Abstract
We developed a series of ligand-inducible riboswitches that control gene expression in diverse species of Gram-negative and Gram-positive bacteria, including human pathogens that have few or no previously reported inducible expression systems. We anticipate that these riboswitches will be useful tools for genetic studies in a wide range of bacteria.
The ability to precisely control gene expression has greatly enhanced the study of microbial genetics and behavior. Common model systems, such as that of Escherichia coli, typically have a variety of genetic control elements available, such as the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac operon (13) and the arabinose-inducible araC promoter (12). However, for many bacteria, simple-to-use methods for inducing gene expression in a ligand-dependent fashion do not exist. While it is possible in principle to transfer the inducible regulatory machinery from one species to another (4), issues including promoter and codon usage, protein folding, and ligand permeability present challenging obstacles. Indeed, with the exception of the tetracycline-inducible expression system (16), most protein-based ligand-inducible expression systems that function well in E. coli have proven difficult to transport into a broad range of bacteria (23, 24). However, bacteria universally employ conserved, RNA-based genetic switching mechanisms to regulate many metabolic pathways (2), suggesting that synthetic riboswitches, which regulate gene expression via small-molecule-RNA interactions, may potentially function as genetic control elements in a broad range of bacterial species.
Over the past decade, synthetic riboswitches have been developed in several model organisms such as E. coli (10, 17), Bacillus subtilis (26), Saccharomyces cerevisiae (3, 29), and Chinese hamster ovary (CHO) cells (30). We reasoned that riboswitches would be particularly amenable to controlling gene expression in a variety of bacterial species because the translational machinery is well conserved and because riboswitches do not require accessory proteins. We recently reported the discovery of several ligand-dependent synthetic riboswitches that strongly activate protein translation in the Gram-negative gammaproteobacterium Escherichia coli (18). These riboswitches produce low levels of background gene expression in the absence of the small molecule theophylline and large (>50-fold) increases in expression in the presence of the ligand. However, attempts to use several of these riboswitches in other gammaproteobacteria, such as Acinetobacter baylyi (9), or the more distantly related firmicute Bacillus subtilis resulted in only modest ligand-dependent changes in gene expression (see the supplemental material). We suspected that this performance differential might result from stricter requirements for complementarity between the ribosome binding site and the 16S rRNA in these organisms (1).
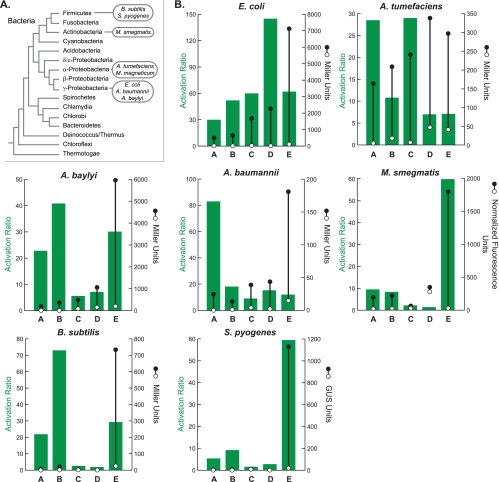
Using a combination of rational design and in vivo screening (9, 17, 27) (see Fig. S1 to S4 and Materials and Methods in the supplemental material), we developed a set of five synthetic riboswitches that enable inducible gene expression in eight diverse bacterial species (Fig. 1A), including organisms that currently have few or no tools with which to titrate gene expression in the laboratory. Three of the organisms are Gram-negative gammaproteobacteria, including E. coli; Acinetobacter baylyi strain ADP1 (28, 32), which is naturally competent but has few methods available for the conditional control of gene expression; and Acinetobacter baumannii (22), which is an opportunistic human pathogen that is often multidrug resistant and can cause severe pneumonia in immunocompromised patients, and, to our knowledge, currently lacks laboratory-inducible genetic control elements. Additionally, we tested the switches in the Gram-negative alphaproteobacterium Agrobacterium tumefaciens (31), which is the causative agent of crown gall disease in dicot plants and is widely used in genetic engineering applications (7). We also tested the switches in three Gram-positive bacteria, including the firmicutes Bacillus subtilis and Streptococcus pyogenes. B. subtilis is a well-studied model organism, while S. pyogenes is a human pathogen that causes several diseases, including pharyngitis (“strep throat”), cellulitis, scarlet fever, and necrotizing fasciitis (6). The development of inducible control elements for S. pyogenes is particularly desirable because the only available expression system typically requires two separate plasmids and is based on nisin, which is itself an antimicrobial agent (11). Finally, we tested our riboswitches in the actinobacterium Mycobacterium smegmatis, which is closely related to the pathogenic Mycobacterium tuberculosis.
FIG. 1.
(A) Bacterial phylogeny. The species studied here are shown in the ovals. (B) Activation ratios and expression levels for riboswitches A to E in each organism. Right axes: expression levels in the absence of theophylline (open circles) and in the presence of theophylline (2 mM; closed circles). Measurements are of β-galactosidase activity in Miller units (20) for all organisms, except for S. pyogenes (β-glucuronidase [GUS] activity in GUS units [14]) and M. smegmatis (normalized fluorescence of GFP [8]). Errors are smaller than the symbol size. Left axes: activation ratios of the riboswitches, which were determined by dividing expression levels in the presence of theophylline by expression levels in the absence of theophylline.
We constructed our riboswitches by subcloning each sequence into a broad-host-range vector, pBAV1K (5), which features a modified form of the pWV01 rolling-circle replicon (see Table S1 and Fig. S1 to S4 in the supplemental material). pBAV1K also includes a T5 promoter and a lac operator sequence that control the transcription of the riboswitch and the lacZ reporter gene. We note that while pBAV1K can transform a variety of bacteria, both its copy number and the efficiency of the T5 promoter vary between hosts (5). For applications in which controlling copy number and/or promoter strength is paramount, it can be helpful to use native promoters and plasmids. Thus, we studied our riboswitches using plasmids specifically adapted for M. smegmatis, S. pyogenes, and Magnetospirillum magneticum. For M. smegmatis, each riboswitch sequence was cloned within the 5′ untranslated region (UTR) of the eGFP gene of pMWS114, which contains the eGFP gene (S65T/F64L) cloned as an EcoRI-HindIII fragment into pMV261 (25). To construct riboswitch plasmids for S. pyogenes, we introduced each sequence by inverse PCR on the template pEU7742 (J. V. Bugrysheva and J. R. Scott, unpublished data), which features the Psag promoter and the gusA reporter gene. A guide for adapting the riboswitch constructs for use in new bacterial species is provided in the supplemental material.
In every organism tested, at least one of the five riboswitches provided a low level of background expression in the absence of the ligand and at least a 25-fold increase in gene expression in the presence of 2 mM theophylline (Fig. 1B). Activation ratios greater than 50-fold were achieved in most organisms, but it is important to note that switches that achieve the highest activation ratios often do so by having the lowest background expression in the absence of a ligand. For applications that demand high levels of gene expression, a switch with a stronger signal (but lower activation ratio) may be desirable. In nearly all organisms studied, there are at least two switches that display comparable activation ratios but substantially different levels of expression (e.g., switches B and E in B. subtilis). In general, both the background and the signal increase moving from switches A to E, which is consistent with the presence of stronger ribosome binding sites or less stable secondary structures in the ligand-free states in switches C to E (see Table S1 in the supplemental material for full details of the ribosome binding site sequences). We note that comparisons of riboswitches within a species are generally more useful than comparisons between species, as the copy number of pBAV1K, the efficiency of the T5 promoter, and the level of codon optimization of the reporter gene vary between species (5). Because expression is dose dependent (see Fig. S5 to S11 in the supplemental material), it should be possible to achieve the desired expression level in a given application by choosing a host-appropriate plasmid and promoter and then titrating the concentration of theophylline.
With synthetic riboswitches validated in seven organisms across four bacterial phyla, we assessed the ability of these switches to control gene expression in an organism not in our initial test set. Magnetospirillum magneticum strain AMB-1 (19) is an aquatic alphaproteobacterium that is able to navigate along Earth's magnetic field lines using a magnetite-containing membrane-bound organelle called the magnetosome. The study of magnetotactic bacteria is providing new insights into the process of biomineralization as well as a better understanding of organelle evolution and biogenesis in prokaryotes. The magnetotactic response of AMB-1 is dependent on the chain organization of the magnetosomes in the cell body, which requires expression of the actin-like cytoskeletal protein MamK (15). Previous work has shown that the chain organization defect observed in a mamK deletion mutant can be restored by the constitutive expression of a green fluorescent protein (GFP)-tagged version of MamK from a plasmid (15). The tunable expression system described here will allow more precise control over MamK levels, thus enabling temporally controlled induction or depletion studies.
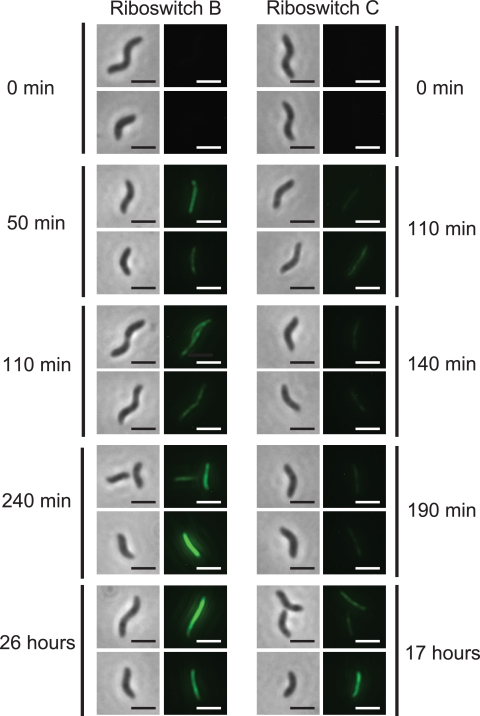
Based on data obtained in studying A. tumefaciens, which is also an alphaproteobacterium and is cultured at a similar temperature (28°C versus 30°C for M. magneticum), we predicted that switch A, B, or C would be best suited for inducing actin-like filament formation in M. magneticum, while switches D and E would likely exhibit high levels of background gene expression in the absence of theophylline. We considered the culture temperature to be important because previous studies have shown that similar riboswitches operate under equilibrium conditions, where temperature impacts the equilibrium (17), and because riboswitch function in E. coli is temperature dependent (see Fig. S12 in the supplemental material). Further consideration of the 16S rRNA sequences of each species suggested that switch A may exhibit higher levels of background and induced gene expression in M. magneticum than in A. tumefaciens due to an additional possible base-pairing interaction between M. magneticum 16S rRNA and the ribosome binding site of switch A (see Fig. S13 in the supplemental material). To test these hypotheses, we subcloned each riboswitch into the previously reported mamK-GFP expression plasmid (15) and imaged cells harboring these constructs at several time points following induction with 1 mM theophylline (see Materials and Methods in the supplemental material). While all five riboswitches showed increases in MamK-GFP expression in the presence of theophylline, switches A, D, and E displayed detectable levels of MamK-GFP expression in the absence of the inducer. Consistent with our predictions, riboswitches B and C produced no visible expression of MamK-GFP in the absence of the inducer. At comparable times postinduction with theophylline (1 mM), riboswitch B produced higher levels of MamK-GFP than riboswitch C, as shown by the appearance of fluorescent filaments extending from pole to pole (Fig. 2), but both riboswitches produced levels of MamK-GFP suitable for magnetosome localization studies (15, 21).
FIG. 2.
Images of riboswitches B and C controlling the expression of a MamK-GFP fusion in M. magneticum in the presence of theophylline (1 mM) as a function of time. Left panels are phase-contrast images; right panels monitor GFP fluorescence emission (2-μm scale bar). All fluorescent images were exposed for 6 s, and the cells were visualized with a 100× objective.
In summary, we developed a series of synthetic riboswitches that function as genetic control elements in a diverse set of Gram-negative and Gram-positive bacteria. Using theophylline, an inexpensive (<$0.20/g) small molecule that is nontoxic at the concentrations used here (10), we observed at least a 25-fold increase in gene expression in all species tested and a greater-than-50-fold increase in two human pathogens. Many unanswered questions surround the mechanisms of A. baumannii and S. pyogenes pathogenesis, and we anticipate that these riboswitches will enable studies that have been hindered by the inability to induce gene expression in conditional knockouts in these and other organisms. Finally, we demonstrated that these riboswitches facilitate genetic complementation studies in M. magneticum, a species with few genetic tools. We expect that the riboswitches presented here will be useful for controlling gene expression in a multitude of bacterial species that lack ligand-inducible expression systems.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants (GM074070 to J.P.G., AI20723 to J.R.S., GM084122 to A.K., and AI51622 to C.R.B.). S.T. and S.K.D. were G.W. Woodruff Fellows. S.T. is an Irvington Institute Fellow of the Cancer Research Institute. A.S. is an NIH K99 Postdoctoral Fellow (K99GM092934). A.W.P. is an NSF Predoctoral Fellow. A.K. is a David and Lucille Packard Foundation Fellow. J.P.G. is an Alfred P. Sloan Fellow and a Camille Dreyfus Teacher-Scholar.
We are grateful to Anton Bryksin and Ichiro Matsumura for plasmids, strains, and helpful discussions. The technical advice of Barbara Froehlich and Julia Bugrysheva for work with S. pyogenes is appreciated.
S.T., C.M.K.R., J.C.S., I.S.G., S.K.D., D.M., A.S., and A.W.P. designed and performed experiments. J.P.G., J.R.S., C.R.B., and A.K. provided oversight. S.T. and J.P.G. conceived the experiments and wrote the paper.
Footnotes
Published ahead of print on 8 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Band, L., and D. J. Henner. 1984. Bacillus subtilis requires a stringent Shine-Dalgarno region for gene expression. DNA 3:17-21. [DOI] [PubMed] [Google Scholar]
- 2.Barrick, J. E., and R. R. Breaker. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, T. S., and C. D. Smolke. 2005. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 23:337-343. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, R., and W. Hillen. 2008. The application of Tet repressor in prokaryotic gene regulation and expression. Microb. Biotechnol. 1:2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryksin, A. V., and I. Matsumura. 8 October 2010. Rational design of a plasmid origin that replicates effectively in both Gram-positive and Gram-negative bacteria. PLoS One 5:e13244. doi: 10.1371/journal.pone.0013244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685-694. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, M., J. E. Fry, S. Pang, H. Zhou, C. M. Hironaka, D. R. Duncan, T. W. Conner, and Y. Wan. 1997. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Desai, S. K. 2008. Ph.D. thesis. Emory University, Atlanta, GA.
- 10.Desai, S. K., and J. P. Gallivan. 2004. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 126:13247-13254. [DOI] [PubMed] [Google Scholar]
- 11.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318-356. [DOI] [PubMed] [Google Scholar]
- 14.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. U. S. A. 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 16.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch, S. A., S. K. Desai, H. K. Sajja, and J. P. Gallivan. 2007. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 14:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch, S. A., and J. P. Gallivan. 2009. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 37:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga, T., T. Sakaguchi, and F. Tadokoro. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 35:651-655. [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 21.Murat, D., A. Quinlan, H. Vali, and A. Komeili. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U. S. A. 107:5593-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassetti, C. M. 2008. Inducible expression systems for mycobacteria. Methods Mol. Biol. 465:255-264. [DOI] [PubMed] [Google Scholar]
- 24.Schofield, D. A., C. Westwater, B. D. Hoel, P. A. Werner, J. S. Norris, and M. G. Schmidt. 2003. Development of a thermally regulated broad-spectrum promoter system for use in pathogenic gram-positive species. Appl. Environ. Microbiol. 69:3385-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 26.Suess, B., B. Fink, C. Berens, R. Stentz, and W. Hillen. 2004. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 32:1610-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topp, S. 2009. Ph.D. thesis. Emory University, Atlanta, GA.
- 28.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. De Baere, A. Nemec, T. Van Der Reijden, E. Carr, I. Tjernberg, and L. Dijkshoorn. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigand, J. E., M. Sanchez, E. B. Gunnesch, S. Zeiher, R. Schroeder, and B. Suess. 2008. Screening for engineered neomycin riboswitches that control translation initiation. RNA 14:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werstuck, G., and M. R. Green. 1998. Controlling gene expression in living cells through small molecule-RNA interactions. Science 282:296-298. [DOI] [PubMed] [Google Scholar]
- 31.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 32.Young, D. M., D. Parke, and L. N. Ornston. 2005. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 59:519-551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.