Abstract
Background
Self/nonself discrimination is an essential feature for pathogen recognition and graft rejection and is a ubiquitous phenomenon in many organisms. Filamentous fungi, such as Neurospora crassa, provide a model for analyses of population genetics/evolution of self/nonself recognition loci due to their haploid nature, small genomes and excellent genetic/genomic resources. In N. crassa, nonself discrimination during vegetative growth is determined by 11 heterokaryon incompatibility (het) loci. Cell fusion between strains that differ in allelic specificity at any of these het loci triggers a rapid programmed cell death response.
Methodology/Principal Findings
In this study, we evaluated the evolution, population genetics and selective mechanisms operating at a nonself recognition complex consisting of two closely linked loci, het-c (NCU03493) and pin-c (NCU03494). The genomic position of pin-c next to het-c is unique to Neurospora/Sordaria species, and originated by gene duplication after divergence from other species within the Sordariaceae. The het-c pin-c alleles in N. crassa are in severe linkage disequilibrium and consist of three haplotypes, het-c1/pin-c1, het-c2/pin-c2 and het-c3/pin-c3, which are equally frequent in population samples and exhibit trans-species polymorphisms. The absence of recombinant haplotypes is correlated with divergence of the het-c/pin-c intergenic sequence. Tests for positive and balancing selection at het-c and pin-c support the conclusion that both of these loci are under non-neutral balancing selection; other regions of both genes appear to be under positive selection. Our data show that the het-c2/pin-c2 haplotype emerged by a recombination event between the het-c1/pin-c1 and het-c3/pin-c3 approximately 3–12 million years ago.
Conclusions/Significance
These results support models by which loci that confer nonself discrimination form by the association of polymorphic genes with genes containing HET domains. Distinct allele classes can emerge by recombination and positive selection and are subsequently maintained by balancing selection and divergence of intergenic sequence resulting in recombination blocks between haplotypes.
Introduction
Self/nonself discrimination is a ubiquitous and essential function of both multicellular and microbial species. In vertebrate species, self/non-self recognition relies on the major histocompatibility complex (MHC); allelic polymorphisms at MHC loci in populations are maintained by balancing selection [1], [2]. Alleles at MHC loci show long-term persistence, such that an allele from one species is often more closely related to an allele in a different species, a pattern that is termed trans-species polymorphisms. Self/nonself or allorecognition determinants involved in histocompatibility reactions are present in the earliest metazoans, including the ascidian, Botryllus schlosseri (Cordate subphylum, Tunicata), and the cnidarium, Hydractinia symbiolongicarpus (Phylum Cnidaria) [3]. In plants, self/non-self recognition during sexual reproduction is mediated by the gametophytic or sporophytic self-incompatibility locus, S, which elicits recognition and rejection of self-pollen. For example, the S locus in crucifers contains two unrelated highly polymorphic recognition genes (SRK and SCR) that are in tight genetic and physical linkage and encode more than 100 specificities [4]. Alleles at the S locus also show trans-species polymorphisms [5].
The filamentous fungal ascomycete species, Neurospora crassa, is an obligately out breeding haploid organism. Nonself recognition and mating between opposite mating types (A and a) initiates sexual reproduction. In filamentous fungi, such as N. crassa, self/nonself discrimination is also important during vegetative growth. For example, germinating asexual spores and mature hyphae of N. crassa colonies undergo cell fusion, which is mediated by self-signaling and chemotropic interactions [6]. However, cell fusion can also occur between colonies of unlike genotypes. If such individuals differ in allelic specificity at nonself recognition loci, the fusion cell is rapidly compartmentalized and undergoes programmed cell death (PCD) (termed heterokaryon incompatibility (HI)) (Figure 1) [7], [8]. HI is suppressed during sexual reproduction in filamentous fungi; genetically different strains are fully inter-fertile and produce viable progeny. HI is analogous to fusion versus fusion-rejection in colonial marine invertebrates [9] where encounters between individuals of the same species can lead to fusion to form a single chimeric colony, or rejection, resulting in a histoincompatibility response in colonial invertebrates or HI in filamentous fungi.
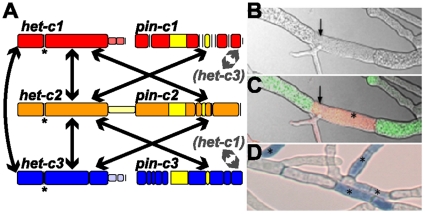
Figure 1. Diagram of the three het-c/pin-c haplotypes in N. crassa and death of fusion cells as a consequence of heterokaryon incompatibility.
A). Cartoon diagram of het-c1/pin-c1, het-c2/pin-c2 and het-c3/pin-c3 haplotypes. Nonself recognition is mediated by genetic interactions between het-c and pin-c from alternate haplotypes (het-c1-pin-c2, het-c1-pin-c3, het-c2-pin-c1, het-c2-pin-c3, het-c3-pin-c1 or het-c3-pin-c2; arrows). Allelic interactions between alternate het-c alleles (het-c1-het-c2, het-c1-het-c3 or het-c2-het-c3) is required for a robust HI response [26], [38]; arrows. The allelic specificity domain of het-c is indicated by an asterisk ‘*’ [38]. Thinner bars indicate intergenic sequence and indels are represented by open spaces. The HET domain of pin-c is shown in yellow. (B–D) Confocal micrographs showing hyphal fusion and heterokaryon formation between a het-c1/pin-c1 strain bearing cytoplasmic GFP and a het-c2/pin-c2 strain carrying a nuclear histone HI dsRED marker (isolates are otherwise isogenic). (B) Differential interference contrast (DIC) micrographs. Arrow indicates cell fusion point. (C) merged image of incompatible fusion [7]. Arrow indicates cell fusion point and asterisk indicates compartmentalized hyphal segment. (D) Hyphae stained with the vital dye Evan's blue [89]. Asterisks show compartmentalized dead hyphal segments.
HI in filamentous fungi is regulated by genetic loci, termed het (for heterokaryon incompatibility) or vic (for vegetative incompatibility) [8], [10]. In filamentous fungi, HI has been shown to reduce the risk of transmission of pathogenic elements, such as infectious virus-like dsRNAs [11], [12], [13], exploitation by aggressive genotypes [14] and has been proposed to function in pathogen recognition [15]. The induction of PCD upon nonself recognition in filamentous fungi is extremely rapid and robust; fusion cells are compartmentalized by septa formation and are dead within ∼30 min post-fusion [10], [11]. Ultrastructural and microscopic phenotypes associated with HI share morphological features with PCD in multicellular metazoans [16], [17], [18], including “apoptotic-like” bodies and TUNEL-positive cells, indicative of nuclear degradation. However, mutational analysis of N. crassa homologs of genes involved in apoptosis in other eukaryotic species showed that HI and its associated cell death was not dependent upon metacaspase homologs or a homolog to apoptosis inducing factor [19]. These data indicate that HI/PCD in N. crassa, and almost certainly in other filamentous fungi, does not occur via a conserved apoptotic mechanism, but likely through a fungal-specific pathway.
Genetic dissection of HI systems has been undertaken in a limited number of fungal species, including N. crassa (for review, see [7]), Podospora anserina (for review, see [8], [15], Aspergillus nidulans [20], [21] and Cryphonectria parasitica [22]. In N. crassa, 11 unlinked loci function in nonself recognition during HI [23]. Two to three allelic specificities occur at each of these loci. Thus, at least 211 different het genotypes are possible in N. crassa populations. Of these eleven het loci, the het-c locus, comprised of two closely linked loci, het-c and pin-c, is the best characterized on a molecular and genetic level. The het-c gene encodes a glycine-rich single-pass plasma membrane protein [24], [25]. The pin-c gene encodes a protein with a HET domain (pfam PF06985 [26]). Three het-c pin-c haplotypes have been identified; het-c1/pin-c1 (formerly known as het-cOR, Oak Ridge), het-c2/pin-c2 (formerly known as het-cPA, Panama) and het-c3/pin-c3 (formerly known as het-cGR, Groveland). Nonself discrimination between strains that differ in het-c/pin-c haplotype requires both allelic (het-c/het-c) and non-allelic interactions (between het-c and pin-c) (Figure 1) [26].
In this study, we examined the evolution of the het-c/pin-c region within ascomycete fungi and asked whether balancing selection extends across the entire het-c/pin-c region. Novel regions of het-c and pin-c, which appear to be under balancing or positive selection, were identified. We subsequently examined hypotheses as to how linkage disequilibrium of the different het-c/pin-c haplotypes may be maintained in populations. Our findings indicate that composition of het loci and the number of alleles found at each het locus within a species is variable and lineage specific, thus shedding new light on the mechanisms by which self/nonself discrimination loci evolve.
Results
Analysis of gene order and the origin of pin-c
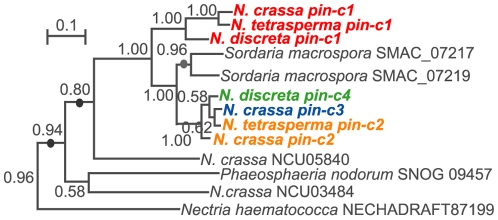
Previously, it was shown that the het-c allelic specificity region (∼200 bp) exhibits trans-species polymorphism [27]. We hypothesized that het-c/pin-c may have evolved as a het locus in Neurospora and closely related genera in the Sordariaceae as a result of a genome rearrangement. To test this hypothesis, we aligned a ∼40 k.bp. region that encompassed het-c from the genome sequences of N. crassa [28], Neurospora tetrasperma, Neurospora discreta, Sordaria macrospora [29], P. anserina [30], Chaetomium globosum, Magnaporthe grisea [31], Gibberella zeae, Sclerotinia sclerotiorum, Botryotinia fuckeliana, Histoplasma capsulatum, Aspergillus niger [32] and Neosartorya fischerii [33] (See Materials and Methods) (Figure 2). With the exception of pin-c (NCU03494), gene order and content in this region was well conserved among filamentous ascomycete species. Synteny analysis supported an ancestral gene order of RNA splicing factor Pad-1 (NCU03491), fatty acid hydroxylase gsl-5 (NCU03492) and protein phosphatase 2C (NCU03495). The placement of het-c (NCU03493) next to gsl-5 (NCU03492) is ancient and occurred during the divergence of the Sordariomycota from the Eurotiomycota (approximately 250–600 MYA [34]). The het-c (NCU03493) and pin-c (NCU03494) genes are linked only in closely related species within the Sordariaceae (Neurospora and Sordaria). In the Neurospora lineage, genes of the pin-c family have undergone at least two duplications from the ancestral gene (Figure 3; See Figure S1 for expanded phylogeny) resulting in NCU05840, NCU03484 and pin-c (NCU03494). These results were surprising because N. crassa does not tolerate gene duplications due to a genome defense mechanism termed Repeat-induced Point (RIP) mutation [35]. Evidence for RIP in NCU05840, NCU03484 and pin-c loci was not detected (data not shown). After the divergence of the Sordariaceae from the rest of the Sordariomycota, the NCU05840 gene duplication and rearrangement led to the current placement of pin-c next to het-c (Figure 2). A comparison of the het-c/pin-c region between N. crassa and the homothallic species, S. macrospora, showed evidence for two tandem duplications [29] resulting in two divergent pin-c alleles and one het-c allele. It is possible that the rearrangements occurred at the tRNA genes found between pin-c (S. macrospora SMAC07229) and PP2C (S. macrospora SMAC07230). In other species, tRNA genes are major sites of rearrangement in repeat poor genomes [28], [36]. In non-Sordariaceae members of the Sordariomycota, the homolog(s) of pin-c are in a different genomic location(s) and are not orthologous to pin-c or to NCU03484. Unlike the phylogeny of het-c homologs in the Pezizomycotina [37], the phylogeny of NCU05840/NCU03494/NCU03484 homologs in filamentous ascomycete fungi was not congruent with species relationships (Figure S1). These observations indicate that multiple rounds of gene duplication and gene loss occurs in this family, as previously observed for HET domain containing genes in species within the Aspergilli [37].
Figure 2. Analysis of gene order shows the recent acquisition of pin-c in the het-c region in Neurospora/Sordaria.
Bars indicate genes and are not drawn to scale. Arrows indicate gene orientation. Lines connect genes that are immediate neighbors on a chromosome or contig. Colored boxes indicate homologous genes as determined by sequence conservation and conserved gene order. Genes not connected to lines are found on a different chromosome or contig than the rest and are therefore unlinked. ‘X’ indicates genome rearrangement break points or indicates presence of intervening gene(s) (not shown). The het-c/pin-c (NCU03493/NCU03494) region in S. macrospora has undergone a gene duplication and rearrangement [29], resulting in one full and one partial het-c paralogs and two complete and divergent pin-c paralogs. The cladogram to the left represents the evolutionary relationship between species and is derived from James et al. [90]. Estimated divergence times are derived from Taylor and Berbee [34].
Figure 3. Bayesian inference of the amino acid sequence of pin-c alleles and related proteins shows three independent duplication events.
Numbers indicate support for nodes based on posterior probability (Bayesian inference). Sequences of pin-c (NCU03494) proteins are in bold and in color, reflecting allelic specificity. Circles on branches indicate possible gene duplication events. An expanded phylogeny is provided as Figure S1.
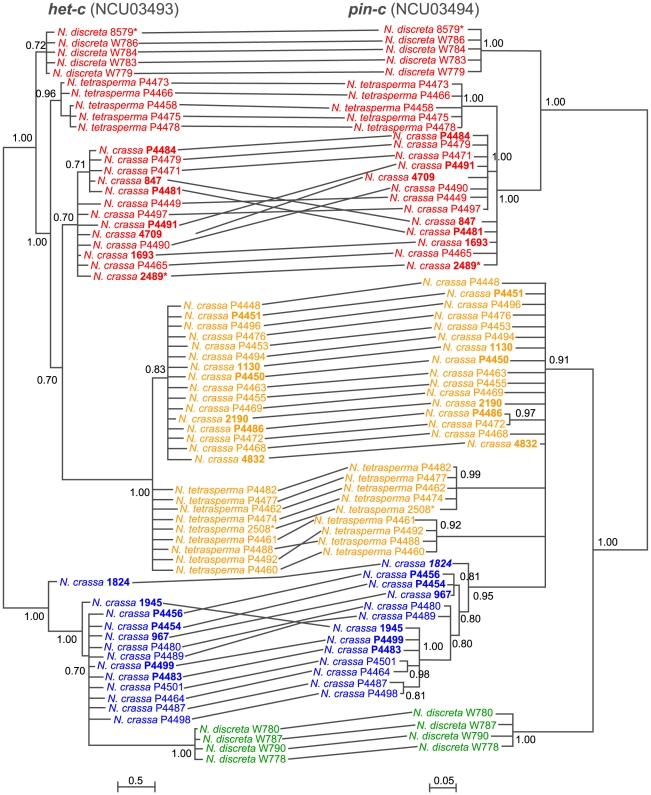
het-c (NCU03493) and pin-c (NCU03494) show trans-species polymorphism and are in severe linkage disequilibrium
Functional analysis of het-c showed that allelic specificity is conferred by a ∼34–48 amino acid region characterized by allele-specific indel patterns that show balancing selection [27], [38], [39]. To extend this study on het-c and to determine whether pin-c is also under balancing selection, we evaluated the het-c specificity domain and a highly variable region of pin-c from 42 strains of N. crassa, 9 strains of N. discreta, and 14 strains of N. tetrasperma (Table S1). All 42 strains of N. crassa contained only one of the three previously characterized het-c alleles, which were present at nearly equal frequency in both global (all strains used in this study) and on a local population scale (Louisiana isolates, all from Franklin, LA) (Figure 4; Figure S2). Analysis of the variable region of pin-c showed an identical pattern (Figure 4; Figure S2) with three classes of pin-c alleles present at nearly equal frequency in both global and local population samples. In contrast to N. crassa, only two allele classes of both het-c and pin-c were recovered from N. discreta and N. tetrasperma isolates (Figure 4). Most strikingly, in all three species, het-c and pin-c alleles were in severe linkage disequilibrium: a particular allele of het-c was associated with the corresponding allele at pin-c. In N. crassa, this corresponds to the het-c1/pin-c1, het-c2/pin-c2 and het-c3/pin-c3 haplotypes (red, yellow and blue, respectively, Figure 4 [26]). In N. tetrasperma, only het-c1/pin-c1 and het-c2/pin-c2 haplotypes were identified, consistent with previous data for het-c [40]. N. discreta also possessed only two haplotypes: a het-c1/pin-c1 haplotype, as well as a haplotype (het-c4/pin-c4) that was distinct (Figure 4; green).
Figure 4. het-c (NCU03493) and pin-c (NCU03494) show trans-species polymorphism and are in severe linkage disequilibrium.
Trees are derived from alignments of partial sequences that correspond to allele specificity domains and were derived by Bayesian analysis [91]. Bayesian posterior probabilities are presented at internal branches. For each clade, alleles are in red, yellow, and blue are for het-c1/pin-c1, het-c2/pin-c2 and het-c3/pin-c3, respectively. Neurospora discreta has two haplotypes, one clearly related to het-c1/pin-c1 (red), but the other is ambiguous relative to het-c2/pin-c2 and het-c3/pin-c3. This haplotype is marked by a green color. In all three Neurospora species, het-c and pin-c alleles show severe linkage disequilibrium (i.e. het-c1 is always with pin-c1, het-c2 is always with pin-c2 and het-c3 is always with pin-c3). Lines connecting alleles between the two trees highlight this correspondence. Crossed lines indicate possible intra-allelic recombination events. Taxa in bold indicate that het-c pin-c allelic specificity was confirmed by functional tests (Table S1).
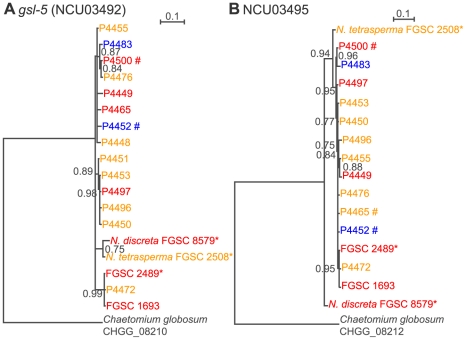
The maintenance of balancing selection in most species is associated with a strong recombination block and extreme sequence divergence [41], [42]. In filamentous fungi, this phenomenon is best demonstrated in the het-6 locus of N. crassa. Like het-c, het-6 HI is controlled by two closely linked genes: het-6 and un-24 [43], with two distinct allele specificities [44]. Alleles at un-24 and het-6 show severe linkage disequilibrium due to a chromosomal inversion that blocks recombination in this region [43], [45]. If het-c and pin-c were in a region that was associated with a chromosomal rearrangement, we hypothesized that alleles at loci linked to het-c/pin-c would show linkage disequilibrium, or possibly evidence of a chromosomal rearrangement(s). Increased genetic diversity at closely linked neutral sites is often observed near loci under balancing selection [46]. We first examined the linkage of loci surrounding het-c and pin-c in all three haplotypes via PCR amplification and DNA sequencing. All three haplotypes showed an identical gene order surrounding het-c/pin-c (data not shown), indicating that a chromosomal rearrangement is not present at the centromere proximal or distal ends of het-c/pin-c. To determine whether alleles at the centromere-proximal locus, gsl-5 (NCU03492; 1.8 kbp from het-c) or at the centromere distal locus, NCU03495 (2 kbp from pin-c), were in linkage disequilibrium with het-c/pin-c haplotypes, we used RNA-Seq data from N. crassa strains used in this study to construct gene phylogenies of gsl-5 (NCU03492) and NCU03495. As shown in Figure 5, a comparison of the topology of gsl-5 (NCU03492) and NCU03495 showed that alleles from neither locus is in linkage disequilibrium with alleles at het-c (NCU03493) or pin-c (NCU03494), nor do they show evidence of trans-species polymorphisms. These data indicate that the recombination block observed in the het-c/pin-c haplotypes is restricted to these two loci and does not extend to surrounding genes.
Figure 5. Gene phylogenies of gsl-5 (NCU03492) or PP2C (NCU03495) show no evidence of linkage disequilibrium with het-c (NCU03493) or pin-c (NCU03494).
Trees were derived from mRNA sequences assembled from RNA-Seq data from isolates shown by Bayesian analysis [91]. Bayesian posterior probabilities are presented at internal branches. Colors represent specificity based on het-c/pin-c haplotype (red: het-c1/pin-c1, yellow: het-c2/pin-c2, blue: het-c3/pin-c3). Alleles without species names are N. crassa. Alleles from sequenced strains are marked with ‘*’. Alleles not found in Figure 4 are marked with ‘#’. Note that allelic differences at NCU03492 and NCU03495 do not group with allelic specificity at NCU03493 (het-c) or NCU03494 (pin-c), indicating that balancing selection and linkage disequilibrium do not extend beyond het-c and pin-c.
To determine whether the divergence in the intergenic region of het-c/pin-c may contribute to a recombination block, we sequenced the intergenic region from 35 strains (22 N. crassa, 4 N. discreta, and 9 N. tetrasperma). Each het-c/pin-c haplotype contained a unique intergenic sequence profile (Figure S3). The intergenic sequences not only varied in length (Table 1), but also in the level of nucleotide substitution (Table 2). For example, a 177 bp insertion of the coding sequence of another gene, NCU08027 (encoding a nucleoside diphosphatase) was detected in the N. crassa het-c2/pin-c2 haplotype (Figure S3). Pair-wise measurements of nucleotide diversity between haplotypes (π [47]) showed a 10-fold difference in nucleotide diversity between versus within haplotypes (Table 2). These data support the hypothesis that recombination between het-c and pin-c is suppressed by DNA sequence divergence, particularly in the het-c/pin-c intergenic region.
Table 1. Length of het-c (NCU03493)/pin-c (NCU03494) intergenic sequences.
| Haplotype | Intergenic sequence length (base pairs) |
| N. crassa het-c1/pin-c1 | 978 |
| N. crassa het-c2/pin-c2 | 1476 |
| N. crassa het-c2/pin-c2 variant (FGSC 4832) | 1047 |
| N. crassa het-c3/pin-c3 | 919 |
| N. discreta het-c1/pin-c1 | 1811 |
| N. discreta het-c4/pin-c4 | 863 |
| N. tetrasperma het-c1/pin-c1 | 973 |
| N. tetrasperma het-c2/pin-c2 | 920 |
Table 2. The average number of nucleotide differences per site (π) for pair wise comparisons of N. crassa het-c (NCU03493)/pin-c (NCU03494) intergenic haplotypes.
| het-c1/pin-c1 | het-c2/pin-c2 | het-c2/pin-c2 variant (FGSC 4832) | het-c3/pin-c3 | |
| het-c1/pin-c1 | 0.04929 | 0.09498 | 0.10672 | 0.16134 |
| het-c2/pin-c2 | - | 0.05876 | 0.05168 | 0.20248 |
| het-c2/pin-c2 variant (FGSC 4832) | - | - | 0 | 0.19775 |
| het-c3/pin-c3 | - | - | - | 0.02966 |
Tests of selection of het-c and pin-c show that both genes are under both balancing and positive selection
Methods used to detect selection on coding sequences fall into two major classes: population methods, based on analyzing the nature and frequency of allele diversity within a species, and codon analysis methods, based on comparing patterns of synonymous and non-synonymous changes in protein coding sequences. High non-synonymous/synonymous substitution ratios have been observed in N. crassa het-c [27] and het-6 [43], as well as het loci from P. anserina [48], [49]. Among population-based methods, Tajima's D is an indicator of coding sequences (CDS) evolving under a non-random process, including directional selection or balancing selection [50]. In a stable population at equilibrium, Tajima's D for a CDS should be close to zero. A positive Tajima's D indicates a contraction in population size or balancing selection, which result in low levels of both rare and high frequency polymorphism, while a negative Tajima's D is associated with positive or diversifying selection, population size expansion or recovery from a selective sweep.
We calculated Tajima's D statistic for het-c (NCU03493) and pin-c (NCU03494) within allele classes and between allele classes for 29 N. crassa strains from Franklin, Louisiana (Table 3). As expected, a positive D statistic was obtained when all three allele classes of het-c were analyzed, consistent for a locus under balancing selection. In contrast, analysis of each individual het-c allele class resulted in a significant negative D score for het-c2 and het-c3 (P-value<0.05, all P-values for Tajima's D scores calculated using assumed beta distribution [50]) and a borderline significant (P-value<0.1) score for het-c1, suggesting that each het-c allele class is also under directional selection. For all pin-c alleles, a significantly positive D score of 2.45 (P-value<0.05) was obtained, providing strong evidence for balancing selection acting at this locus. For individual allele classes of pin-c, although variation in the D statistic was observed, it was not statistically significant (all P-values>0.1).
Table 3. Tajima's D statistic for coding regions within and between het-c (NCU03493) and pin-c (NCU03494) allele classes.
| Allele | Tajima's D statistic | Probability |
| het-c , all alleles | 1.33965 | P>0.10 |
| het-c1 alleles | −1.53263 | 0.10>P>0.05 |
| het-c2 alleles | −1.90353 | P<0.05* |
| het-c3 alleles | −2.23236 | P<0.01* |
| pin-c , all alleles | 2.44820 | P<0.05* |
| pin-c1 alleles | 1.15021 | P>0.10 |
| pin-c2 alleles | −1.04141 | P>0.10 |
| pin-c3 alleles | 0.09330 | P>0.10 |
* = significant value.
To test for regions of selection in het-c (NCU03493) and pin-c (NCU03494), Tajima's D was re-calculated on a sliding window of size 3 alignment with a step of 3 columns (corresponding to each codon). The ratio of the rate of non-synonymous substitutions (Ka) to the rate of synonymous substitutions (Ks) or ω was calculated for each codon in a multiple alignment using an evolutionary codon model [51], which enabled calculating ω at each codon site using a maximum-likelihood (ML) approach [52]. For het-c alleles, it was clear that balancing selection was confined to two regions of the coding sequence (Figure 6A, E, F). The first of these regions is the well-known specificity domain (codons/amino acids 194–236) [39] (Figure 6E). However, a second region of het-c showing balancing selection was also identified (codons/amino acids 521–599) (Figure 6F). These data are consistent with deletion analysis of het-c, where constructs missing the specificity domain still retained a low level of incompatibility [38]. Consistent with a negative Tajima's D statistic, Figure 6B–D also show that specific codons of het-c appear to be under strong positive selection. These data suggest that selection may be occurring for het-c alleles that differ in function, either for restricting induction of PCD between het-c pin-c alleles of the same haplotype, or increasing severity of PCD via interactions between haplotypes.
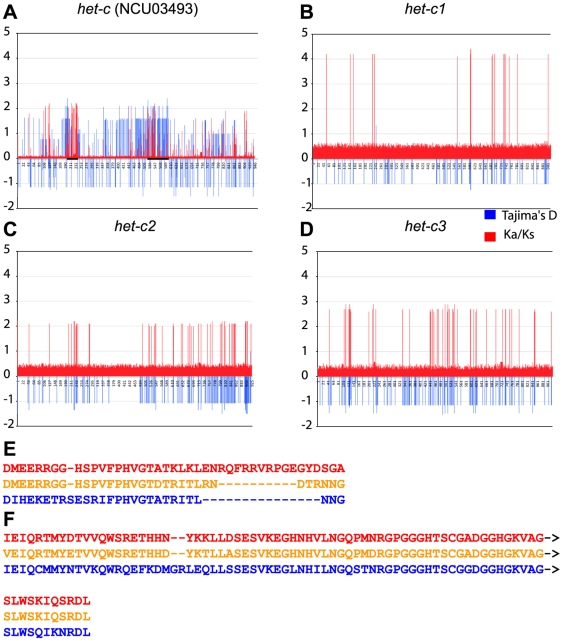
Figure 6. Measure of Ka/Ks ratios and Tajima's D on the coding region of het-c show evidence for balancing selection between, but not within allele classes.
A) Ka/Ks ratios (ω, ratio of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates; red) calculated for each codon for all het-c (NCU03493) alleles shows significant positive selection at positions 126, 215, 219, 232–235, 451 and 546 (P = 0.001) by likelihood ratio test (red).Tajima's D (blue) calculated for each codon shows borderline significant balancing selection (P<0.1) at positions 194–236 (which includes the het-c (NCU03493) allelic specificity region [27], [38]). Significant balancing selection (P<0.05) is also seen at positions 521–599. Both regions are shown as bold bars on (A). Evaluation of Ka/Ks (red) and Tajima's D (blue) within a het-c (NCU03493) allele class shows evidence for positive selection, but not balancing selection: B) het-c1 alleles, C) het-c2 alleles and D) het-c3 alleles. Note evidence for positive selection at different sites for all three het-c (NCU03493) alleles. E and F) Amino acid alignments of known (E) and new putative (F) specificity regions of het-c based on Tajima's D analysis. Bold bars on the x-axis of panel A indicate the locations of these regions.
For pin-c, two regions (codons/amino acids 8–78 and 190–390) appear to be under balancing selection and are likely to be specificity determinants for non-allelic interactions with het-c (Figure 7A, E, F). The second and larger of these two regions overlaps with the HET domain (codons/amino acids 335–533). Similar to full gene analyses above, codon-specific analysis of Tajima's D and ω indicated that the three pin-c allele classes are under different evolutionary pressures or have significantly different evolutionary histories. A region in the 5′ end of the coding region of pin-c1 alleles appears to be under balancing selection, with three different allele types (Figure 7B) and which was different from the two regions identified from the comparison of all pin-c alleles. These allele variants were not observed in pin-c2 or pin-c3 alleles. Both pin-c2 and pin-c3 have a large number of codons with large ω. In pin-c2, these codons are mirrored by a negative Tajima's D indicating a high rate of low frequency polymorphism (Table 3; Figure 7C). It seems likely that either many codons in pin-c2 are under positive or diversifying selection or pin-c2 alleles have had a recent population expansion. Analysis of pin-c3 alleles showed an extremely high ω with three regions that were highly polymorphic (Figure 7D). However, these codons have a D statistic close to zero indicating a balance between mutation and genetic drift (Table 3). These data are consistent with pin-c3 alleles being at equilibrium, with many codons under highly relaxed selection.
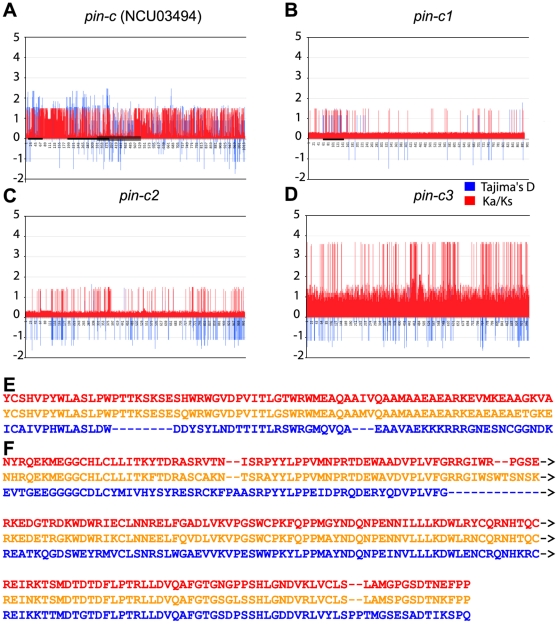
Figure 7. Measure of Ka/Ks ratios and Tajima's D on the coding region of pin-c (NCU03494) show evidence for balancing selection between allele classes.
Ka/Ks ratios (ω, ratio of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates; red) were calculated for each codon for all pin-c alleles and significant positive selection between positions 77–746 (p = 0.001) by likelihood ratio test was detected. These positions include the HET domain (codons 335–533) of pin-c (NCU03494)(large bar on x-axis). Tajima's D calculated for each codon for all pin-c alleles shows significant balancing selection (P<0.1) at positions 8–78 and 190–390 (smaller bars on x-axis; see E and F). Evaluation of Ka/Ks (red) and Tajima's D (blue) within a pin-c allele class shows variable selection: B) A region from codon 60 to 150 shows evidence of balancing selection in pin-c1 alleles (shown as a bar on the x axis) C) pin-c2 D) pin-c3. pin-c3 alleles show an extremely high ω with three very polymorphic regions (codons 448–491, 569–639 and 738–781). E and F) Amino acid alignments of putative allelic specificity regions of pin-c (the locations of these regions are indicated by bars on the x-axis of panel A).
As was noted for the recombination block characteristic of het-c/pin-c haplotypes, the unusual selective pressure evident from Ka/Ks ratios and Tajima's D test does not extend to loci surrounding het-c and pin-c. For example, a similar analysis of Ka/Ks and Tajima's D performed on gsl-5 (NCU03492) showed no evidence of selection (Figure S4).
Both het-c and pin-c show phylogenetic discordance
In N. crassa there should be strong selection against rare recombinants between het-c/pin-c haplotypes due to production of self-incompatible progeny. However, initial phylogenetic analysis of the different exons of het-c and pin-c resulted in inconsistent topologies between trees, suggesting that recombination does occur (Figure 4). We evaluated phylogenetic discordance in N. crassa using a test for recombination within alignments of het-c and pin-c using the likelihood-based selection genetic algorithm (GARD) [53]. GARD identified 3 potential recombination break points with four distinct phylogenetic trees that corresponded to different regions of het-c. Only one of the four (at position 491 of the alignment) was determined to be significant by the Kishino-Hasegawa test (KH test) (P-value = 0.00060) [54](Figure S5). In pin-c, nine potential recombination break points with 10 distinct phylogenetic trees were identified (Figure S6); all 9 breakpoints were significant (P-value = 0.00180).
All phylogenetic trees constructed from both het-c (NCU03493) and pin-c (NCU03494) showed distinct het-c1/pin-c1 and het-c3/pin-c3 haplotypes (Figures S5 and S6). However, the het-c2 and pin-c2 alleles showed phylogenetic discordance between trees. Most strikingly, het-c2 alleles were more closely related to het-c1 alleles, while pin-c2 alleles were more closely related to pin-c3 alleles. A close examination of tree topologies revealed that in the first region of het-c (nucleotide positions 1–490), het-c2 alleles form a single unresolved clade that clustered with het-c3 alleles (Figure S5). In the second region of het-c (positions 491–1376), which includes the het-c specificity region, the het-c2 alleles formed a resolved cluster with het-c1 alleles. This pattern holds through the third region of het-c (positions 1377–1899). The fourth region of het-c (positions 1900–2832) was poorly resolved. The third and fourth partitions of het-c are delineated by poorly supported break points and likely to be evidence of rate variation rather that topological incongruence. However, the sudden topological switch of het-c2 in regions 1 and 2 indicates that a recombination event occurred between het-c1 and het-c3 alleles in the evolution of the het-c2 allele (Figure 8).
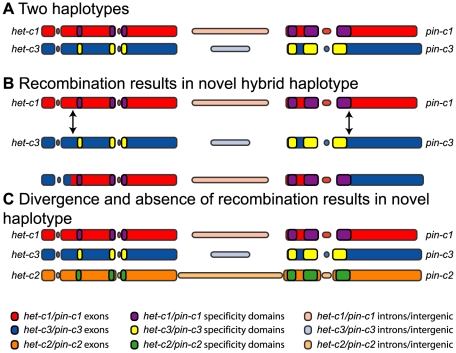
Figure 8. Model of the recombination-based emergence of het-c2/pin-c2 haplotype.
Colored bars indicate the coding and non-coding regions of each haplotype. Red indicates het-c1/pin-c1, blue indicates het-c3/pin-c3, and yellow indicates het-c2/pin-c2 haplotypes. Intergenic regions and introns are indicated by thinner bars and muted shades. Indels are show as spaces. The known and putative specificity regions of each haplotype are shown by purple, yellow, and green bars for het-c1/pin-c1, het-c3/pin-c3, and het-c2/pin-c2, respectively. All bars are drawn to scale.
The first and third partitions of pin-c2 (nucleotide positions 1–222 and 961–1233), cluster with pin-c1 alleles (Figure S6), consistent with the clustering of het-c2 with het-c1 in regions 2–3 (Figure S5). This result was also consistent with the observation that the intergenic regions of het-c1/pin-c1 and het-c2/pin-c2, while highly divergent, showed higher levels of sequence identity (Table 2) and were more closely related to each other than to the intergenic sequence of het-c3/pin-c3 (Figure S7). The second partition of pin-c showed a clearly resolved clade for pin-c1, pin-c2 and pin-c3. Interestingly, these partitions overlap closely with the regions determined to be under balancing selection in pin-c (Figure 7A). Partition 4 of pin-c is largely consistent with partitions 1 and 3 (pin-c2 clusters with pin-c1), but shows some recombination between pin-c2 and pin-c3. However, a sudden topological switch in partition 5 of pin-c2 (positions 983–1949), and which continues throughout the rest of the gene, results in the clustering of all pin-c2 alleles with pin-c3 alleles (Figure S6). This topological shift between regions 3/4 and 5 of pin-c is concordant with a similar shift between regions 1 and 2 of het-c, consistent with a recombination event occurring between het-c1/pin-c1 and het-c3/pin-c3 haplotypes in the evolution of the het-c2/pin-c2 haplotype (Figure 8).
Our data support a model in which the het-c2/pin-c2 allele has formed recently by recombination between existing het-c1/pin-c1 and het-c3/pin-c3 alleles. By this model, het-c2/pin-c2 diverged from het-c1/pin-c1 and het-c3/pin-c3 quite recently as compared to their divergence time from each other. To explore this further, we estimated divergence times for het-c and pin-c alleles both from homologous genes in other species and from each other. These estimates were obtained by fitting trees by maximum likelihood (Langely-Fitch) [55] to a molecular clock as implemented in the r8s software [56]. We assumed a constant molecular clock calibrated by a previous estimate of the divergence of the Eurotiomycetes from the Sordariomycetes at 200 million years ago (mya) [34]. Our results indicated a divergence of Neurospora from Sordaria approximately 21.1 to 31.3 mya and divergence of N. crassa and N. tetrasperma approximately 2.6 to 2.8 mya. These estimates are very similar to previous estimates of divergence between these groups (36 mya for the divergence of Neurospora and Sordaria [27] and 3.5 to 5.8 mya for N. crassa and N. tetrasperma [57]). Consistent with our hypothesis, the het-c1/pin-c1 and het-c3/pin-c3 haplotypes diverged from each other approximately ∼16 to 20 mya, whereas the het-c2/pin-c2 haplotype only appears 3 to 12 mya (Figure S8). This result indicates that the het-c/pin-c HI locus was initially bi-allelic. These data also support the gain of het-c2/pin-c2 haplotype in an ancestor of N. crassa and N. tetrasperma rather that the gain of het-c2/pin-c2 in an ancestor of N. crassa and N. discreta with subsequent loss of the het-c2/pin-c2 haplotype in N. discreta.
Our analysis above indicated that recombination was rare between het-c/pin-c haplotypes. Our expectation was that recombination within haplotypes would be unrestricted. To test this hypothesis, we evaluated tree topologies within specific allelic classes for both het-c (NCU03493) and pin-c (NCU03494) for both coding regions and intergenic sequence. We did not find evidence of recombination within any het-c allele class (data not shown), possibly due to the high conservation of het-c, which makes any signal of recombination difficult to detect. Similarly, support for recombination within pin-c1, or within intergenic regions of a single haplotype, was not detected (data not shown). However, strong evidence for recombination within the pin-c2 allele class and within the pin-c3 allele class was detected (Figures S9 and S10); pin-c2 and pin-c3 allele classes both contain highly variant strains, which provided robust evidence for recombination.
Discussion
Here we investigated the evolution and diversity of a nonself recognition locus in N. crassa. We show that the het-c/pin-c haplotypes that mediate nonself recognition and HI in N. crassa have evolutionary features in common with nonself recognition systems in other eukaryotic species, including extreme polymorphism, low recombination frequencies, frequency dependent selection and trans-species polymorphism. Our study shows that the het-c/pin-c haplotypes evolved as a consequence of a gene duplication/genomic rearrangement event, whereby pin-c was inserted near the het-c locus in the ancestor of Sordaria/Neurospora. We show that a third het-c/pin-c haplotype (het-c2/pin-c2) was generated via recombination in an ancestor of N. crassa/N. tetrasperma. The three het-c/pin-c haplotypes subsequently diverged via mutation and reduced recombination associated with extreme divergence of the intergenic sequences between het-c and pin-c. The ability to discriminate nonself via genetic differences at het-c/pin-c is a gain-of-function consequence of this genome rearrangement/divergence, rather than a disruption of het-c or pin-c function, as deletion mutants of het-c and pin-c are phenotypically wild type [26].
pin-c alleles are extremely polymorphic (pin-c alleles are ∼50% identical and have numerous indels). The pin-c gene encodes a cytoplasmic ∼900 amino acid protein with a HET domain (pfam PF06985) [26], [37]. The HET domain is a ∼150 amino acid region that is common in predicted genes in filamentous ascomycete genomes and appears to be uniquely found in these species. The HET domain has no identified function other than for nonself recognition and HI [37]. Six of the seven molecularly characterized het interactions involve proteins with predicted HET domains (N. crassa het-6, tol and pin-c [26], [43], [58] and P. anserina het-D, het-E and het-R [59], [60]. HET domains are death effectors; over-expression of just the HET domain causes HI and cell death, no matter what the genetic background [61] (our unpublished results). Our data suggest that the gene duplication/genome rearrangement to form het-c pin-c haplotypes enabled pin-c to function as a death effector upon nonself recognition, perhaps mediated by physical interactions between HET-C and PIN-C proteins. Previously, we showed that physical interaction between alternate HET-C proteins (HET-C1/HET-C2, HET-C1/HET-C3 and HET-C2/HET-C3) was dependent upon the allelic specificity domain [25], [39]. Codon-based analysis of the coding sequence of het-c and pin-c confirms that the previously identified het-c specificity region is under balancing selection (amino acids 194–236). However, we also identified a novel region of het-c that was also under balancing selection, and which is consistent with previous experimental results suggesting this region has a role in HI [38]. We also identified two regions under strong balancing selection in pin-c (amino acid positions 8–78 and 190–390). These data will enable further experimentation to determine whether these regions are the pin-c allelic specificity determinants and whether they mediate protein-protein interactions between HET-C and PIN-C.
Analysis of gene order at het-c (NCU03493) has shown that the ancestral state of het-c was not as a HI locus. This hypothesis is supported by studies in other species in which het-c is not polymorphic [62], [63], [64]. For example, analysis of the het-c homolog in P. anserina, hch, showed that 11 isolates possessed identical alleles. However, the introduction of the N. crassa het-c2 allele via transformation induced an HI-like response [64]. Similarly, the introduction of the het-c2 allele into Aspergillus niger also induced an HI-like response, even though no natural polymorphisms existed at the A. niger het-c locus [62].
Our analysis of recombination both within and between alleles at het-c/pin-c strongly supports a model in which recombination between the existing het-c1/pin-c1 and het-c3/pin-c3 haplotypes resulted in the creation of the novel het-c2/pin-c2 haplotype. Tajima's D was negative for pin-c2 alleles (−1.04141) supporting the hypothesis that pin-c2 alleles have had a recent population expansion, perhaps following the creation of this novel specificity via recombination. Our analyses indicated that the het-c2/pin-c2 haplotype emerged after the origin of the het-c1/pin-c1 haplotype in the Sordariaceae. Once the rare viable het-c2/pin-c2 hybrid haplotype existed, this strain would be incompatible with every other Neurospora strain, which presumably conferred a strong selective advantage, until it settled into equilibrium with the other het-c/pin-c haplotypes. This is exactly the case today, with each het-c/pin-c haplotype found in roughly 30% of the population (Figure S2). The generation of novel specificities by recombination appears to function in other self/nonself recognition systems and is well characterized in the orchestrated site-specific V(D)J system that generates variability in immunoglobulin and T cell receptor proteins in vertebrate systems [65]. In plant species, recombination to produce new specificities at the S-locus was first proposed by Fisher [66]. Although recombination has been detected between the kinase domain of SRK and SCR [67], [68] and Kusaba et al provided evidence that recombination occurs between alleles of the stigma-specific S glycoprotein (SLG = SCR) in Brassica [69], novel allelic specificities and co-evolution of the S receptor kinase (SRK) was not determined. Similarly, in the Solanaceae, sequence support for intragenic recombination at the S-RNase gene was observed in Petunia inflata [70]. In N. crassa, the generation of novel specificities at het-c/pin-c is a very similar situation to the evolution of S-locus. Both involve co-evolution of multiple genes or the result may be HI, as in the case of N. crassa, or self-fertility, as in the case of the S-locus in plants. The recombination event that resulted in the het-c2/pin-c2 haplotype appears to meet this requirement as it involved regions of both het-c and pin-c to generate a novel specificity.
By genetic analyses, N. crassa is predicted to have at least 11 unlinked het loci [23]. However, N. crassa has 52 HET domain genes, far more than are predicted to function as nonself recognition loci. Our analyses of pin-c-like HET domain genes in the genomes of filamentous fungi show multiple gene duplication events, rapid diversification and gene loss (Figure S1). Muirhead et al predicted through simulation that given a strong selection for the maintenance of HI, the maximum number of loci possible will function as het loci [71]. This simulation also predicted that unless the number of loci with the potential to function in HI was saturated, then each locus would contain two alleles. For example 3 loci with 2 alleles each provides more diversity than 2 loci with 3 alleles each (23<32). Our understanding of the selective advantage conferred by the maintenance of HI in fungi, as a barrier to the spread of infectious mycoviruses and the prevention of resource plundering by less fit genotypes, would fit a model in which the most fit individual would be one that was incompatible with all other individuals in the population, except with itself. Given such a selective pressure and a large population size, fitness is conferred by maximizing het diversity on a genome-wide scale. The recent emergence (in an evolutionary time scale) of the het-c/pin-c haplotype subsequent to gene duplication and rearrangement can be interpreted as selection for the maximum number of het loci. The rapid expansion of the het-c2 haplotype to create a tri-allelic het-c/pin-c system would seem to indicate that the number of het loci in N. crassa is near saturation. This hypothesis is supported by other tri-allelic het loci in N. crassa [72]. It is possible that in filamentous fungi, het loci are born out of dynamic genomic regions that allow for gene duplication and re-arrangement beyond the genomic norm. N. crassa provides an excellent model by which to investigate the birth, diversification and death of self/nonself recognition loci. The genomic resources available in the filamentous fungi, including genome sequences of a number of fungi related to N. crassa, well defined species, population genetics and the recent availability of RNA-Seq data for >100 N. crassa individuals from two populations will enable a full genome analysis of evolution and diversification of all nonself recognition loci encoded within a genome.
Methods
Strains and culture conditions
In this study, the laboratory strain (FGSC 2489) and 41 additional wild N. crassa strains, 9 wild N. discreta strains, and 13 wild N. tetrasperma strains were used (Table S1). Twenty-six of the N. crassa strains and all of the N. tetrasperma strains originated from Franklin, Louisiana. The remaining strains originated from collection sites in Hawaii, Louisiana (not Franklin), Montana, New Mexico, Haiti, Ivory Coast, Liberia, Pakistan, and Panama. All of the N. crassa and N. tetrasperma strains used in this study are part of the Perkins collection [73] administered by the Fungal Genetics Stock Center (FGSC) or from FGSC's own collection. The strains of N. discreta used in this study are from the personal collection of David Jacobsen and were acquired from the John Taylor Laboratory at UC Berkeley. All strains from which genomic DNA was sequenced were cultivated on Vogel's minimal media (VMM) [74] on slants and plates at 22° or 34° C by standard methods [74]. Strains from which RNA was sequenced by RNA-Seq were grown on a VMM plate at 25°C for 25 hours under constant light. A plug of hyphae was cut from each plate and transferred to a Bird's media [75] plate overlaid with cellophane. The culture was incubated at 25°C under constant light.
RNA extraction
Mycelia was harvested and immediately added to 1 mL of TRIzol reagent (Invitrogen Life Technologies) and Zirconia/silica beads (0.2 g, 0.5-mm diameter; Biospec Products). Cells were disrupted using a MiniBeadBeater instrument (Biospec Products) at maximum speed for 30 seconds twice in succession. Total RNA was extracted according to the manufacturer's protocol for TRIzol (Invitrogen). Total RNA was quantified by bioanalyzer (Agilent).
cDNA synthesis
For polyA RNA purification, 10 µg of total RNA was bound to dynal oligo(dT) magnetic beads (Invitrogen 610.02) two times, using the manufacturer's instructions. Purified polyA RNA was fragmented by metal-ion catalysis [76] using fragmentation reagents from Ambion (AM12450). For first strand cDNA synthesis 1 µg fragmented polyA RNA was incubated with 3 µg random hexamers (Invitrogen 48190-011), and incubated at 65°C for 5 minutes and then transferred to ice. 1st strand buffer (Invitrogen 18064-014) was added to 1× final concentration (4 µL). Dithiothreitol (DTT), dNTPs and RNAseOUT (Invitrogen 10777-019) were added to 100 mM, 10 mM, and 20 U/20 µL respectively, the sample incubated at 25°C for 2 minutes. 200 U of Superserscript II (Invitrogen 18064-014) was added and the sample incubated at 25°C for 10 minutes, 42°C for 50 minutes and 70°C for 15 minutes.
For second strand synthesis, 51 µL of H2O, 20 µL of 5× second strand buffer (Invitrogen 10812-014), and dNTPs (10 mM) were added to the first strand cDNA synthesis mix and incubated on ice for 5 minutes. RNaseH (2 U) (Invitrogen 18021-014), DNA pol I (50 U) (Invitrogen 18010-017) were then added and the mixture was incubated at 16°C for 2.5 hours.
Library construction
End-repair was performed by adding 45µL of H2O, T4 DNA ligase buffer with 10 mM ATP (NEB B0202S) (10 µL), dNTP mix (10 mM), T4 DNA polymerase (15 U) (NEB M0203L), Klenow DNA polymerase (5 U) (NEB M0210S), and T4 PNK (50 U) (NEB M0201L) to the sample and incubating at 20°C for 30 minutes. A single base was added each cDNA fragment by adding Klenow buffer (NEB M0212L), dATP (1 mM), and Klenow 3′ to 5′ exo- (15 U) (NEB M0212L). The mixture was then incubated at 37°C in for 30 minutes.
Standard Illumina adapters (FC-102-1003) were ligated to the cDNA fragments using 2× DNA ligase buffer (Enzymatics L603-HC-L), 1 µL of adapters, and DNA ligase (5 U) (Enzymatics L603-HC-L). The sample was incubated at 25°C for 15 minutes.
The sample was purified in a 2% low-melting point agarose gel, and a slice of gel containing 200-bp fragments was removed and the DNA purified. The polymerase chain reaction (PCR) was used to enrich the sequencing library. A 10 µL aliquot of purified cDNA library was amplified by PCR. PCR cycling conditions were: a denaturing step at 98°C for 30 seconds, 14 cycles of 98°C for 10 seconds, 65°C for 30 seconds, 68°C for 30 seconds, and a final extension at 68°C for 5 minutes.
All libraries were sequence using an Illumina Genome Analyzer-II (Vincent J. Coates Genomic Sequencing Laboratory, UC Berkeley) using standard Illumina operating procedures.
Sequencing
Genomic DNA was extracted from all strains by the standard method of grinding in liquid nitrogen followed by phenol/chloroform purification [77]. Regions spanning the het-c/pin-c region were amplified by polymerase chain reaction (PCR) using standard methods. Primer sequences are available upon request. The resulting PCR fragments were purified and directly sequenced using ABI dye terminator chemistry using standard methods. Sequences for NCU03492 were amplified by PCR and sequenced by standard methods and by RNA-seq. All sequences have been submitted to Genbank under accession numbers HQ396330-HQ396439.
Phylogenetic analyses
Sequences used to construct phylogeny of pin-c and related genes for Figure 3 and Figure S1 were acquired from GenBank. Gene names are listed in Figure S1. Strain and gene information used to construct Figure 2 are summarized in Table S2. Sequences used for the construction of Figures 4 and 5 were generated in this study. Information on strain/gene name and accession numbers are listed in Table S3.
Alignments were performed using T-coffee [78]. Regions of the alignment containing gaps were removed from the analysis. Models of molecular evolution were selected using the Akaike Information Criterion (AIC) implemented in ProtTest 2.4 [79]. Bayesian AA analysis employed the Whelan-Goldman AA substitution protein matrix [80]. Using MrBayes 3.1.1 [91], two runs were carried out for 1 M generations. Each run included four chains, and trees were sampled every 100 generations. A burnin sample of 1000 trees was discarded. The remaining trees were used to estimate posterior probabilities and branch lengths with the sumt command in MrBayes [91].
Analysis of Recombination
The Genetic Algorithm Recombination Detection (GARD) method of Kosakovsky Pond et al was used to determine recombination break point [53]. Substitution models used in the GARD analysis were chosen for each analysis by comparison to the general time reversible model (GTR) [81] using the information criterion of Akaike [82]. Models used were Tamura-Nei (TrN) [83] for het-c (NCU03493) and Hasegawa, Kishino and Yano 1985 (HKY85) [84] for pin-c (NCU03494). For het-c1/pin-c1 intra-allele analyses, sequences used were from strains FGSC 1945 (AF196305.1, DQ309558.1), FGSC 1824 (HQ396342), FGSC 967 (HQ396387), P4452 (HQ396347), P4454 (HQ396333), P4456 (HQ396338), P4464 (HQ396350), P4480 (HQ396432), P4483 (HQ396377), P4487 (HQ396362), P4489 (HQ396422), FGSC 2489 (XM_950787.2, XM_950788.1), P4471 (HQ396391), P4479 (HQ396433), P4484 (HQ396388), P4490 (HQ396421), P4491 (HQ396383), FGSC 2190 (AF195874.1, DQ309557.1), FGSC 1130 (HQ396336), FGSC 4832 (HQ396394), P4448 (HQ396397), P4451 (HQ396357), P4455 (HQ396413), P4476 (HQ396340), P4486 (HQ396363), and P4496 (HQ396379) for het-c (NCU03493) and pin-c (NCU03494). FGSC 2489 (XM_950787.2, MX_950788.1), P4471 (HQ396391), P4479 (HQ396433), P4484 (HQ396388), P4490 (HQ396421) and P4491 (HQ396383) were used. For het-c2/pin-c2 intra-allele analyses, sequences from FGSC 2190 (AF195874.1, DQ309557.1), FGSC 1130 (HQ396336), FGSC 4832 (HQ396394), P4448 (HQ396397), P4451 (HQ396357), P4455 (HQ396413), P4476 (HQ396340), P4486 (HQ396363) and P4496 (HQ396379) were used. For the het-c3/pin-c3 intra-allele analyses, sequences from FGSC 1945 (AF196305.1, DQ309558.1), FGSC 1824 (HQ396342), FGSC 967 (HQ396387), P4452 (HQ396347), P4454 (HQ396333), P4456 (HQ396338), P4464 (HQ396350), P4480 (HQ396432), P4483 (HQ396377), P4487 (HQ396362), and P4489 (HQ396422) were used. The version of GARD used was that implemented on the Datamonkey web server [85] with no site-to-site rate variation and 2 rate classes.
Coalescent Analysis
We used the Langley-Fitch (LF) [55] method to fit phylogenetic trees for het-c (NCU03493) and pin-c (NCU03494) to geologic time. We used the LF implementation found in the r8s suit of programs [56]. We assumed a consistent molecular clock across the tree with the root dated at 200 million years ago, the estimated divergence time of the crown Ascomycota [34]. Trees for het-c and pin-c used in the coalescent analysis were calculated by maximum likelihood (ML) in PAUP* [86]. Alignments were performed using T-coffee [78]. Regions of the alignment containing gaps were removed from the analysis. Models of molecular evolution were selected using the Akaike Information Criterion (AIC) implemented in jModeltest [87]. The ML analysis employed the HKY85 model of sequence evolution was used with the gamma distribution.
Calculation of Ka/Ks and Tajima's D
Tajima's D [50] statistics and Ka/Ks were calculated for both het-c (NCU03493) and pin-c (NCU03494) both between and within allele classes for 29 strains of N. crassa. Aligned coding sequences of het-c (NCU03493) and pin-c (NCU03494) from FGSC 967 (het-c3/pin-c3, HQ396387), FGSC 1130 (het-c2/pin-c2, HQ396336), FGSC 1824 (het-c3/pin-c3, HQ396342), FGSC 1945 (het-c3/pin-c3, AF196305.1, DQ309558.1), FGSC 2190 (het-c2/pin-c2, AF195874.1, DQ309557.1), FGSC 2489 (het-c1/pin-c1, DQ309556.1, XM_950787.2), FGSC 4832 (het-c2/pin-c2, HQ396394), P4448 (het-c2/pin-c2, HQ396397), P4451 (het-c2/pin-c2, HQ396357), P4452 (het-c3/pin-c3, HQ396347), P4454 (het-c3/pin-c3, HQ396333), P4455 (het-c2/pin-c2, HQ396413), P4456 (het-c3/pin-c3, HQ396338), P4464 (het-c3/pin-c3, HQ396350), P4471 (het-c1/pin-c1, HQ396391), P4476 (het-c2/pin-c2, HQ396340), P4479 (het-c1/pin-c1, HQ396433), P4480 (het-c3/pin-c3, HQ396432), P4481 (het-c1/pin-c1, HQ396401), P4483 (het-c3/pin-c3, HQ396377), P4484 (het-c1/pin-c1, HQ396388), P4486 (het-c2/pin-c2, HQ396363), P4487 (het-c3/pin-c3, HQ396362). P4489 (het-c3/pin-c3, HQ396422), P4490 (het-c1/pin-c1, HQ396421), P4496 (het-c2/pin-c2, HQ396379), and P4501 (het-c3/pin-c3, HQ396427) were used for both analyses. The D statistic in all cases was calculated by the method of Tajima as implemented in DnaSP [88]. All segregating sites were considered. Two-tailed confidence limits of D were determined assuming a beta distribution as recommended by Tajima. Full length alignments were used to calculate D both within and between alleles. A 3 bp sliding window was used for codon by codon calculations of D.
The ratio of the rate of non-synonymous substitutions (Ka) to the rate of synonymous substitutions (Ks) was calculated for full length het-c (NCU03493) and pin-c (NCU03494) sequences from the same strains of N. crassa used to calculated the D statistic. The Ka/Ks ratio or ω was calculated for each codon in a multiple alignment using an evolutionary codon model [51], which enabled calculating ω at each codon site using a maximum-likelihood (ML) approach. The method used was that of Stern et al as implemented in Selecton 2.2 [52].
Supporting Information
Expanded phylogeny of pin-c and related genes.
(1.20 MB TIF)
Diversity at het-c/pin-c amongst Louisiana and global isolates of Neurospora crassa.
(1.05 MB TIF)
Representation of het-c/pin-c intergenic sequences in Neurospora.
(1.15 MB TIF)
Measures of Ka/Ks ratios and Tajima's D on the coding region of gsl-5 (NCU03492) do not show evidence for balancing or positive selection.
(1.37 MB TIF)
Analysis of recombination at het-c in N. crassa.
(1.60 MB TIF)
Analysis of recombination at pin-c in N. crassa.
(3.40 MB TIF)
Analysis of recombination in the het-c/pin-c intergenic region in N. crassa.
(1.25 MB TIF)
Estimated divergence dates of het-c and pin-c alleles from related HET domain genes.
(0.90 MB TIF)
Analysis of recombination within haplotypes at pin-c2 in N. crassa.
(1.30 MB TIF)
Analysis of recombination within haplotypes at pin-c3 in N. crassa.
(1.61 MB TIF)
Table of strains used in this study.
(0.14 MB DOC)
Sequence accessions used in figures 4 (gsl-5 and NCU03495) and 5 (het-c and pin-c).
(0.11 MB DOC)
Acknowledgments
We thank Dr. Monty Slatkin and Chris Ellison for their careful reading and comments on the manuscript and Dr. John W. Taylor for helpful discussions. We thank Dr. David Jacobson for providing strains and Dr. Kevin McCluskey at the FGSC for facilitating obtaining strains from the Perkins Collection.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this work is supported by an R01 grant from the National Institutes of Health to N.L.G. (GM60468) and a collaborative R24 grant from the National Institutes of Health to Drs. John W. Taylor, Rachel Brem (UCB) and N.L.G. (GM081597). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Figueroa F, Gunther E, Klein J. MHC polymorphisms pre-dating speciation. Nature. 1988;335:265–271. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- 2.Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 3.Dishaw LJ, Litman GW. Invertebrate allorecognition: the origins of histocompatibility. Curr Biol. 2009;19:R286–288. doi: 10.1016/j.cub.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasrallah J. Recognition and rejection of self in plant reproduction. Science. 2004;296:305–308. doi: 10.1126/science.296.5566.305. [DOI] [PubMed] [Google Scholar]
- 5.Charlesworth D. Multi-allelic self-incompatibility polymorphisms in plants. Bioessays. 1995;17:31–38. [Google Scholar]
- 6.Fleissner A, Leeder AC, Roca MG, Read ND, Glass NL. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci USA. 2009;106:19387–19392. doi: 10.1073/pnas.0907039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass NL, Dementhon K. Non-self recognition and programmed cell death in filamentous fungi. Curr Op Microbiol. 2006;9:553–558. doi: 10.1016/j.mib.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosberg RK, Hart MW. Mate selection and the evolution of highly polymorphic self/nonself recognition genes. Science. 2000;289:2111–2114. doi: 10.1126/science.289.5487.2111. [DOI] [PubMed] [Google Scholar]
- 10.Glass NL, Kaneko I. Fatal attraction: Nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2:1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biella S, Smith ML, Aist JR, Cortesi P, Milgroom MG. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc Royal Soc London - Series B: Biol Sci. 2002;269:2269–2276. doi: 10.1098/rspb.2002.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortesi P, McCulloch CE, Song H, Lin H, Milgroom MG. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectric parasitica. Genetics. 2001;159:107–118. doi: 10.1093/genetics/159.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debets F, Yang X, Griffiths AJ. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–119. doi: 10.1007/BF00313797. [DOI] [PubMed] [Google Scholar]
- 14.Debets A, Griffiths JF. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol Res. 1998;102:1343–1349. [Google Scholar]
- 15.Paoletti M, Saupe SJ. Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays. 2009;31:1201–1210. doi: 10.1002/bies.200900085. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson DJ, Beurkens K, Klomparens KL. Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet Biol. 1998;23:45–56. doi: 10.1006/fgbi.1997.1020. [DOI] [PubMed] [Google Scholar]
- 17.Newhouse JR, MacDonald WL. The ultrastructure of hyphal anastomoses between vegetatively compatible and incompatible virulent and hypovirulent strains of Cryphonectria parasitica. Can J Bot. 1991;69:602–614. [Google Scholar]
- 18.Marek SM, Wu J, Glass NL, Gilchrist DG, Bostock RM. Nuclear DNA degradation during heterokaryon incompatibility in Neurospora crassa. Fungal Genet Biol. 2003;40:126–137. doi: 10.1016/s1087-1845(03)00086-0. [DOI] [PubMed] [Google Scholar]
- 19.Hutchison E, Brown S, Tian C, Glass NL. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiol. 2009;155:3957–3970. doi: 10.1099/mic.0.032284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anwar MM, Croft JH, Dales RB. Analysis of heterokaryon incompatibility between heterokaryon-compatibility (h-c) groups R and GL provides evidence that at least eight het loci control somatic incompatibility in Aspergillus nidulans. J Gen Microbiol. 1993;139:1599–1603. doi: 10.1099/00221287-139-7-1599. [DOI] [PubMed] [Google Scholar]
- 21.Pal K, van Diepeningen AD, Varga J, Hoekstra RF, Dyer PS, et al. Sexual and vegetative compatibility genes in the aspergilli. Stud Mycol. 2007;59:19–30. doi: 10.3114/sim.2007.59.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortesi P, Milgroom MG. Genetics of vegetative incompatibility in Cryphonectria parasitica. Appl Environ Microbiol. 1998;64:2988–2994. doi: 10.1128/aem.64.8.2988-2994.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins DD. Main features of vegetative incompatibility in Neurospora crassa. Fungal Genet Newsl. 1988;35:44–46. [Google Scholar]
- 24.Saupe SJ, Kuldau GA, Smith ML, Glass NL. The product of the het-C heterokaryon incompatibility gene of Neurospora crassa has characteristics of a glycine-rich cell wall protein. Genetics. 1996;143:1589–1600. doi: 10.1093/genetics/143.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar S, Iyer G, Wu J, Glass NL. Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility. EMBO J. 2002;21:4841–4850. doi: 10.1093/emboj/cdf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko I, Dementhon K, Xiang Q, Glass NL. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics. 2006;172:1545–1555. doi: 10.1534/genetics.105.051490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Saupe SJ, Glass NL. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc Natl Acad Sci USA. 1998;95:12398–12403. doi: 10.1073/pnas.95.21.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 29.Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, et al. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:e1000891. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 32.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 33.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 35.Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends Genetics. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 37.Fedorova N, Badger J, Robson G, Wortman J, Nierman W. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics. 2005;6:177. doi: 10.1186/1471-2164-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saupe SJ, Glass NL. Allelic specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa is determined by a highly variable domain. Genetics. 1997;146:1299–1309. doi: 10.1093/genetics/146.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Glass NL. Identification of specificity determinants and generation of alleles with novel specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa. Mol Cell Biol. 2001;21:1045–1057. doi: 10.1128/MCB.21.4.1045-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell AJ, Jacobson DJ, Natvig DO. Allelic diversity at the het-c locus in Neurospora tetrasperma confirms outcrossing in nature and reveals an evolutionary dilemma for pseudohomothallic ascomycetes. J Mol Evol. 2001;52:94–102. doi: 10.1007/s002390010138. [DOI] [PubMed] [Google Scholar]
- 41.Kamau E, Charlesworth D. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr Biol. 2005;15:1773–1778. doi: 10.1016/j.cub.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 42.Dobzhansky T, Epling C. The suppression of crossing over in inversion heterozygotes of Drosophila pseudoobscura. Proc Natl Acad Sci USA. 1948;34:137–141. doi: 10.1073/pnas.34.4.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Micali CO, Smith ML. A nonself recognition gene complex in Neurospora crassa. Genetics. 2006;173:1991–2004. doi: 10.1534/genetics.106.057562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mir-Rashed N, Jacobson DJ, Dehghany MR, Micali OC, Smith ML. Molecular and functional analyses of incompatibility genes at het-6 in a population of Neurospora crassa. Fungal Genet Biol. 2000;30:197–205. doi: 10.1006/fgbi.2000.1218. [DOI] [PubMed] [Google Scholar]
- 45.Powell AJ, Jacobson DJ, Natvig DO. Ancestral polymorphism and linkage disequilibrium at the het-6 region in pseudohomothallic Neurospora tetrasperma. Fungal Genet Biol. 2007;44:896–904. doi: 10.1016/j.fgb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saupe S, Turcq B, Bégueret J. Sequence diversity and unusual variability at the het-c locus involved in vegetative incompatibility in the fungus Podospora anserina. Curr Genet. 1995;27:466–471. doi: 10.1007/BF00311217. [DOI] [PubMed] [Google Scholar]
- 49.Chevanne D, Saupe SJ, Clavé C, Paoletti M. WD-repeat instability and diversification of the Podospora anserina HNWD non-self recognition gene family. BMC Evol Biol. 2010;10:134. doi: 10.1186/1471-2148-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 52.Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, et al. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 54.Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 55.Langley CH, Fitch WM. An examination of the constancy of the rate of molecular evolution. J Mol Evol. 1974;3:161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- 56.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 57.Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiu PKT, Glass NL. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics. 1999;151:545–555. doi: 10.1093/genetics/151.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Espagne E, Balhadere P, Penin ML, Barreau C, Turcq B. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics. 2002;161:71–81. doi: 10.1093/genetics/161.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chevanne D, Bastiaans E, Debets A, Saupe SJ, Clavé C, et al. Identification of the het-r vegetative incompatibility gene of Podospora anserina as a member of the fast evolving HNWD gene family. Curr Genet. 2009;55:93–102. doi: 10.1007/s00294-008-0227-5. [DOI] [PubMed] [Google Scholar]
- 61.Paoletti M, Clavé C. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot Cell. 2007;6:2001–2008. doi: 10.1128/EC.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Diepeningen AD, Pal K, van der Lee TA, Hoekstra RF, Debets AJ. The het-c heterokaryon incompatibility gene in Aspergillus niger. Mycol Res. 2009;113:222–229. doi: 10.1016/j.mycres.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Kerenyi Z, Olah B, Jeney A, Hornok L, Leslie JF. The homologue of het-c of Neurospora crassa lacks vegetative compatibility function in Fusarium proliferatum. Appl Environ Microbiol. 2006;72:6527–6532. doi: 10.1128/AEM.01543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saupe SJ, Clavé C, Sabourin M, Bégueret J. Characterization of hch, the Podospora anserina homolog of the het-c heterokaryon incompatibility gene of Neurospora crassa. Curr Genetics. 2000;38:39–47. doi: 10.1007/s002940000130. [DOI] [PubMed] [Google Scholar]
- 65.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 66.Fisher R. A model for the generation of self-sterility alleles. J Theoret Biol. 1961;1:411–414. [PubMed] [Google Scholar]
- 67.Edh K, Widen B, Ceplitis A. The evolution and diversification of S-locus haplotypes in the Brassicaceae family. Genetics. 2009;181:977–984. doi: 10.1534/genetics.108.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takuno S, Fujimoto R, Sugimura T, Sato K, Okamoto S, et al. Effects of recombination on hitchhiking diversity in the Brassica self-incompatibility locus complex. Genetics. 2007;177:949–958. doi: 10.1534/genetics.107.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kusaba M, Nishio T, Satta Y, Hinata K, Ockendon D. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc Natl Acad Sci USA. 1997;94:7673–7678. doi: 10.1073/pnas.94.14.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Hughes AL, Tsukamoto T, Ando T, Kao T. Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 2001;125:1012–1022. doi: 10.1104/pp.125.2.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muirhead CA, Glass NL, Slatkin M. Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics. 2002;161:633–641. doi: 10.1093/genetics/161.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howlett B, Leslie JF, Perkins DD. Putative multiple alleles at the vegetative (heterokaryon) incompatibility loci het-c and het-8 in Neurospora crassa. Fungal Genet Newsl. 1993;40:40–42. [Google Scholar]
- 73.Perkins DD, Turner BC, Barry EG. Strains of Neurospora collected from nature. Evolution. 1976;30:281–313. doi: 10.1111/j.1558-5646.1976.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 74.Vogel HJ. A convenient growth medium for Neurospora. Microbiol Genet Bull. 1956;13:42–46. [Google Scholar]
- 75.Metzenberg RL. Bird Medium: an alternative to Vogel Medium. Fungal Genet Newsl. 2004;51:19–20. [Google Scholar]
- 76.Browne KA. Metal ion-catalyzed nucleic acid alkylation and fragmentation. J Am Chem Soc. 2002;124:7950–7962. doi: 10.1021/ja017746x. [DOI] [PubMed] [Google Scholar]
- 77.Cambareri EB, Kinsey JA. An ultra-fast method of DNA extraction from Neurospora. Fungal Genet Newsl. 1993;40:22–23. [Google Scholar]
- 78.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 79.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 80.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 81.Tavare S. Some probabilistic and statistical problems in the analysis of DNA sequence. Providence, RI: Amer Math Soc; 1986. [Google Scholar]
- 82.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19 [Google Scholar]
- 83.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 84.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 85.Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 86.Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 87.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 88.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 89.Gaff DF, Okong'O-Ogola O. The use of non-permeating pigments for testing the survival of cells. J Exp Bot. 1971;22:756–758. [Google Scholar]
- 90.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 91.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded phylogeny of pin-c and related genes.
(1.20 MB TIF)
Diversity at het-c/pin-c amongst Louisiana and global isolates of Neurospora crassa.
(1.05 MB TIF)
Representation of het-c/pin-c intergenic sequences in Neurospora.
(1.15 MB TIF)
Measures of Ka/Ks ratios and Tajima's D on the coding region of gsl-5 (NCU03492) do not show evidence for balancing or positive selection.
(1.37 MB TIF)
Analysis of recombination at het-c in N. crassa.
(1.60 MB TIF)
Analysis of recombination at pin-c in N. crassa.
(3.40 MB TIF)
Analysis of recombination in the het-c/pin-c intergenic region in N. crassa.
(1.25 MB TIF)
Estimated divergence dates of het-c and pin-c alleles from related HET domain genes.
(0.90 MB TIF)
Analysis of recombination within haplotypes at pin-c2 in N. crassa.
(1.30 MB TIF)
Analysis of recombination within haplotypes at pin-c3 in N. crassa.
(1.61 MB TIF)
Table of strains used in this study.
(0.14 MB DOC)
Sequence accessions used in figures 4 (gsl-5 and NCU03495) and 5 (het-c and pin-c).
(0.11 MB DOC)