hensley 2004). This study found that infection generally resulted in the differential regulation of genes involved in immune response, IFN response, coagulation, and acute phase response. Notably, gene expression profiles from Zaire EBOV- and MARV-infected cells were more similar than were the profiles of Zaire EBOV- versus Reston EBOV-infected cells or MARV- versus Reston EBOV-infected cells. Zaire EBOV and MARV activated fewer IFN-inducible genes, relative to Reston EBOV, and Zaire EBOV and MARV inhibited cellular responses to exogenously added IFN-α to a greater extent than did Reston EBOV (Kash and others 2006). These studies suggest that Reston EBOV, which is generally thought to be attenuated in humans relative to other filoviruses, may have a diminished capacity to counter IFN-α/β responses relative to other filoviruses. Whether this difference reflects a decreased capacity of Reston EBOV gene products to block cellular responses to infection is not yet clear. An alternate hypothesis would be that Reston EBOV exhibits reduced replication kinetics, such that factors suppressing IFN responses might be produced at lower efficiency in EBOV Reston infections than in other filovirus infections. Although different viruses exhibited variable levels of IFN inhibition, these studies do support the hypothesis that different filoviruses influence cellular signaling pathways in similar but non-identical ways. They also suggest that the ability of filoviruses to modulate innate immune response pathways, particularly IFN-related pathways, may influence pathogenesis.
A separate study compared responses of a different human liver cell line, HepG2, to infection with wild-type Zaire EBOV versus a virus with a point mutation in VP35, the viral suppressor of IFN-α/β production (discussed in detail later) (Hartman and others 2008). This study differed from a number of earlier studies in that purified virus, rather than unpurified virus preparations that might contain products released from infected cells, was used for the infections. The purified wild-type virus induced in the HepG2 cells far fewer gene expression changes than had been seen in earlier studies of wild-type virus infection. Interestingly, the virus with the mutant VP35 induced a more vigorous activation of IFN-α/β-responsive genes than wild-type virus. Surprisingly, this IFN response in mutant-infected cells was not apparent in the gene expression profile at 24 h postinfection but was dramatically apparent at 48 h postinfection. This is despite the fact that intracellular levels of viral RNA copy number only increased a further 3-fold between 24 and 48 h postinfection (Hartman and others 2008). Since viral RNA replication products are presumably the triggers of IFN-α/β production in infected cells (Habjan and others 2008), it will be of interest to determine why the cellular IFN response seems to lag relative to the time viral RNAs accumulate in the VP35 mutant-infected cells. The relative quiescence of cells infected with purified preparations of wild-type virus is also of interest. It raises the possibility that many of the changes attributed to EBOV infection in earlier studies might not reflect responses to infection itself, but rather may reflect responses to other substances (eg, cytokines) in the inocula used. As proinflammatory responses appear to play such a critical role during in vivo filovirus infection, this will be an important issue to resolve. This study does, however, support the fact that EBOVs can effectively inhibit host IFN-α/β responses and that VP35 contributes to this capability.
Macrophages and dendritic cells (DCs) are thought to play critical roles in EBOV pathogenesis. This conclusion is partly based upon the observation that these cell types are infected in vivo, and upon the observation that in vitro EBOV productively infects these cells and induces several phenomena that correlate with processes seen as Ebola hemorrhagic fever progresses (Feldmann and others 1996; Geisbert and others 2000; Hensley and others 2002; Bosio and others 2003; Geisbert and others 2003c; Mahanty and others 2003). Among these, in vitro infection of monocytes or macrophages induces proinflammatory cytokine production, promotes endothelial leakage, induces tissue factor expression, and stimulates bystander apoptosis of lymphocytes (Feldmann and others 1996; Geisbert and others 2000; Hensley and others 2002; Geisbert and others 2003b, 2003c). Based on these in vitro observations and upon studies of in vivo infections, models have been developed hypothesizing that macrophages are central to EBOV pathogenesis (reviewed in Bray and Geisbert (2005)). In these models, factors secreted by infected macrophages will attract to sites of infection additional monocytes and macrophages, thus promoting infection of new cells. Macrophage cytokines are also proposed to influence, by a variety of mechanisms, endothelial permeability, ultimately promoting circulatory collapse (Bray and Geisbert 2005). Infection also promotes, in vivo and in vitro, expression on macrophages of cell surface tissue factor, a type II cytokine receptor that interacts with factors VIIa and X in the circulation to trigger coagulation. Activation of coagulation cascades promotes the hemorrhagic manifestations of EBOV infection, but could also exert additional pleiotropic effects (Geisbert and others 2003c; Geisbert and Hensley 2004). Notably, tissue factor, as a cytokine-like receptor, can signal intracellularly, and activation of coagulation cascades triggers signaling through protease-activated receptors (PARs) (Ruf 2004). The consequences of such signaling for EBOV pathogenesis remains uncertain, but therapeutic interventions designed to interrupt the coagulopathy that occurs in infected monkeys do have a clinical benefit (Geisbert and others 2003a; Hensley and others 2007). Given the prominence of coagulation in primate EBOV infection, a recent study is of note (Niessen and others 2008). This study demonstrates a critical role for DC PAR signaling in sustaining systemic inflammation during mouse models of sepsis, a syndrome (like viral hemorrhagic fever) where coagulation and systemic inflammation appear to be coupled (Niessen and others 2008).
The available data indicate that infection of monocytes or macrophages with live EBOV or MARV results in cytokine and chemokine production as early as 3–6 h postinfection (Gupta and others 2001; Stroher and others 2001; Hensley and others 2002); and inactivated (UV- or γ-irradiated) viruses also induce cytokines/chemokines (Stroher and others 2001; Hensley and others 2002). In the inactivation experiments, the inactivation was sufficient to prevent detectable virus protein expression (Stroher and others 2001). Therefore, productive virus replication is not required for the monocyte and macrophage proinflammatory responses. However, sustained cytokine production (over the course of 48 h and longer) only occurs with live virus infection, suggesting that this response requires sustained virus replication (Gupta and others 2001; Hensley and others 2002). Interestingly, most in vitro studies of EBOV-infected monocytes, macrophages, or peripheral blood mononuclear cells (PBMC) report little IFN-α/β response in the infected cultures (Gupta and others 2001; Stroher and others 2001). In the 1 study in which significant IFN-α was produced, viral titers continued to increase significantly even after IFN-α was detected (Hensley and others 2002). The latter observation is consistent with the presence in EBOV of mechanisms to evade the antiviral effects of IFN-α.
Infection in vitro of monocyte-derived DCs with Zaire EBOV yields a somewhat different picture than that seen in macrophages. In DCs, Zaire EBOV infection does not result in full activation of the cells, despite productive infection of DCs (Bosio and others 2003; Mahanty and others 2003). In 1 study, infected monocyte-derived DCs did not secrete proinflammatory cytokines, did not up-regulate co-stimulatory molecules, and poorly stimulated T cells (Mahanty and others 2003). In a second study, monocyte-derived DCs underwent an “aberrant maturation” characterized by an absence of cytokine secretion, increased CD40 and CD80 levels but little increase in HLA-DR and CD86, and unchanged CD11c, CD83, and CCR5 levels. Infection also suppressed allogeneic T-cell stimulation as measured by [3H]-thymidine incorporation (Bosio and others 2003). Interestingly, the latter study also demonstrated that infection suppresses DC ability to produce IFN-α (Bosio and others 2003). Consistent with these in vitro studies, in infected nonhuman primates, DCs did not appear to be activated, as evidenced by unchanged levels of CD80 and CD86 levels in overall leukocyte populations between days 1 and 3 postinfection (Reed and others 2004). Based on these observations, it has been argued that EBOV suppression of DC function prevents initiation of adaptive immune responses and facilitates uncontrolled, systemic virus replication (Bray and Geisbert 2005). Why EBOV should trigger cytokine secretion by monocytes and macrophages but fail to induce DC cytokine secretion and maturation is not clear but clearly deserves further study. Given the observation that purified virus induces much reduced responses in HepG2 cells, it will be of interest to compare monocyte and macrophages responses to purified versus unpurified virus. It will also be of interest, given that most studies of monocyte and macrophage infection have not carefully monitored efficiencies of infection, to determine to what extent cytokine production represents a direct response to infection versus a bystander effect in which infected cells signal to uninfected cells such that the latter produce cytokines. Finally, it will be of significant interest to determine to what extent EBOV-encoded inhibitors of IFN responses may modulate monocyte, macrophage, and DC responses to infection.
Filoviruses Evade IFN Responses
Consistent with the microarray studies cited earlier, Zaire EBOV infection renders human umbilical vein endothelial cells, PBMCs, and human monocytes-derived macrophages refractory to polyI:polyC-induced IFN-α production (Harcourt and others 1998; Gupta and others 2001). For example, EBOV infection of human PBMC and macrophages elicits proinflammatory chemokines and cytokines but does not elicit measurable IFN-α or IFN-β until 3 days postinfection, and infection suppressed IFN-α/β production induced by poly(I:C) (Gupta and others 2001). Zaire EBOV infection also inhibits responses of HUVECs to IFNs (Harcourt and others 1998). Infected cells exhibit reduced formation of IFN-α/β- or IFN-γ-induced transcription factor complexes, as assessed by electrophoretic mobility shift assays and reduced the IFN-α- or IFN-γ-induced expression of IRF-1 and 2′,5′-oligoadenylate synthetase (Harcourt and others 1999). This inhibition was IFN-specific as infection did not prevent formation of IL-1β -induced NF-κB transcription factor complexes or IL-1β-induced gene expression (Harcourt and others 1999).
The VP35 Protein Inhibits IFN-α/β Production
To date, 1 EBOV protein, VP35 has been shown to inhibit IFN-α/β production. Expression of VP35 was initially found to functionally substitute, in trans, for the influenza virus NS1 protein, a protein known to inhibit the IFN-α/β response to infection (Basler and others 2000). Specifically, when VP35 was expressed in 293T cells, it could complement the growth defect of an influenza virus that lacked the NS1 protein, whereas expression of other EBOV proteins did not rescue growth of this mutant influenza virus (Basler and others 2000; Garcia-Sastre and others 1998). Consistent with the interpretation that VP35 complemented growth of the mutant influenza virus by suppressing cellular IFN responses, other viral inhibitors of IFN responses, including the influenza A virus NS1 protein, the herpes simplex virus type 1 γ134.5 protein, and the vaccinia virus E3L protein, also enhanced mutant influenza virus growth (Basler and others 2000). The ability of VP35 to substitute for NS1 correlated with its ability to suppress production of IFN-β mRNA and to inhibit activation of the IFN-β promoter or the ISG54 promoter induced by transfected polyI:polyC, by mutant influenza virus infection, or by Sendai virus infection (Basler and others 2000). Consistent with its ability to inhibit activation of both the IFN-β promoter and the ISG54 promoter, VP35 can prevent the activation of interferon regulatory factor 3 (IRF-3) (Basler and others 2003). VP35 expression inhibited the Sendai virus-induced nuclear accumulation of an IRF-3–GFP fusion protein, inhibited the Sendai virus-induced dimerization of IRF-3, and blocked the virus-induced hyperphosphorylation of IRF-3 that triggers its activation (Basler and others 2003; Cardenas and others 2006). These observations, made upon overexpression of VP35, are consistent with data obtained from EBOV-infected cells. For example, little IRF-3 nuclear accumulation was detected in WT Zaire EBOV-infected cells (Basler and others 2003; Hartman and others 2006). However, infection of cells with a Zaire EBOV that encodes a mutated VP35 resulted in enhanced nuclear accumulation of IRF-3, when this was measured 24 h postinfection (Hartman and others 2006).
Inhibition of the RIG-I Pathway
The cellular RNA helicases RIG-I and MDA-5 are intra-cellular sensors of virus infection that can trigger IFN-α/β production. Several negative-strand RNA viruses, including influenza virus, rabies virus, and vesicular stomatitis virus, trigger IFN-α/β induction via RIG-I (Hornung and others 2006; Kato and others 2006; Pichlmair and others 2006). That filoviruses will also trigger IFN-α/β by a RIG-I pathway is suggested by the fact that purified Zaire EBOV RNA activated the IFN-β promoter in a RIG-I-dependent manner upon transfection into 293T cells (Habjan and others 2008). VP35 is able to block RIG-I-induced expression of IFN-α/β (Cardenas and others 2006). When RIG-I was overexpressed in 293T cells, IFN-β activation could be seen; but coexpression of VP35 blocked this activation. Similarly, when MAVS, IKK-ɛ, or TBK-1 were overexpressed, IFN-β promoter activation was detected, but coexpression of VP35 was sufficient to impair this activation (Cardenas and others 2006). The inhibition of IFN-β promoter activation induced by RIG-I correlated with the ability of VP35 to impair IRF-3 phosphorylation and to inhibit production of IFN-β from the endogenous cellular genes (Cardenas and others 2006). Because VP35 does not detectably inhibit induction of gene expression by a constitutively active IRF-3 mutant (IRF-35D), a mutant which is active without exogenous inducers, these data strongly suggest that VP35 specifically inhibits the phosphorylation of IRF-3 by IKK-ɛ and/or TBK-1 (Basler and others 2003).
VP35 Interacts With the Kinase Domain of IKK-ɛ and TBK-1 to Block IRF-3/IRF-7 Activation
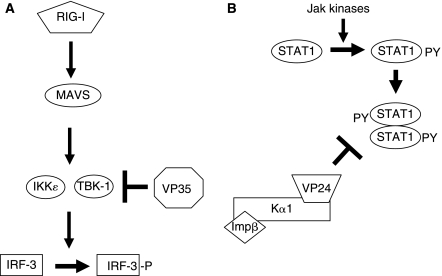
Recent data demonstrates that VP35 can serve as an alternative substrate for IKK-ɛ/TBK-1, impairing the ability of these kinases to interact with IRF substrates (Fig. 1A). Co-immunoprecipitation experiments demonstrated an interaction of VP35 with each kinase; and the N-terminal kinase domains, which are relatively homologous to one another, were sufficient to interact with VP35. In vitro kinase reactions demonstrated that recombinant IKK-ɛ purified from transfected 293T cells is able to phosphorylate VP35 in vitro, whereas an irrelevant control protein, glutathione S-transferase, was not phosphorylated by IKK-ɛ (Prins and others 2009). Mass spectroscopy on VP35 purified from both the in vitro kinase assays and cells in which both VP35 and IKK-ɛ were overexpressed has identified several VP35 serines apparently phosphorylated by IKK-ɛ (unpublished observation). It remains to be determined whether this phosphorylation modulates VP35 function. In addition to resulting in its phosphorylation, VP35 interaction with IKK-ɛ or TBK-1 can disrupt their ability to interact with IRF-3 or IRF-7. This was demonstrated by performing co-immunoprecipitation experiments with either IKK-ɛ or TBK-1. It was found that IRF-3, like VP35, interacts with the kinase domain of IKK-ɛ; and VP35 can impair the interaction of both IRF-3 and IRF-7 with IKK-ɛ and TBK-1 (Prins and others 2009). In support of this model, when in vitro kinase assays were performed using lysates of cells overexpressing IKK-ɛ as a source of kinase and a GST-IRF-3 construct as substrate, the presence of VP35 results in decreased IRF-3 phosphorylation (Prins and others 2009). IKK-ɛ not only interacts with IRF-3 and IRF-7 but also with the upstream signaling molecule MAVS (Hiscott and others 2006). It is therefore of interest that expression of high levels of VP35 also impaired IKK-ɛ–MAVS interaction (Prins and others 2009). In sum, these data support a model in which VP35 interacts with IKK-ɛ and TBK-1 through their kinase domains resulting in impaired interaction with IRF-3 and IRF-7. This model does not, however, exclude other mechanisms by which VP35 might also suppress IFN-α/β production. This model also does not exclude the possibility that VP35 will affect additional signaling pathways. Because VP35 interacts with the kinase domains of IKK-ɛ and TBK-1, it will be of interest to determine whether it may also interact with the NF-κB-activating IKK-α and IKK-β, the kinase domains of which share homology with IKK-ɛ and TBK-1.
FIG. 1.
Function of Ebola virus (EBOV)-encoded IFN-antagonist proteins. (A) EBOV VP35 inhibits IFN-α/β production by interacting with the IRF-3 kinases IKK-ɛ and TBK-1. This prevents efficient phosphorylation of IRF-3, blocking activation of the IFN-β promoter. (B) EBOV VP24 prevents nuclear accumulation of activated, tyrosine-phosphorylated STAT1 through an interaction with NPI-1 family karyopherin-α proteins, which otherwise import dimerized STAT1 into the nucleus.
VP35 Recruits Host SUMO Modification Machinery to Inhibit IRF-7 Function
Recently, VP35 has also been found to interact with IRF-7, Ubc9, and PIAS1 (Chang and others 2009). Ubc9 and PIAS1 are E2 and E3 SUMO ligases, respectively. They, in cooperation with an E1 protein, covalently conjugate SUMO proteins to target proteins, including many cellular transcription factors. Virus-induced SUMOylation of IRF-3 and IRF-7 has been found to contribute to negative feedback regulation of IFN-α/β production (Kubota and others 2008). VP35 was found to promote IRF-7 SUMOylation of IRF-3 and IRF-7, thus impairing IFN-β gene transcription (Chang and others 2009). It will be of interest to define the relative contributions of IRF-3/-7 SUMOylation versus IKK-ɛ/TBK-1 inhibition versus dsRNA sequestration to inhibition of IFN-α/β production in EBOV-infected cells. It will also be of interest to determine whether the interaction with PIAS1 and Ubc9 influences other functions of VP35.
Other Functions of VP35
VP35 also inhibits the activation of the dsRNA-activated protein kinase (PKR), an IFN-induced, dsRNA-activated, cellular serine/threonine kinase. PKR has long been known to inhibit virus replication by impairing translation by phosphorylating the α-subunit of the translation initiation factor eIF-2 (Gale and Katze 1998). Virus-encoded inhibitors of PKR, including the adenovirus VA1 RNA, the vaccinia virus E3L protein, and the influenza A virus NS1 protein, were among the first viral products found to suppress the antiviral effects of IFNs (Kitajewski and others 1986; Chang and others 1992; Lu and others 1995).
VP35 was shown to inhibit PKR using a recombinant mutant herpes simplex virus 1 (HSV-1) that lacked the γ134.5 gene. γ134.5 encodes a protein that recruits protein phosphatase 1, a cellular protein, to dephosphorylate eIF-2α; HSV-1 mutants lacking this function display increased sensitivity to the antiviral effects of IFN. The presence of VP35 preserved the protein expression and replication of the γ134.5 deletion virus following treatment of the cells with IFN-α, whereas a parental γ134.5 null virus lacking VP35 was sensitive to the antiviral effects of IFN-α (Feng and others 2007). Restoration of virus replication by VP35 correlated with suppression of PKR activation and reduced eIF-2α phosphorylation. Interestingly in this study, an R312A VP35 mutant, which lacks dsRNA-binding activity (Cardenas and others 2006; Feng and others 2007), still inhibited PKR. In contrast, another study suggests that dsRNA binding mutants of VP35 cannot block PKR activation (Schümann and others 2009).
VP35 can also suppress, as can NS1 and E3L, RNA silencing (Haasnoot and others 2007). The function is dependent upon VP35 dsRNA-binding activity (Haasnoot and others 2007). RNA silencing serves an antiviral function in plants and insects, and viruses that infect plants and insects often encode suppressors of RNA silencing (reviewed in Voinnet (2005)). The gene products of several mammalian viruses can also inhibit RNA silencing, including the adenovirus VAI RNA, the influenza virus NS1 protein, the vaccinia virus E3L protein, and the HIV Tat protein (Bucher and others 2004; Li and others 2004; Lu and Cullen 2004; Bennasser and others 2005). Likewise, wild-type VP35 impaired knockdown of a transfected luciferase reporter gene by a short-hairpin RNA, but dsRNA-binding-defective VP35 mutants did not exert this inhibitory activity (Haasnoot and others 2007). Additionally, VP35 could complement a Tat-minus HIV-1 mutant (Haasnoot and others 2007). However, although the possibility that RNA silencing serves an antiviral function in mammalian cells remains an intriguing possibility, there is as yet no definitive demonstration that this is the case (Voinnet 2005). Therefore, the relevance of this function for EBOV infection requires further inquiry.
Biochemical and Structural Studies of VP35
Recently, structural and biochemical studies have begun to shed light on how VP35 interacts with dsRNA and suppresses IFN-α/β production. VP35 contains an N-terminal oligomerization domain and a C-terminal dsRNA-binding domain, each of which appear to be critical for maximal inhibition of IFN-α/β production (Hartman and others 2004; Reid and others 2005; Cardenas and others 2006; Hartman and others 2006; Hartman and others 2008) (Fig. 2A). Mutations designed to disrupt a predicted coiled-coil domain within the N-terminus of the MARV VP35 impair its ability to function as a component of the viral RNA polymerase complex, and similar mutations in the N-terminal region of Zaire EBOV VP35 diminishes the ability of VP35 to antagonize the IRF-3 pathway and to inhibit transcription of IFN genes (Hartman and others 2004; Hartman and others 2006; Hartman and others 2008). However, tethering of an exogenous oligomerization domain with the VP35 C-terminus partially restores IFN inhibition (Reid and others 2005), suggesting that the IFN inhibitory domain (IID) is located near the C-terminus of VP35 (Moller and others 2005; Reid and others 2005) (Fig. 2A). Further supporting a critical role for the VP35 C-terminus in suppression of IFN production, deletion of the C-terminal 40 residues of VP35 eliminates IFN inhibition and leads to loss of dsRNA binding (Hartman and others 2004). The structure of the EBOV VP35 IID was recently solved and the structure reveals that the VP35 IID forms a unique fold that binds dsRNA (Fig. 2B). Comparison of the VP35 IID structure with other IFN antagonist such as influenza NS1A/NS1B revealed that their structures are distinct from one another (Leung and others 2009) (Fig. 2).
FIG. 2.
Structure of the VP35 interferon inhibitory domain (IID). (A) Representation of the full-length VP35 with the IID indicated by the box. The α-helical and β-sheet subdomains are indicated by pink and green coloring, respectively. (B) Ribbon representation of the IID. (C and D) Electrostatic surface representations of the IID. Red, white, and blue represent negative, neutral, and positive electrostatic potential.
The structure of EBOV VP35 IID consists of 2 subdomains, both of which are required to maintain the overall fold and therefore required for function. The α-helical subdomain contains a 4 helix bundle that spans ∼70 residues, where the helices are arranged in an antiparallel manner. The β-sheet subdomain contains a 4-stranded mixed β-sheet, an α-helix, and a left-handed type II polyproline helix (Fig. 2B). The inter-subdomain interface is largely formed by residues from helices 2 and 4 and β-sheets 3 and 4, and the polyproline region. The residues within these structural elements are nearly identical in all Ebola VP35 isolates sequenced thus far (Leung and others 2009). Previous studies have shown that mutation of basic residues R305, K309, and R312 lead to loss of dsRNA binding and IFN inhibition (Hartman and others 2004; Cardenas and others 2006; Hartman and others 2006). These residues were initially identified through a sequence similarity match with influenza NS1A protein during a bioinformatics search of viral antagonists of host IFN system (Hartman and others 2004). In the structure, these basic residues form an extended basic patch, which contains other highly conserved basic residues K319, R322, K339 (see Fig. 2C and 2D). Unlike the sequences surrounding residues R305, K309, and R312, the sequences near these additional basic residues do not show sequence similarities to influenza NS1 or other viral antagonists (Leung and others 2009). But, all these basic residues are important for dsRNA binding suggesting that interactions between dsRNA and these basic residues are important for VP35-mediated functions (Leung and others 2009). Together, these data suggest that VP35 likely targets multiple host elements through dsRNA-dependent mechanisms, which are mediated by a cluster of basic residues centered on R312 in EBOV VP35 IID.
The availability of a high-resolution structure coupled with multiple functions performed by Ebola VP35 IID provides an opportunity for structure-based antiviral design strategies. In particular, examination of the structure suggests that there are several surfaces in the VP35 IID that form cavities, which can be exploited as drug-binding sites. Moreover, mutation of non-basic residues in these interfaces also leads to loss of structure and function (GKA, DWL, CFB, unpublished observations), suggesting that these residues and areas surrounding them can also be targeted by inhibitors. Interestingly, the C-terminal region of Zaire Ebola VP35 shows a very high degree of sequence similarity with other EBOV (eg, 88% identical and 95% similar, at the amino acid level to the Reston VP35 C-terminus). Therefore, compounds identified through potential in silico and in vitro screening processes are likely to identify compounds that could potentially function as pan-filo antivirals.
While the structure of the EBOV VP35 IID has provided a wealth of structural information and identified several key residues important for dsRNA-dependent functions (eg, central basic patch), it also raises a series of yet unanswered questions. For example, the C-terminal IID is clearly monomeric in solution, but forms an oligomer in the full-length form through the N-terminus. Moreover, existence of several different oligomerization interfaces during crystallization (eg, structure was a dimer in the crystallographic asymmetric unit) suggests that the C-terminus is likely to support additional interactions in the context of the homo-trimer. It is also unclear if VP35 recognition of dsRNA is sequence- and/or length-dependent and if dsRNA interactions with the oligomeric form of the full-length VP35 molecule can induce additional protein–protein interactions that play important roles in IFN inhibition and in other VP35-mediated functions such as genome replication and nucleocapsid formation. Given the ability of Ebola VP35 to antagonize IRF-3 phosphorylation in a dsRNA-dependent and -independent manner, coupled with its ability to interact with TBK-1 and IKK-ɛ kinases suggests that structural characterization of the full-length protein and complexes with nucleic acids will be necessary to address these issues.
The EBOV VP24 Protein Inhibits IFN-α/β and IFN-γ Signaling
As noted earlier, EBOV infection inhibits not only IFN-α/β production, but also cellular responses to IFN-α/β and IFN-γ (Harcourt and others 1999; Kash and others 2006). A systematic screen of Zaire EBOV proteins identified the EBOV VP24 protein as a capable of preventing gene expression induced by IFN-α/β or IFN-γ (Reid and others 2006). Expression of VP24 was also able to counteract the antiviral effects of IFN-β (Reid and others 2006). This latter function was demonstrated by transfecting Vero cells with expression plasmids, including a VP24 plasmid, treating the transfected cells with IFN-β and then infecting with a Newcastle disease virus encoding GFP (Park and others 2003). While IFN induced an antiviral state, suppressing GFP expression from NDV in empty vector-transfected cells, expression of VP24 rescued NDV replication and GFP expression. Because VP24 expression impaired cellular responses to both IFN-α/β and IFN-γ, further experiments focused on STAT1, a transcription factor activated by both pathways (Reid and others 2006). When IFN-α/β is added to cells, STAT1 and STAT2 typically becomes tyrosine-phosphorylated, resulting in formation of STAT1:STAT2 dimers, via phosphotyrosine–SH2 domain interactions. When IFN-γ is added to cells, STAT1 is the principal STAT to be tyrosine-phosphorylated, resulting in STAT1 homodimers. In either case, STAT dimerization leads to nuclear accumulation of the STAT dimers (reviewed in McBride and Reich (2003)). When VP24 was expressed in cells, addition of IFN resulted in STAT1 phosphorylation at levels comparable to untransfected cells or to cells expressing the VP35 protein (Reid and others 2006). However, phospho-STAT1 failed to accumulate in the nucleus of VP24-expressing cells. In contrast, phospho-STAT1 rapidly accumulated in the nucleus of cells not expressing VP24 (Reid and others 2006). Importantly, EBOV-infected cells exhibited the same phenomenon; STAT1 was tyrosine-phosphorylated in response to IFN-β, but it remained cytoplasmic (Reid and others 2006). Thus, VP24, by virtue of its ability to prevent nuclear accumulation of phospho-STAT1, likely renders EBOV-infected cells resistant to IFNs.
The capacity of VP24 to prevent nuclear accumulation of phospho-STAT1 correlates with its ability to interact with select karyopherin-α (also known as importin-α) proteins (Fig. 1B). Karyopherin-α proteins act as adaptors, which mediate nuclear import of cargo. Karyopherin-αs interact with both the nuclear localization signal (NLS) present on a cargo protein to be imported into the nucleus, and with karyopherin-β (Adam and Adam 1994; Imamoto and others 1995). Karyopherin-β mediates docking and subsequent movement through the nuclear pore complex (NPC) (Moroianu and others 1995a, 1995b; Gorlich and others 1996). There are 6 karyopherin-α family members in mammalian cells. One, karyopherin-α1, was previously demonstrated to mediate the nuclear import of phospho-STAT1 (Sekimoto and others 1997; McBride and others 2002; Melen and others 2003). When VP24 was initially tested for the ability to interact with karyopherins-α1, -α2, -α3, and -α4, it interacted specifically with -α1 (Sekimoto and others 1997; McBride and others 2002; Melen and others 2003). Expression in cells of VP24 was further found, in co-immunoprecipitation experiments, to disrupt interaction of phopho-STAT1 with karyopherin-α1 (Reid and others 2006).
The 6 members of the karyopherin-αs family can be classified, based upon sequence similarity, into 3 distinct subfamilies (Kohler and others 1997): the Rch1 subfamily (karyopherin-α2) (Cuomo and others 1994; Weis and others 1995), the Qip1 subfamily (karyopherin-α3 and karyopherin-α4) (Kohler and others 1997; Miyamoto and others 1997; Seki and others 1997; Nachury and others 1998), and the nucleoprotein interactor 1 (NPI-1) subfamily (karyopherin-α1, karyophrein-α5, and karyopherin-α6) (Cortes and others 1994; Moroianu and others 1995a; O'Neill and Palese 1995; Kohler and others 1997; Kohler and others 1999). Of particular interest is the NPI-1 subfamily, which was so named because of the founding member, karyopherin-α1, was found to interact with and mediate the nuclear import of the influenza virus nucleoprotein (O'Neill and Palese 1995; Cros and others 2005). Within the NPI-1 subfamily, karyopherin-α1 shares greater than 80% sequence similarity with karyopherin-α5 and karyopherin-α6 (Melen and others 2003). Further experimentation with VP24 demonstrated that it can interact not only with karyopherin-α1 but also with the other members of the NPI-1 group, karyopherins-α5 and -α6. All 3 NPI-1 family members were also able to co-immunoprecipitate with phospho-STAT1 (Reid and others 2007). Because a separate study also identified an interaction between karyopherin-α6 and activated STAT1 (Ma and Cao 2006), it seems likely that all 3 NPI-1 family members can mediate nuclear import of activated STAT1. As was the case with karyopherin-α1, VP24 prevented interaction of karyopherins-α5 and -α6 with phospho-STAT1 (Reid and others 2007).
The basis by which VP24 inhibits karyopherin-α–phospho-STAT1 interaction has also been addressed. Karyopherin-α proteins possess 10 armadillo (arm) repeats located in the center of the protein (reviewed in Goldfarb and others (2004)). These are flanked by an N-terminal karyopherin-β-binding domain (importin-β-binding domain) and a C-terminal domain. Toward the C-terminus, the tenth ARM domain binds the cellular apoptosis susceptibility (CAS), exportin an interaction that mediates the nuclear export of karyopherin-αs. Structural studies indicate that karyopherin-α can accommodate 2 monopartite basic NLSs or 1 bipartite basic NLS, via interaction with arm repeats 2–4 or 7–9 (Goldfarb and others 2004). However, the atypical STAT1 NLS interacts with a C-terminal region of karyopherin-α1 that is distinct from the more central region recognized by typical multibasic amino acid mono- and bipartite NLSs (McBride and Reich 2003). Specifically, previous studies defined karyopherin-α1 residues 425–538 as critical for phospho-STAT1 binding and, within this region arm repeats 8 and 9 were found to be important (Sekimoto and others 1997; Melen and others 2003). Mapping studies identified a similar region of karyopherin-α as required for VP24 interaction (Reid and others 2007). Specifically, the requirement for karyopherin-α1 residues 425–538 for STAT1 interaction was confirmed in co-immunoprecipitation experiments. VP24 interaction was found to require karyopherin-α1 residues 458–504. Thus, the VP24-binding site appears to lie within the STAT1-binding site on karyopherin-α1 (Reid and others 2007). One possible mechanism by which VP24 may inhibit STAT1 activation would be for VP24 to bind to the STAT1-binding site on karyopherin-α, effectively competing with STAT1. The available data is consistent with such a model. Transfection of cells with increasing amounts of VP24 expression plasmid resulted in a dose-dependent decrease in detectable karyopherin-α-phospho-STAT1 interaction; and in vitro-binding experiments demonstrated a similar ability of VP24 to impair karyopherin-α binding to phospho-STAT1 (Reid and others 2007). However, other mechanisms, for example an overall disruption of normal karyopherin-α folding that leads to a loss of phospho-STAT1 binding cannot be excluded. It also remains unclear whether inhibition of STAT1–karyopherin interaction is the sole mechanism by which VP24 inhibits STAT1 nuclear import. It is possible, for example, that VP24 may also disrupt normal trafficking of the NPI-1 karyopherin-αs.
Defining the mechanism by which VP24 affects nuclear import will further our understanding of its full impact on the host cell. If VP24 alters trafficking of the NPI-1 karyopherin-αs, then this might affect nuclear trafficking of numerous cellular factors. Because VP24 interacts with the C-terminal region of karyopherin-αs, while basic NLSs interact with the central regions of karyopherin-αs, VP24's impact on the nuclear import of other proteins is not certain. Nor it is clear whether or not VP24 can influence interaction of karyopherin as with other host cell proteins, such as importin-β1. In addition to clarifying the mechanisms by which EBOVs modulate IFN signaling, a fuller characterization of the interplay between VP24, karyopherin-αs, and cellular proteins may also shed light on fundamental issues regarding nuclear import. Although a number of examples have been described, there is a paucity of data, in general, on the identities of proteins that undergo nuclear import by select karyopherin-α family members. However, it is clear from studies on Drosophila development that altering karyopherin-α expression is important for neural differentiation (reviewed in Terry and others (2007)). Thus, selective disruption of select nuclear import pathways by VP24 might have broad effects upon the host cell. Studies also suggest that selective interaction of basic NLSs with different karyopherin-αs is determined both by the sequence of the NLS as well as its context in the cargo protein, but study of this issue remains limited (Friedrich et al. MCB). There are very few examples of cargo proteins that interact with the C-terminal region of karyopherin-αs such as is seen with activated STAT1 and VP24. Identifying proteins whose interaction with the NPI-1 karyopherin-αs is disrupted by VP24 might identify proteins that interact with the karyopherin-α C-terminus and might suggest a functional significance for this mode of nuclear import.
Inhibition of IFN Signaling by Marburg Virus
As noted earlier, a study by the Katze and Mühlberger laboratories suggested that filoviruses suppress cellular antiviral responses, with Zaire EBOV and MARV better suppressing expression of host cell antiviral genes than Reston EBOV, a virus thought to be less virulent in humans (Kash and others 2006). This study also identified a fundamental difference in the way EBOV and the related MARV counteract IFN signaling (Kash and others 2006). When infected cells were treated with IFN-α, all viruses suppressed cellular responses relative to IFN-α-treated, uninfected cells. However, Zaire EBOV did not prevent STAT1 or STAT2 phosphorylation, consistent with the model of EBOV VP24 function described earlier. However, in MARV-infected cells to which IFN-β was added, phosphorylation of both STAT1 and STAT2 was suppressed, suggesting that MARV encodes an alternate means of suppressing IFN signaling (Kash and others 2006). Therefore, it will be important to define how MARV blocks STAT1 and STAT2 activation in response to IFNs and to determine how the different mechanisms may influence the outcome of EBOV versus MARV infections.
Conclusions and Future Directions
Ebola and MARV are among the most deadly viruses described. Their extreme virulence likely stems from the fact that they are zoonotic pathogens that have not adapted to primate hosts. In apparent consequence, these viruses “accidentally” trigger robust innate responses that, in most cases, fail to clear or effectively control the infection. The result is an infection and accompanying host response that frequently proves fatal. The capacity of these viruses to suppress IFN responses appears to be critical for these unrelenting infections to become established and maintained. The studies described earlier provide insight into the mechanisms by which filovirus IFN-antagonist proteins function. Future studies should also take advantage of the data obtained thus far to devise strategies to treat these infections.
Acknowledgments
C.F.B. is supported by NIH funds including AI059536 and the Northeast Biodefense Center (AI057158-Lipkin). G.K.A. is supported by NIH grant R01AI081914, a Developmental Grant from the Midwest Regional Center of Excellence (U54AI057160-Stanley[PI]), and funding from Carver Charitable Trust (#09-3271).
References
- Adam EJ. Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF. Mikulasova A. Martinez-Sobrido L. Paragas J. Muhlberger E. Bray M. Klenk HD. Palese P. Garcia-Sastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF. Wang X. Muhlberger E. Volchkov V. Paragas J. Klenk HD. Garcia-Sastre A. Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y. Le SY. Benkirane M. Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bosio CM. Aman MJ. Grogan C. Hogan R. Ruthel G. Negley D. Mohamadzadeh M. Bavari S. Schmaljohn A. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188:1630–1638. doi: 10.1086/379199. [DOI] [PubMed] [Google Scholar]
- Bray M. Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bucher E. Hemmes H. de Haan P. Goldbach R. Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- Cardenas WB. Loo YM. Gale M., Jr Hartman AL. Kimberlin CR. Martinez-Sobrido L. Saphire EO. Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH. Kubota T. Matsuoka M. Jones S. Bradfute SB. Bray M. Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW. Watson JC. Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes P. Ye ZS. Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros JF. Garcia-Sastre A. Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. 2005;6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- Cuomo CA. Kirch SA. Gyuris J. Brent R. Oettinger MA. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H. Bugany H. Mahner F. Klenk HD. Drenckhahn D. Schnittler HJ. Filovirus-induced endothelial leakage triggered by infected monocytes/macrophages. J Virol. 1996;70:2208–2214. doi: 10.1128/jvi.70.4.2208-2214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. Cerveny M. Yan Z. He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M., Jr Katze MG. Molecular mechanisms of interferon resistance mediated by viral- directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Egorov A. Matassov D. Brandt S. Levy DE. Durbin JE. Palese P. Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Geisbert TW. Hensley LE. Ebola virus: new insights into disease aetiopathology and possible therapeutic interventions. Expert Rev Mol Med. 2004;6:1–24. doi: 10.1017/S1462399404008300. [DOI] [PubMed] [Google Scholar]
- Geisbert TW. Hensley LE. Gibb TR. Steele KE. Jaax NK. Jahrling PB. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab Invest. 2000;80:171–186. doi: 10.1038/labinvest.3780021. [DOI] [PubMed] [Google Scholar]
- Geisbert TW. Hensley LE. Jahrling PB. Larsen T. Geisbert JB. Paragas J. Young HA. Fredeking TM. Rote WE. Vlasuk GP. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet. 2003a;362:1953–1958. doi: 10.1016/S0140-6736(03)15012-X. [DOI] [PubMed] [Google Scholar]
- Geisbert TW. Hensley LE. Larsen T. Young HA. Reed DS. Geisbert JB. Scott DP. Kagan E. Jahrling PB. Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003b;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW. Young HA. Jahrling PB. Davis KJ. Kagan E. Hensley LE. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J Infect Dis. 2003c;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- Goldfarb DS. Corbett AH. Mason DA. Harreman MT. Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Gorlich D. Henklein P. Laskey RA. Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gupta M. Mahanty S. Ahmed R. Rollin PE. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology. 2001;284:20–25. doi: 10.1006/viro.2001.0836. [DOI] [PubMed] [Google Scholar]
- Haasnoot J. de Vries W. Geutjes EJ. Prins M. de Haan P. Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M. Andersson I. Klingstrom J. Schumann M. Martin A. Zimmermann P. Wagner V. Pichlmair A. Schneider U. Muhlberger E. Mirazimi A. Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt BH. Sanchez A. Offermann MK. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology. 1998;252:179–188. doi: 10.1006/viro.1998.9446. [DOI] [PubMed] [Google Scholar]
- Harcourt BH. Sanchez A. Offermann MK. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1beta, in endothelial cells. J Virol. 1999;73:3491–3496. doi: 10.1128/jvi.73.4.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL. Dover JE. Towner JS. Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL. Ling L. Nichol ST. Hibberd ML. Whole genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL. Towner JS. Nichol ST. A C-terminal basic amino acid motif of Zaire Ebola virus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hensley LE. Stevens EL. Yan SB. Geisbert JB. Macias WL. Larsen T. Daddario-DiCaprio KM. Cassell GH. Jahrling PB. Geisbert TW. Recombinant human activated protein C for the postexposure treatment of Ebola hemorrhagic fever. J Infect Dis. 2007;196(Suppl 2):S390–S399. doi: 10.1086/520598. [DOI] [PubMed] [Google Scholar]
- Hensley LE. Young HA. Jahrling PB. Geisbert TW. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol Lett. 2002;80:169–179. doi: 10.1016/s0165-2478(01)00327-3. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Lacoste J. Lin R. Recruitment of an interferon molecular signaling complex to the mitochondrial membrane: Disruption by hepatitis C virus NS3-4A protease. Biochem Pharmacol. 2006;72:1477–1484. doi: 10.1016/j.bcp.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Hornung V. Ellegast J. Kim S. Brzozka K. Jung A. Kato H. Poeck H. Akira S. Conzelmann KK. Schlee M. Endres S. Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Imamoto N. Shimamoto T. Takao T. Tachibana T. Kose S. Matsubae M. Sekimoto T. Shimonishi Y. Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC. Muhlberger E. Carter V. Grosch M. Perwitasari O. Proll SC. Thomas MJ. Weber F. Klenk HD. Katze MG. Global suppression of the host antiviral response by Ebola- and Marburg viruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Sato S. Yoneyama M. Yamamoto M. Matsui K. Uematsu S. Jung A. Kawai T. Ishii KJ. Yamaguchi O. Otsu K. Tsujimura T. Koh CS. Reis e Sousa C. Matsuura Y. Fujita T. Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kitajewski J. Schneider RJ. Safer B. Munemitsu SM. Samuel CE. Thimmappaya B. Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- Kohler M. Ansieau S. Prehn S. Leutz A. Haller H. Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Kohler M. Speck C. Christiansen M. Bischoff FR. Prehn S. Haller H. Gorlich D. Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T. Matsuoka M. Chang TH. Tailor P. Sasaki T. Tashiro M. Kato A. Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW. Ginder ND. Fulton DB. Nix J. Basler CF. Honzatko RB. Amarasinghe GK. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci USA. 2009;106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX. Li H. Lu R. Li F. Dus M. Atkinson P. Brydon EW. Johnson KL. Garcia-Sastre A. Ball LA. Palese P. Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Wambach M. Katze MG. Krug RM. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- Ma J. Cao X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell Signal. 2006;18:1117–1126. doi: 10.1016/j.cellsig.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Mahanty S. Hutchinson K. Agarwal S. McRae M. Rollin PE. Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170:2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- McBride KM. Banninger G. McDonald C. Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 2002;21:1754–1763. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM. Reich NC. The ins and outs of STAT1 nuclear transport. Sci STKE. 2003;2003:RE13. doi: 10.1126/stke.2003.195.re13. [DOI] [PubMed] [Google Scholar]
- Melen K. Fagerlund R. Franke J. Kohler M. Kinnunen L. Julkunen I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem. 2003;278:28193–28200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y. Imamoto N. Sekimoto T. Tachibana T. Seki T. Tada S. Enomoto T. Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Moller P. Pariente N. Klenk HD. Becker S. Homooligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J. Blobel G. Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995a;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J. Hijikata M. Blobel G. Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta sub-unit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995b;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV. Ryder UW. Lamond AI. Weis K. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen F. Schaffner F. Furlan-Freguia C. Pawlinski R. Bhattacharjee G. Chun J. Derian CK. Andrade-Gordon P. Rosen H. Ruf W. Dendritic cell PAR1-S1P3 signalling couples coagulation and inflammation. Nature. 2008;452:654–658. doi: 10.1038/nature06663. [DOI] [PubMed] [Google Scholar]
- O'Neill RE. Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- Park M-S. Shaw ML. Munoz-Jordan J. Cros JF. Nakaya T. Bouvier N. Palese P. Garcia-Sastre A. Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A. Schulz O. Tan CP. Naslund TI. Liljestrom P. Weber F. Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Prins KC. Cardenas WB. Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases, IKKɛ and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DS. Hensley LE. Geisbert JB. Jahrling PB. Geisbert TW. Depletion of peripheral blood T lymphocytes and NK cells during the course of Ebola hemorrhagic Fever in cynomolgus macaques. Viral Immunol. 2004;17:390–400. doi: 10.1089/vim.2004.17.390. [DOI] [PubMed] [Google Scholar]
- Reid SP. Cardenas WB. Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the Ebola virus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP. Leung LW. Hartman AL. Martinez O. Shaw ML. Carbonnelle C. Volchkov VE. Nichol ST. Basler CF. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP. Valmas C. Martinez O. Sanchez FM. Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf W. Emerging roles of tissue factor in viral hemorrhagic fever. Trends Immunol. 2004;25:461–464. doi: 10.1016/j.it.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Schümann M. Gantke T. Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009 doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T. Tada S. Katada T. Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- Sekimoto T. Imamoto N. Nakajima K. Hirano T. Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher U. West E. Bugany H. Klenk HD. Schnittler HJ. Feldmann H. Infection and activation of monocytes by Marburg and Ebola viruses. J Virol. 2001;75:11025–11033. doi: 10.1128/JVI.75.22.11025-11033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ. Shows EB. Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Weis K. Mattaj IW. Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]