Abstract
Repulsive guidance molecule c (RGMc) plays a critical role in iron metabolism. Inactivating mutations cause juvenile hemochromatosis, a severe iron overload disorder. Understanding mechanisms controlling RGMc biosynthesis has been hampered by minimal information about the RGMc gene. Here we define the structure, examine the evolution, and establish mechanisms of regulation of the mouse RGMc gene. RGMc is a 4-exon gene that undergoes alternative RNA splicing to yield 3 mRNAs with 5′ different untranslated regions. Gene transcription is induced during myoblast differentiation, producing all 3 mRNAs. We identify 3 critical promoter elements responsible for transcriptional activation in skeletal muscle, comprising paired E-boxes, a putative Stat and/or Ets element, and a MEF2 site, and muscle transcription factors myogenin and MEF2C stimulate RGMc promoter function in non-muscle cells. As these elements are conserved in RGMc genes from multiple species, our results suggest that RGMc has been a muscle-enriched gene throughout its evolutionary history.
Keywords: Repulsive guidance molecule (RGM), RGMc, hemojuvelin, juvenile hemochromatosis, transcriptional regulation, molecular evolution
Introduction
Iron-related metabolic and hematologic disorders affect millions of individuals worldwide. Iron plays a critical role in numerous cellular processes ranging from oxygen exchange and energy metabolism [1] to nucleic acid synthesis and DNA repair [2], yet too much or too little iron can cause severe tissue and organ damage [3]. As a result, iron levels are tightly regulated in humans and other mammalian species [4], with primary control being exerted at the level of absorption from the small intestine [4; 5]. Hemojuvelin (HJV) or repulsive guidance molecule c (RGMc) is a recently identified gene that was initially linked to systemic iron metabolism by the discovery that mutations in humans caused the rapidly progressive and severe iron overload disorder, juvenile hemochromatosis (JH) [6; 7]. This relationship was strengthened when mice engineered to lack RGMc also developed iron overload [8; 9]. As RGMc/HJV appears to indirectly regulate the expression of hepcidin [10; 11; 12], a peptide hormone made in the liver that negatively controls intestinal iron absorption in the duodenum [5; 13], it is thus a component of a homeostatic pathway that regulates iron uptake [3; 8; 9; 10; 12].
RGMc was discovered not only as a gene mutated in JH [10], but also was identified as a novel transcript expressed during skeletal muscle differentiation [14], and as a member of a conserved three-gene family that receives its name from the axonal guidance molecule RGMa [15; 16; 17; 18]. Unlike RGMa or RGMb, RGMc is not expressed in the central nervous system, but rather is produced by striated muscle and by hepatocytes in the liver [14; 15; 18]. During development RGMc transcripts are expressed first in the somites in both mice and zebrafish [14; 17; 19], and then later in skeletal muscle, as well as in the embryonic heart and liver [9; 14]. This unique pattern of RGMc expression in striated myocytes and hepatocytes is maintained in the adult (Fig. 1A). To date, the responsible molecular mechanisms for tissue-specific gene expression have not been elucidated, and very little is known about RGMc gene regulation in any species, as no promoter has been characterized. Here we define the structure of the mouse RGMc gene and identify the DNA elements responsible for its high-level gene transcription in skeletal muscle. Further analysis reveals that these cis-acting muscle-specifying DNA elements are highly conserved in RGMc genes from multiple mammalian species, supporting the hypothesis that RGMc has been a muscle-enriched gene throughout its evolutionary history.
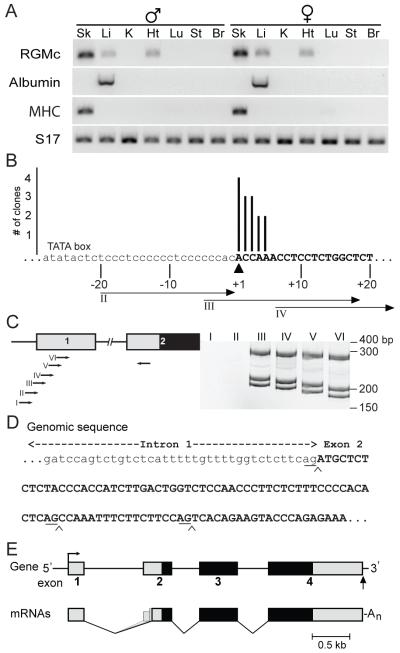
Figure 1. Establishing mouse RGMc gene structure.
A. RGMc mRNA is expressed in striated muscle and in the liver. Results of RT-PCR experiments for RGMc, albumin, skeletal muscle myosin heavy chain polypeptide 3 (MHC), and S17 mRNAs using RNA from tissues (Sk, skeletal muscle (gastrocnemius); Li, liver; K, kidney; Ht, heart; Lu, lung; St, stomach; Br, brain) of adult male (left) and female (right) mice. B. Mapping the 5′ end of the mouse RGMc gene by 5′ RACE using mouse skeletal muscle RNA. The number of clones is graphed on the y-axis above the corresponding location of the 5′ residue on the x-axis. The putative transcription start site is denoted as +1 (arrowhead), with exon 1 in upper case letters. A potential TATA box is labeled, and primers II - IV used in (C) are indicated below the sequence. C. Mapping the 5′ end of the mouse RGMc gene by RT-PCR with cDNA from mouse skeletal muscle RNA and overlapping PCR primers located in different parts of RGMc exon 1, as seen on the gene map to the left (see Supplemental Table 1 for DNA sequences of primers). Exons 1 and 2 are depicted as boxes, with the 5′ UTR in gray and the protein coding region in black, and introns and flanking DNA as horizontal lines. Results are seen to the right, and molecular weight markers are indicated (see Supplemental Fig. 1A for results with heart and liver RNA). In addition to mapping the 5′ end of exon 1, the results also show that alternative RNA splicing occurs between exons 1 and 2. D. DNA sequence of the junction between intron 1 and exon 2 of the mouse RGMc gene. Exon 2 is in upper case letters; the locations of alternative RNA splicing are noted by chevrons, with the –AG splice-acceptor residues underlined. E. Organization of the mouse RGMc gene and mRNAs. The gene contains 4 exons (boxes) and three introns (thin lines). The transcription start site is denoted as a bent arrow, and the polyadenylation site as a vertical arrow. The three RGMc mRNAs are diagramed below, and result from use of alternative splice acceptor sites at the 5′ end of exon 2.
Results
Defining RGMc gene structure
Analyses of genomic databases suggest that RGMc is a 4-exon gene in mice, humans, and several other species [20], but the 5′ end of exon 1 has not been established in any species, and the promoter has not been characterized. We thus mapped the transcription start site for mouse RGMc as a means to first identify and then functionally dissect the promoter. RGMc mRNA is expressed in adult male and female mouse skeletal and cardiac muscle and in the liver with no apparent gender differences in transcript abundance, at least in the C57Bl6 strain (Fig. 1A). By 5′ RACE we mapped the RGMc transcription start site (TSS) in skeletal muscle, and found that the 5′ end of 14/15 independent cDNA clones clustered within a 5-nucleotide region that was located ~25 nucleotides 3′ to a putative TATA box [21] in genomic DNA (Fig. 1B). We obtained similar results by RT-PCR with overlapping exon 1-specific primers and mouse muscle RNA (Fig. 1C), which validated the same TSS with RNA from mouse heart and liver (Supplemental Fig. 1).
The RT-PCR experiments designed to map the 5′ end of RGMc exon 1 used a common primer located in exon 2 (Fig. 1C), and results consistently yielded 3 distinct cDNAs that differed in length by 18 to 77 nucleotides (Fig. 1C and Supplemental Fig. 1). By DNA sequencing, all three classes of cDNAs contained identical parts of RGMc exon 1, but differed in the extent of exon 2 (all sequences matched mouse RGMc genomic DNA, Fig. 1D and Supplemental Fig. 1). We interpret these results to indicate that the mouse RGMc gene undergoes alternative RNA splicing to generate transcripts with varying lengths of exon 2, a hypothesis supported by evidence for splice acceptor sites at each of the three putative junctions between intron 1 and exon 2 (AG nucleotides underlined in Fig. 1D). Additional support comes from an expressed mouse sequence tag in GenBank (AI196626), which matches the intermediate-sized version of exon 2. A similar intermediate-sized exon 2 has been identified for human RGMc (EST numbers: DA762328 and DA764726). Also, comparative analyses of 10 mammalian species reveals sufficiently similar genomic DNA sequences to suggest that alternative RNA splicing is a common feature of many RGMc genes (Supplemental Fig. 1). Taken together, our results show that mouse RGMc is a 4-exon gene with a discrete TSS in exon 1 and alternative RNA splicing involving exon 2 that leads to three distinct transcripts that vary in the length of the 5′ untranslated region (Fig. 1E).
RGMc gene transcription is induced during skeletal muscle differentiation
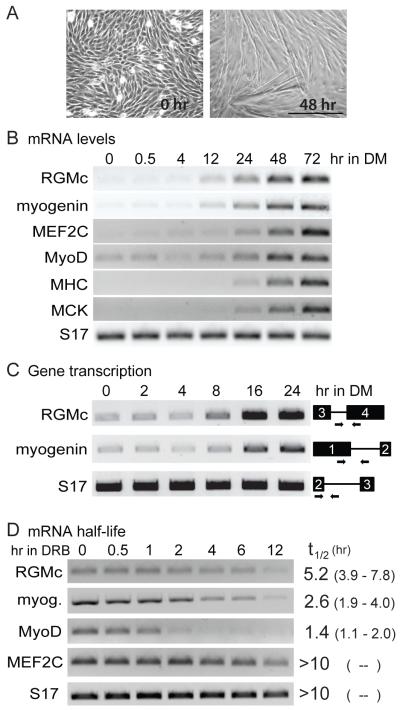
We next examined RGMc gene expression during skeletal muscle differentiation, using the C2 myoblast line as a model [22; 23; 24] (Fig. 2A). RGMc mRNA was detected within 12 hr after onset of C2 cell differentiation, and its abundance increased progressively during the subsequent 60 hr in a pattern similar to myogenin, a critical transcription factor that is expressed early in muscle differentiation [25; 26] (Fig. 2B). Accumulation of RGMc mRNA in differentiating C2 myoblasts appeared to be a secondary to induction of RGMc gene transcription, as measured by stimulation of nascent nuclear RGMc RNA beginning at ~8 hr after addition of DM, a pattern that was temporally similar to myogenin gene activation (Fig. 2C). We also examined RGMc mRNA stability in myoblasts after 48 hr of differentiation [27], and found that RGMc is a moderately long-lived mRNA, with a half-life of ~5.2 hrs, more than twice that of myogenin, and nearly four times as long as MyoD (Fig. 2D). Taken together, results in Fig. 2 demonstrate that induction of RGMc gene transcription is a critical regulatory step responsible for accumulation of RGMc mRNA in differentiating muscle cells.
Figure 2. RGMc gene transcription is induced during skeletal muscle differentiation.
A. Myotube formation occurs during C2 myoblast differentiation, as illustrated by phase contrast microscopy (200X magnification) at confluent cell density (0 hr) and after incubation in DM for 48 hr. Scale bar is 250 μm. B. Time course of gene expression for RGMc, myogenin, MEF2C, MyoD, myosin heavy chain (MHC), muscle creatine kinase (MCK), and S17 during C2 myoblast differentiation measured by RT-PCR. C. Time course of RGMc, myogenin, and S17 gene transcription during C2 myoblast differentiation, as measured by accumulation of nascent nuclear transcripts. Gene maps are to the right, and show approximate locations of intron-exon primer pairs (see Supplemental Table 1 for DNA sequences of primers). Exons appear as black boxes (exon sizes are not to scale). D. Measurement of mRNA half-life for RGMc, myogenin, MyoD, MEF2C, and S17 mRNAs in differentiating C2 myoblasts. The T½ for each mRNA is listed to the right of a representative experiment (the 95% confidence interval is in parentheses). See ‘Materials and Methods’ for details.
Analysis of RGMc promoter function in muscle differentiation
To investigate RGMc promoter function in myoblasts, we first showed that a genomic fragment containing ~4.1 kb of 5′ flanking DNA plus 118 nt of exon 1 could stimulate luciferase reporter activity in differentiating C2 myoblasts and in 10T½ mesenchymal stem cells expressing the muscle determination factor, MyoD, by infection with a recombinant adenovirus [28]. In each cell line, RGMc promoter activity was enhanced by ~35-fold after induction of muscle differentiation to levels ~10% of a myogenin promoter - reporter plasmid, whose activity also was stimulated during differentiation (Fig. 3A). In contrast, neither RGMc nor myogenin were activated in 10T½ cells in the absence of MyoD (data not shown), providing further support for the idea that RGMc gene transcription is induced during skeletal muscle differentiation.
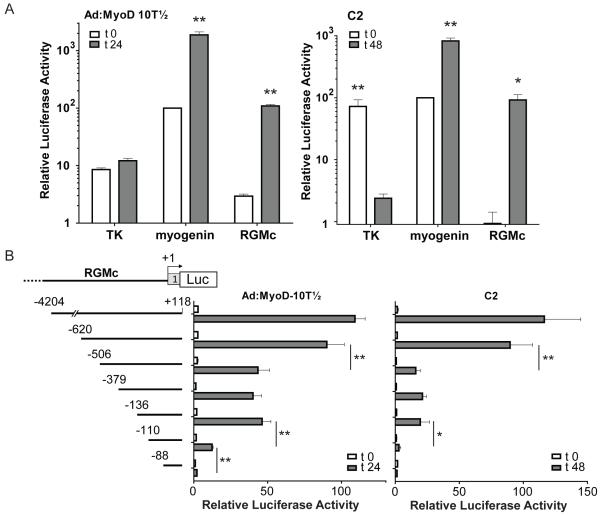
Figure 3. Mapping the regions of the RGMc promoter that are induced during muscle differentiation.
Results of luciferase assays in differentiating Ad:MyoD-10T½ cells (left panels) and C2 myoblasts (right panels) that were transiently transfected with reporter genes containing (A) the minimal thymidine kinase (TK) promoter, the mouse myogenin promoter, or the mouse RGMc promoter (coordinates −4204 to +118), and (B) reporter genes containing a series of 5′ truncations of the mouse RGMc promoter. Cells were incubated in DM for 0 (white bars), or 24 or 48 hr (gray bars). The graphs summarize results of 3 - 5 independent experiments (mean ± S.E.), each performed in duplicate (* - p < 0.05, ** - p < 0.005). Myogenin promoter values at t 0 have been set to 100 in each graph (average measurements at t 0 were 7.8 × 104 (Ad-MyoD-10T½ cells) or 7.3 × 103 (C2 cells) light units/μg total protein/sec).
To identify the DNA elements responsible for the transcriptional activity of RGMc during muscle differentiation we analyzed a series of 5′ promoter deletions. Three major regions were identified based on declines in reporter gene activity when each segment was eliminated: nucleotides −620 to −506, −136 to −110, and −110 to −88 (Fig. 3B and Supplemental Fig. 2).
We looked for additional transcriptional control regions that might be active in muscle differentiation, and evaluated 3 regions that spanned the entire body of the mouse RGMc gene and 3′ flanking DNA (Fig. 4A). As none of these DNA fragments altered induction of RGMc promoter activity during muscle differentiation (Fig. 4B), the results indicate no other muscle transcriptional enhancers (or repressors) are located outside of the RGMc proximal promoter.
Figure 4. Analyzing the RGMc gene for potential transcriptional enhancers.
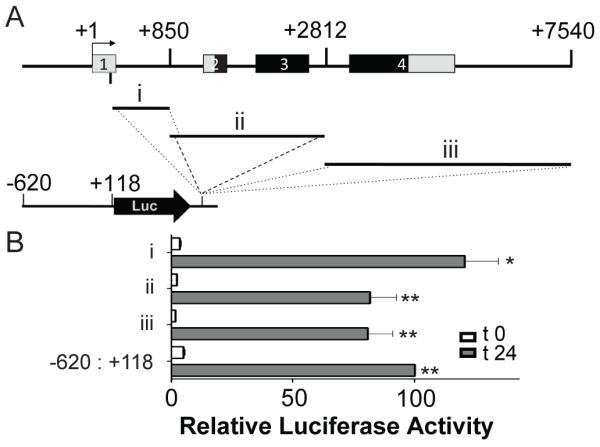
A. Map of mouse RGMc gene showing regions that were fused downstream of firefly luciferase (Luc) and the RGMc promoter (coordinates −620 to +118) to test for enhancer activity in differentiating Ad-MyoD-10T½ cells. B. Graphs depict results of luciferase assays after incubation in DM for 0 (white bars) or 24 hr (gray bars) (mean ± S.E. of 3 experiments, each performed in duplicate; values for the RGMc promoter at 24 hr were set to 100 (* - p < 0.01, ** - p < 0.001, vs. t 0)).
Identifying proximal promoter elements responsible for RGMc transcriptional activity during muscle differentiation
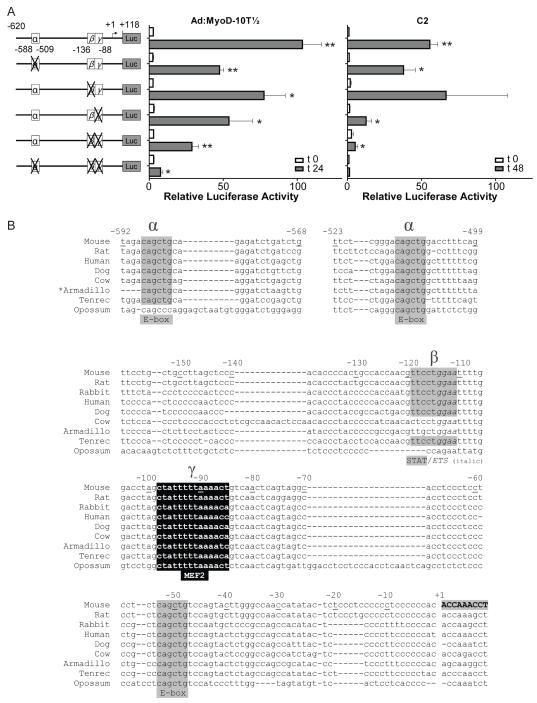
We introduced inactivating nucleotide substitutions into DNA sequences potentially responsible for RGMc promoter activity during muscle differentiation (Fig. 3). Mutation of two E-boxes (putative binding sites for myogenic basic helix-loop-helix (bHLH) transcription factors, including MyoD and myogenin, with the consensus sequence, CANNTG) in the segment from −620 to −506 (α-element, Fig. 5) resulted in a 50% decrease in promoter activity in MyoD-expressing 10T½ cells, and a 25% decline in C2 myoblasts (Fig. 5). Disruption of a putative myocyte enhancer factor 2 (MEF2) sequence from −110 to −88 (γ, Fig. 5) caused a 50% reduction in MyoD 10T½ cells, and a ~75% decrease in C2 myoblasts (Fig. 5). By contrast, elimination of a potential Stat binding site (TTCN3GAA [29; 30; 31]) and/or Ets element (GGA(A/T) [32; 33; 34]) from −136 to −110 (β, Fig. 5), was less effective, and led to only a ~25% decline in promoter activity in MyoD 10T½ cells, and had no effect in C2 myoblasts, although when combined with mutation of the γ region, RGMc promoter activity was decreased by ~75 - 90% (Fig. 5A). Mutation of all three elements reduced reporter gene expression to basal levels, results that we interpret to demonstrate that together these three proximal promoter sites are responsible for RGMc transcriptional activity during skeletal muscle differentiation. As depicted in Fig. 5B, both of the E-boxes and the MEF2 site are highly conserved in putative RGMc gene promoters from 9 mammalian species, while the postulated Stat/Ets composite element is less conserved.
Figure 5. Characterizing promoter elements that control RGMc gene transcription during muscle differentiation.
A. Results are depicted of luciferase assays in differentiating Ad:MyoD-10T½ cells (left panel) and C2 myoblasts (right panel) transiently transfected with reporter genes containing substitution mutations of the mouse RGMc promoter (illustrated on the maps to the far left; details are in (B) and ‘Materials and Methods’). Cells were incubated in DM for 0 (white bars), or 24 or 48 hr (gray bars) before analysis. The graphs depict results of 3 - 10 independent experiments (mean ± S.E.), each performed in duplicate (* - p < 0.05, ** - p < 0.001, vs. t 0). B. Comparative mapping of RGMc promoter elements from different species. DNA sequence alignment of part of the proximal RGMc promoter from 9 mammalian species. Highlighted regions include paired E-boxes at −588 to −583 and −514 to −509 (α-site), the β-site from −120 to −110, and the γ site, a putative MEF2 element, from −98 to −85. Another E-box also is indicated at −53 to −48. Mouse exon 1 is in upper case and bold letters.
To directly test the role of MEF2 proteins and myogenic bHLH transcription factors in regulating RGMc promoter function, we performed co-transfection experiments in non-muscle 10T½ cells. Constitutively active MEF2C (MEF2C-VP16) was able to stimulate RGMc promoter activity by ~10-fold over an empty vector control, but had little effect on an RGMc promoter-reporter gene with an inactivating mutation in the MEF2 site (Fig. 6A). Similarly, myogenin was able to increase RGMc promoter activity by > 20-fold, but this was reduced by only ~50% for the α-element mutant (Fig. 6B), possibly because another conserved E-box is found in the RGMc proximal promoter at −53 to −48 (Fig. 5B). In contrast to these results, constitutively active Stat5b was not able to stimulate RGMc promoter activity (data not shown).
Figure 6. Stimulation of RGMc promoter activity by myogenin and MEF2C.
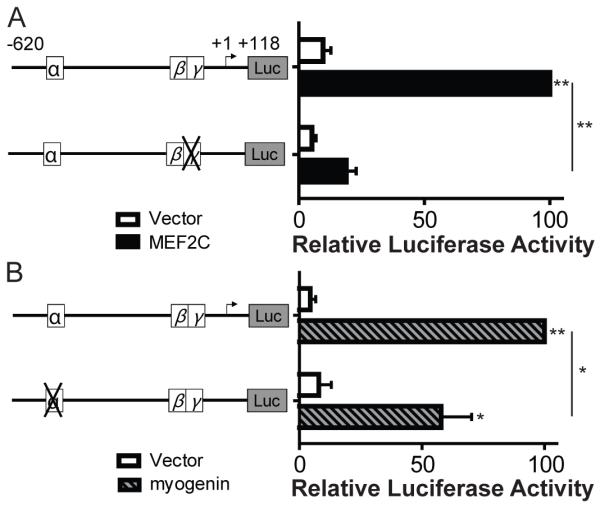
Effects of MEF2C (A) or myogenin (B) on the activity of wild-type (wt) or mutant (depicted as an X) RGMc promoters in 10T½ cells. A. Results of luciferase assays after co-transfection with an expression plasmid for constitutively active MEF2C (MEF2C-VP16) or pcDNA3 (vector) and either wt RGMc promoter or the γ-mutant. B. Results of luciferase assays after co-transfection of a myogenin or EGFP (vector) expression plasmids with either wt RGMc promoter or the α-mutant. For A and B the graphs represents results of 3 independent experiments (mean ± S.E.), each performed in duplicate (* - p < 0.05, ** - p < 0.005).
Little activity of the RGMc promoter in liver cell lines
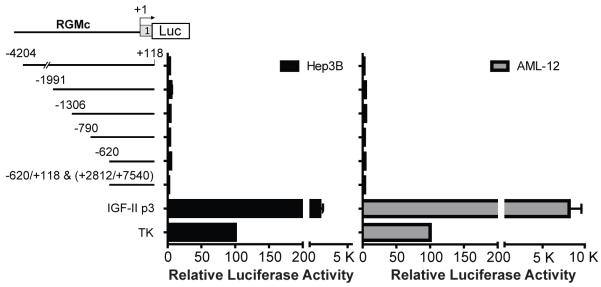
RGMc may be one of the only genes expressed exclusively in striated muscle and in hepatocytes. To investigate RGMc promoter function in liver cells, we performed a series of transfection experiments with mouse RGMc promoter-luciferase fusion genes into human Hep3B liver cancer cells, which have been reported to produce RGMc mRNA [12], and into mouse AML-12 hepatocytes. None of the 5 promoter deletions tested were active, as luciferase values were < 10% of what was measured with a thymidine kinase promoter-reporter plasmid, and were < 1% of the activity seen with mouse Igf2 promoter 3 (Fig. 7). Similarly negative results were seen when the region from +2812 to +7540 of the mouse RGMc gene was added to the promoter-reporter plasmid (Fig. 7), or when the RGMc promoter-luciferase fusion genes were transfected into human HepG2 hepatocarcinoma cells (Supplemental Fig. 3). Based on these results, it appears that the regulatory domains responsible for RGMc transcriptional activity in liver cells do not map to the promoter or the body of the gene, or that the transcription factors necessary for RGMc expression are not produced in Hep3B, AML-12, or HepG2 cells.
Figure 7. The RGMc promoter is not active in the Hep3B or AML-12 liver cell lines.
The graphs summarize results of luciferase assays using Hep3B human hepatoma cells (left) or mouse AML-12 hepatocytes (right) transiently transfected with reporter genes containing different regions of mouse the RGMc promoter and gene. Cells were incubated in growth media for 24 hr before analysis (mean of 2 independent experiments in duplicate for RGMc constructs and 5 for control plasmids, IGF-II promoter (p) 3 - luciferase and TK-luciferase). Values for TK-luciferase were set to 100 relative luciferase activity units (average measurements were 3 × 104 (Hep3B cells) or 8 × 103 (AML-12 cells) light units/μg total protein/sec).
Evolutionary conservation of the RGMc gene promoter
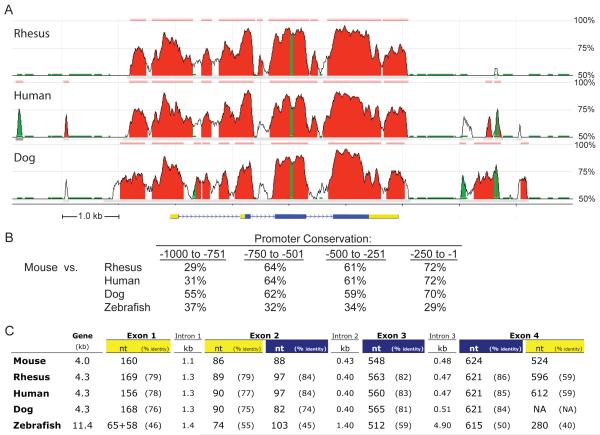
An assumption in genomics is that evolutionarily well-conserved regions of the genome are more likely to be functionally important than are segments that have diverged. Although counter examples exist where ultra-conserved DNA when deleted in transgenic mice showed little effect on phenotype [35], there are numerous individual cases where evolutionary conservation has provided critical insight into gene function [36; 37]. The mouse RGMc promoter elements identified here as being responsible for RGMc gene regulation in muscle are highly conserved among mammals, although are less so in zebrafish (Fig. 5B), and the 750 bp of the proximal promoter 5′ to the TSS is nearly as conserved in mammals as RGMc exon 1, although it is less than the coding exons (Fig. 8A and C). Of note, even though the putative RGMc promoter is more divergent in zebrafish, islands of conserved sequences for potential muscle-specific transcriptional elements are found adjacent to the putative TSS (see Supplemental Fig. 4 and discussion below), which is consistent with the hypothesis that RGMc expression in muscle is evolutionarily ancient.
Figure 8. Evolutionary conservation of the RGMc gene and promoter.
A. Graphs depicting nucleotide conservation between mouse RGMc and either rhesus macaque, human, or dog RGMc/HJV. The horizontal length of each conserved block on the x-axis corresponds to the length of the alignment, and the y-axis depicts the percent identity over a 100 bp sliding window for each block between mouse and the corresponding species listed (results derived from tools in Ref. [66]). Regions with >70% identity over 100 bp are shown in red and highlighted above the alignment, with repetitive elements in green within these blocks. Regions below 50% conservation are not pictured. Below the graphs is a diagram of mouse RGMc (non-coding regions in yellow, protein coding sequences in blue). B. RGMc promoter conservation between mouse and rhesus macaque, human, dog, or zebrafish presented in 250 nt increments from 1 to 1000 nt 5′ to the TSS. Alignments were created using the EMBOSS Needleman-Wunsch (GLOBAL) alignment algorithm utilizing the EDNAFULL substitution matrix from the European Bioinformatics institute (EBI) with a Gap Penalty of 10.0 and Extension Penalty of 0.5. NCBI reference sequences include mouse, NT_039240.7; rhesus macaque, NW_001108926; human, NT_167185.1; dog, NW_876264.1; zebrafish, NW_001877680.2. C. Characteristics of the RGMc gene in mouse, rhesus macaque, human, dog, and zebrafish. Exons are numbered with lengths in bp (and introns in kb), with non-coding regions in yellow and coding sequences in blue as in (A), and % identity in relation to mouse RGMc noted in parentheses. Exon 2 splice variants are not shown for simplicity (for additional details see Fig. 1). Only the smallest exon 2 is compared. There are no full-length cDNAs in public databases for rhesus macaque or dog RGMc, and the lengths of exon 1 have been derived based on similarity with mouse and human exon 1. Similarly, there is no information available for the 3′ UTR of dog RGMc mRNA that would correspond to the non-coding part of exon 4 (listed as NA in the figure). There is a 138 nt repetitive DNA element in dog RGMc exon 3, which has been eliminated to allow comparisons with the other species (the actual length of dog exon 3 is 703 nt). The zebrafish RGMc gene contains 5 exons. Exons 1 and 2 are listed together as 65+58 nt (exon 1 is 65 nt, exon 2 is 58 nt, with a 2.0 kb intron separating them).
Discussion
Experiments presented in this manuscript delineate the topography of the mouse RGMc gene and define mechanisms of RGMc gene regulation. Key findings include the demonstration of alternative RNA splicing between exons 1 and 2, which leads to expression in muscle and liver of three distinct RGMc transcripts that vary in the length of the 5′ UTR, and the identification and characterization of DNA elements in the proximal RGMc promoter responsible for RGMc gene transcription in skeletal muscle. Additionally, we show that a 4 kb segment of the RGMc promoter is insufficient to direct transcription in three liver cell lines. Comparative analysis of RGMc genes from 10 mammalian species further supports the hypothesis that both differential RNA processing and at least two of the three the muscle-specific promoter elements have been evolutionarily conserved.
Many genes undergo alternative RNA splicing [38; 39; 40; 41], which can lead to transcripts encoding different protein species [38; 42] or containing distinct regulatory properties, such as differential stability or translatability [43]. The precise mechanisms that control alternative splicing are complex, and full understanding of how splice-site selection is determined remains incomplete [39]. For RGMc the three mRNAs characterized here vary in the length of the 5′ UTR, and appear to be expressed at fairly equivalent levels in muscle and liver. Further studies will be needed to determine if there are unique functional properties for each transcript.
Like several other genes expressed in skeletal muscle, RGMc gene transcription is controlled by a combinatorial interplay of muscle-restricted and more broadly expressed transcription factors, including members of the myogenic bHLH family, which includes Myf-5, MyoD, myogenin, and MRF4 [44; 45; 46], and the MEF2-family, MEF2A - D [47; 48]. MEF2C has been shown to be directly activated by and to cooperate functionally with myogenic bHLH proteins during muscle differentiation in vitro [49], and during skeletal muscle development in vivo [50]. Our results demonstrate that a set of paired E-boxes (α-element) and a MEF2 site (γ-element) are critical for RGMc promoter activity in muscle cells, and suggest that myogenin and MEF2C may be key transcription factors acting at these sites. We also have identified a third region in the proximal RGMc promoter, termed the β-element, that also is necessary for full transcriptional activity in differentiating muscle cells, but have not yet defined the responsible transcription factors. Leading candidates include members of the Stat and Ets families, although our preliminary studies rule out Stat5b. Both Stat3 and several Ets factors will need to be tested, as each has been shown to positively regulate gene expression during muscle differentiation [51; 52; 53].
RGMc is also expressed in cardiac muscle and in hepatocytes (Fig. 1A, and [9; 14; 15; 17; 18; 19]). To our knowledge there are no other genes that exhibit this pattern of gene expression, placing RGMc in a unique position to provide insights into distinct tissue-specific transcriptional regulatory mechanisms. Unfortunately our initial studies did not reveal any information on transcriptional control in the liver, as the RGMc promoter was minimally functional in the three hepatic cell lines tested. Of note, RGMc mRNA is not produced in the liver of mice lacking the liver-enriched transcription factor HNF4α [54], and ChIP-seq studies with human liver chromatin have identified a peak of HNF4α binding near the human RGMc promoter (personal communication from M. D.Wilson of the Department of Oncology, University of Cambridge, UK; also see ref [37]). Clearly, future studies should focus on defining mechanisms responsible for RGMc gene activity in both hepatocytes and cardiac myocytes.
The results presented in this manuscript represent the first analysis of promoter function for any member of the RGM family. As both RGMa and RGMb have completely different profiles of gene expression than RGMc, being produced in distinct parts of the central nervous system, and not in muscle or liver [15; 16; 17; 18; 55; 56], it is likely that the critical transcriptional control elements will be different. However, comparative genomic analysis of RGMa and RGMb genes indicates the presence of several well conserved E-boxes and MEF2 sites in their putative promoters (data not shown), which could be regulated by neuronal specific bHLH proteins such as N-twist [57] in conjunction with MEF2 family members. Future studies will be needed to define the specific mechanisms of regulation of RGMa and RGMb gene transcription.
RGMc mRNA has been detected in skeletal muscle from several vertebrates ranging from mice to zebrafish [14; 17; 19; 20]. While mouse and zebrafish RGMc promoters are not well-conserved across the entire region upstream of the first known exon (Fig. 8B and C), a bioinformatics analysis reveals the presence of several E-boxes, MEF2 sites, and Ets/Stat elements in the putative zebrafish promoter (Supplemental Fig. 4), suggesting that similar transcriptional mechanisms to those identified for the mouse RGMc gene in skeletal muscle may be active in zebrafish muscle. In addition, a single RGM gene has been identified in the urochordate, Ciona intestinalis (sea squirt), based on genomic data [20; 58]. The RGM gene in Ciona appears to contain 4 exons, and numerous MEF2, E-box, and Stat/Ets-sites surround the putative promoter (Supplemental Fig. 4), although it is not known whether Ciona RGM is expressed in muscle. Understanding the mechanisms controlling expression of the three RGM genes in vertebrates and the single RGM in a model organism like Ciona will help define the evolutionary history of the entire RGM family and discern how each member acquired its distinct tissue-specific regulatory modules.
Materials and Methods
Materials
Restriction enzymes, buffers, ligases, and polymerases were purchased from New England Biolabs (Beverly, MA), BD Biosciences (Clontech, Mountain View, CA), and Fermentas (Hanover, MD). The BCA protein assay kit was from Pierce (Thermo Scientific Life Sciences, Rockford, IL), and the QuikChange XL site-directed mutagenesis kit from Stratagene (La Jolla, CA). TransIT LT-1 was from Mirus Corp (Madison, WI). Dulbecco’s modified Eagle’s medium (DMEM), Superscript III first-strand synthesis kit, Trizol, and horse serum were from Life Technologies (Carlsbad, CA). Fetal bovine serum (FBS) and newborn calf serum (NCS) were from Hyclone (Logan, UT). Luciferase assay reagent was purchased from Promega (Madison, WI). DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) was from Sigma (St. Louis, MO), and DNA purification reagents from Qiagen (Valencia, CA). Oligonucleotides were synthesized at the OHSU DNA Services Core. All other chemicals were reagent grade and were purchased from commercial suppliers.
Construction of RGMc promoter-reporter plasmids
Mouse RGMc genomic DNA was isolated from BAC clone RP24-136I19 (Children’s Hospital Oakland Research Institute BACPAC resource center (http://bacpac.chori.org/), Oakland, CA). All RGMc DNA sequences characterized in this manuscript match what is present in mouse genome databases [59] except for a 33-bp region of intron 2, which is not present in the BAC DNA. DNA fragments generated by restriction enzyme digestion or PCR were purified after preparative agarose gel electrophoresis by ion-exchange chromatography (Qiaexx II gel extraction kit, Qiagen), and sub-cloned into the pGL3-basic firefly luciferase vector (Promega) by standard methods. Mutations in the RGMc promoter were introduced by site-directed mutagenesis with the following oligonucleotide primers (top strand is shown, mutations are in lower case): E-box at −588 to −583: 5′-GGTGGAGAGAGAGTAGAgctagcCAGAGATCTGATCTGGGC-3′; E-box at −514 to −509: 5′-GCTCTCGGATTTCTCGGGAgggcccGACCTTTCAGCTTCTG-3′; β-site at −119 to −111: 5′-GCTCCCACACCCCACTGCCACCAACGcgtCTGcccTTTTGGACCTAG-3′; MEF2 site at −98 to −84: 5′-CCTGGAATTTTGGACCTAGtggccaTTAgAAtTcTCAACTCAGTAGGCACCTCCCTCCTCC-3′. All DNA modifications were confirmed by sequencing.
Cell culture
Cells were incubated at 37°C in humidified air with 5% CO2. C2 myoblasts (passages 4 to 10) were grown on gelatin-coated tissue culture dishes in DMEM with 10% heat-inactivated FBS and 10% NCS. At confluent density cells were washed and low-serum differentiation medium was added (DM = DMEM with 2% horse serum). Cell images were captured by phase contrast microscopy using a Nikon Eclipse T300 microscope with an attached Roper Scientific Cool Snap FX CCD camera. C3H10T½ mouse embryonic fibroblasts (#CCL-226, ATCC, Manassas, VA) were maintained between passages 12 and 18, and incubated on gelatin-coated dishes in DMEM plus 10% FBS. They were converted to myoblasts by infection with a recombinant adenovirus for mouse MyoD [60], followed by incubation in DM as above. Mouse AML12 cells (ATTC #CRL-2254) were incubated in a 1:1 mixture of DMEM and Ham’s F12 medium supplemented with ITS (Final concentrations: 10 μg/ml insulin (I), 5.5 μg/ml transferrin (T), 6.7 ng/ml selenium (S)), 0.1 μM dexamethasone, and 10% FBS. Human hepatoma cell lines HepG2 (ATTC #HB-8065) and Hep3B (ATTC #HB-8064) were cultured in Eagle’s MEM plus 10% FBS.
Recombinant adenoviruses
Recombinant adenoviruses for MyoD (Ad-MyoD) and β-galactosidase (Ad-β-gal) were prepared as described [60].
Analysis of gene transcription by promoter-reporter gene assays
C2 and 10T½ cells on gelatin-coated 12-well plates were transfected with individual RGMc promoter-reporter plasmids or with controls [(thymidine-kinase (TK) - Luc [61], mouse myogenin - Luc [28], mouse IGF-II promoter 3 - Luc [62], 4xE-box TK - Luc [28]] at 50% or 25% of confluent density, respectively (0.7 μg of plasmid DNA per well for C2 cells, 0.4 μg for 10T½ cells). C2 cell extracts were harvested either one day later (undifferentiated), or after DM was added for an additional 48 hr (differentiated). For 10T½ cells, one day after transfection cells were infected with Ad-MyoD or Ad-β-gal, and extracts were collected following another day in growth medium (undifferentiated), or after DM was added for 24 hr (differentiated). Cell extracts from individual experiments were stored at −80°C, and were assayed together for luciferase activity, as described [28], and results were normalized to cellular protein concentrations. At least 3 experiments were performed for each promoter - reporter plasmid using duplicate transfections per experiment. To assess effects of myogenin or MEF2 on RGMc promoter activity, co-transfection experiments were performed in 10T½ cells with selected RGMc promoter - reporter genes, and expression plasmids for mouse myogenin (myogenin-IRES-EGFP or EGFP [28]), or constitutively active MEF2C (MEF2C-VP16 - from J. Molkentin [47; 63]) in pcDNA3 or empty vector.
Animal studies
Male and female C57Bl6 mice were housed at the OHSU Animal Care Facility on a 12 hr light/dark schedule with free access to food and water, and received care according to National Institutes of Health guidelines. At 3 months of age, mice were euthanized by cervical dislocation and tissues were harvested, flash-frozen in liquid nitrogen, and pulverized prior to RNA isolation. Animal studies were approved by the OHSU Animal Care and Use Committee.
RNA isolation and analysis
Total cellular RNA was isolated from cells and tissues using Trizol, followed by sodium acetate-ethanol precipitation and suspension in RNAse-free de-ionized water. Nuclear RNA was isolated from cells as described [62]. RNA concentrations were determined spectrophotometrically at 260 nm, and quality assessed by agarose gel electrophoresis. RNA (5 μg) was reverse transcribed in a final volume of 20 μl, with either oligo-dT primers (for total RNA) or random hexamers (for nuclear RNA), and PCR was performed with 0.1 μl of cDNA and the primers listed in Supplemental Table 1. The linear range of product amplification was established in pilot studies for each primer pair, and the cycle number that reflected the approximate midpoint was used in final experiments. This varied from 18 - 30 cycles for total RNA and from 25 - 30 cycles for nuclear RNA. Results were visualized after electrophoresis through 1.0 - 1.8% agarose or 10% PAGE gels.
RNA half-life
Confluent C2 cells were incubated in DM for 48 hr, washed, and DRB [75 μM] was added in DM for 0.5 to 12 hr. Total cellular RNA was isolated, and used in RT-PCR experiments, as above. Half-life was determined by averaging results of two independent experiments using non-linear regression fit to a one-phase decay equation, Y = Y0 • e−kX.
Mapping the 5′ end of the mouse RGMc gene
The 5′ RACE method was employed with mouse skeletal muscle RNA. First strand cDNA was prepared using specific primers complementary to portions of mouse RGMc exons 2 and 4 (Supplemental Table 1). Subsequent steps were as described [64; 65], with primers for second strand cDNA synthesis from RGMc exon 1 (Supplemental Table 1), and for PCR from RGMc exon 1 and a poly T adaptor (Supplemental Table 1). Gel-purified PCR products were cloned into the pGEM-T Easy vector (Promega), and the DNA was sequenced. A total of 15 independent clones were analyzed. A nested RT-PCR-based method also was used to map the 5′ extent of RGMc exon 1 from mouse muscle, liver, and heart RNA. Primers are listed in Supplemental Table 1. A total of 83 independent clones were characterized by restriction enzyme mapping and/or DNA sequencing.
Data Analysis
Results were graphed and analyzed using Prism (GraphPad Software, San Diego, CA) or Excel (Microsoft, Redmond, WA). All differences were assessed using Student’s t-test with p <0.05 as a cut-off for significance.
Computational Analysis
We manually compared putative RGMc promoter sequences from other species with mouse RGMc, and used information from the UCSC Genome Browser [59] and the LLNL-ECR Browser [66] to perform alignments of RGMc exons and introns.
Additional information on genomic alignments may be found in Ref. [20]. Blocks of DNA sequence were compared using EBI “Align” algorithm written by Alan Bleasby (see references [67; 68]). Transcription factor sites were analyzed using Dcode [69], rVista 2.0 [70], the TRANSFAC database [71], and JASPAR [72], and were individually confirmed with original primary references, as cited within this manuscript.
Supplementary Material
Acknowledgements
We thank Lisa Wilson and David Kuninger for assistance with preliminary experiments, Dr. Melissa Wong of OHSU for mouse tissues, and other members of our laboratory for helpful comments during the development of this work. These studies have been supported in part by National Institutes of Health grants T32 HL007781 (Training Grant in Molecular Hematology) and F30 HL095327 (to C. J. S.), and by R01 DK042748-21 (to P. R.).
Abbreviations
- Ad
adenovirus
- BAC
Bacterial Artificial Chromosome
- BCA
bicinchoninic acid
- bHLH
basic helix-loop-helix
- DM
Differentiation Media
- DMEM
Dulbecco’s modified Eagle’s medium
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole
- E
embryonic day
- EST
expressed sequence tag
- ETS
E26 avian retrovirus Transformation-Specific
- FBS
fetal bovine serum
- HJV
hemojuvelin
- JH
juvenile hemochromatosis
- MADS
MCM1 (minichromosome maintenance)–agamous–deficiens–serum response factor
- MCK
creatine kinase-muscle
- MEF2
myocyte enhancer factor 2
- MHC
myosin heavy chain (myh3)
- MyoD
myogenic differentiation-1
- Myog
myogenin
- NCS
newborn calf serum
- RACE
rapid amplification of cDNA ends
- RGM
Repulsive Guidance Molecule
- RLU
relative luciferase units
- RT-PCR
reverse transcription polymerase chain reaction
- STAT
signal transducers and activators of transcription
- TFBS
transcription factor binding sites
- TSS
transcription start site
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have report no conflicts of interest that would influence the work presented in this manuscript.
References
- [1].Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- [2].Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IKO, Spies M. The Iron-containing Domain Is Essential in Rad3 Helicases for Coupling of ATP Hydrolysis to DNA Translocation and for Targeting the Helicase to the Single-stranded DNA-Double-stranded DNA Junction. J. Biol. Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- [3].Pietrangelo A. Hereditary hemochromatosis--a new look at an old disease. N Engl J Med. 2004;350:2383–97. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- [4].Andrews NC, Schmidt PJ. Iron Homeostasis. Annual Review of Physiology. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- [5].Zhang D-L, Hughes RM, Ollivierre-Wilson H, Ghosh MC, Rouault TA. A Ferroportin Transcript that Lacks an Iron-Responsive Element Enables Duodenal and Erythroid Precursor Cells to Evade Translational Repression. Cell Metabolism. 2009;9:461–473. doi: 10.1016/j.cmet.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pietrangelo A, Caleffi A, Henrion J, Ferrara F, Corradini E, Kulaksiz H, Stremmel W, Andreone P, Garuti C. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128:470–9. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- [7].Gehrke SG, Pietrangelo A, Kascak M, Braner A, Eisold M, Kulaksiz H, Herrmann T, Hebling U, Bents K, Gugler R, Stremmel W. HJV gene mutations in European patients with juvenile hemochromatosis. Clin Genet. 2005;67:425–8. doi: 10.1111/j.1399-0004.2005.00413.x. [DOI] [PubMed] [Google Scholar]
- [8].Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–91. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–6. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- [11].Lin L, Nemeth E, Goodnough JB, Thapa DR, Gabayan V, Ganz T. Soluble hemojuvelin is released by proprotein convertase-mediated cleavage at a conserved polybasic RNRR site. Blood Cells Mol Dis. 2008;40:122–31. doi: 10.1016/j.bcmd.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- [13].Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- [14].Kuninger D, Kuzmickas R, Peng B, Pintar JE, Rotwein P. Gene discovery by microarray: identification of novel genes induced during growth factor-mediated muscle cell survival and differentiation. Genomics. 2004;84:876–89. doi: 10.1016/j.ygeno.2004.07.013. [DOI] [PubMed] [Google Scholar]
- [15].Niederkofler V, Salie R, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci. 2004;24:808–18. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schmidtmer J, Engelkamp D. Isolation and expression pattern of three mouse homologues of chick Rgm. Gene Expr Patterns. 2004;4:105–10. doi: 10.1016/s1567-133x(03)00144-3. [DOI] [PubMed] [Google Scholar]
- [17].Samad TA, Srinivasan A, Karchewski LA, Jeong SJ, Campagna JA, Ji RR, Fabrizio DA, Zhang Y, Lin HY, Bell E, Woolf CJ. DRAGON: a member of the repulsive guidance molecule-related family of neuronal- and muscle-expressed membrane proteins is regulated by DRG11 and has neuronal adhesive properties. J Neurosci. 2004;24:2027–2036. doi: 10.1523/JNEUROSCI.4115-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oldekamp J, Kramer N, Alvarez-Bolado G, Skutella T. Expression pattern of the repulsive guidance molecules RGM A, B and C during mouse development. Gene Expr Patterns. 2004;4:283–8. doi: 10.1016/j.modgep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- [19].Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, Schaper K, Segerdell E, Song P, Sprunger B, Taylor S, Van Slyke CE, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–5. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Severyn CJ, Shinde U, Rotwein P. Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem J. 2009;422:393–403. doi: 10.1042/BJ20090978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. Journal of Molecular Biology. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- [22].Tureckova J, Wilson EM, Cappalonga JL, Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J Biol Chem. 2001;276:39264–70. doi: 10.1074/jbc.M104991200. [DOI] [PubMed] [Google Scholar]
- [23].Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–7. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- [24].Yaffe D, Saxel O. A myogenic cell line with altered serum requirements for differentiation. Differentiation. 1977;7:159–66. doi: 10.1111/j.1432-0436.1977.tb01507.x. [DOI] [PubMed] [Google Scholar]
- [25].Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- [26].Nabeshima Y, Hanaoka K, Hayasaka M, Esuml E, Li S, Nonaka I, Nabeshima Y.-i. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- [27].Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- [28].Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J Biol Chem. 2006;281:29962–71. doi: 10.1074/jbc.M605445200. [DOI] [PubMed] [Google Scholar]
- [29].Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- [30].Leonard WJ, O’Shea JJ. JAKS AND STATS: Biological Implications*. Annual Review of Immunology. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- [31].Horvath CM, Wen Z, Darnell JE., Jr. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–94. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- [32].Shore P, Whitmarsh AJ, Bhaskaran R, Davis RJ, Waltho JP, Sharrocks AD. Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol Cell Biol. 1996;16:3338–49. doi: 10.1128/mcb.16.7.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. The International Journal of Biochemistry & Cell Biology. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- [34].Gutierrez-Hartmann A, Duval DL, Bradford AP. ETS transcription factors in endocrine systems. Trends in Endocrinology & Metabolism. 18:150–158. doi: 10.1016/j.tem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- [35].Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of Ultraconserved Elements Yields Viable Mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chia DJ, Varco-Merth B, Rotwein P. Dispersed Chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J Biol Chem. 2010;285:17636–47. doi: 10.1074/jbc.M110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-Vertebrate ChIP-seq Reveals the Evolutionary Dynamics of Transcription Factor Binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–9. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- [39].Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of Messenger RNA Precursors. Annual Review of Biochemistry. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- [41].Black DL. MECHANISMS OF ALTERNATIVE PRE-MESSENGER RNA SPLICING. Annual Review of Biochemistry. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- [42].Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–84. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- [43].Kozak M. Regulation of Translation in Eukaryotic Systems. Annual Review of Cell Biology. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- [44].Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- [45].Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–90. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- [46].Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- [47].Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–40. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- [49].Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–36. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- [50].Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–32. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- [51].Wang K, Wang C, Xiao F, Wang H, Wu Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem. 2008;283:34029–36. doi: 10.1074/jbc.M803012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sun L, Ma K, Wang H, Xiao F, Gao Y, Zhang W, Wang K, Gao X, Ip N, Wu Z. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol. 2007;179:129–38. doi: 10.1083/jcb.200703184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].De Val S, Anderson JP, Heidt AB, Khiem D, Xu S-M, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Developmental Biology. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- [54].Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4{alpha} orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proceedings of the National Academy of Sciences. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, Mueller BK. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–5. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- [56].Matsunaga E, Tauszig-Delamasure S, Monnier PP, Mueller BK, Strittmatter SM, Mehlen P, Chedotal A. RGM and its receptor neogenin regulate neuronal survival. Nat Cell Biol. 2004;6:749–55. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- [57].Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, Cripps RM, Black BL. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002;249:174–90. doi: 10.1006/dbio.2002.0753. [DOI] [PubMed] [Google Scholar]
- [58].Camus LM, Lambert LA. Molecular evolution of hemojuvelin and the repulsive guidance molecule family. J Mol Evol. 2007;65:68–81. doi: 10.1007/s00239-006-0241-5. [DOI] [PubMed] [Google Scholar]
- [59].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wilson EM, Hsieh MM, Rotwein P. Autocrine growth factor signaling by insulin-like growth factor-II mediates MyoD-stimulated myocyte maturation. J Biol Chem. 2003;278:41109–13. doi: 10.1074/jbc.C300299200. [DOI] [PubMed] [Google Scholar]
- [61].Woelfle J, Billiard J, Rotwein P. Acute Control of Insulin-like Growth Factor-I Gene Transcription by Growth Hormone through Stat5b. J. Biol. Chem. 2003;278:22696–22702. doi: 10.1074/jbc.M301362200. [DOI] [PubMed] [Google Scholar]
- [62].Kou K, Rotwein P. Transcriptional activation of the insulin-like growth factor-II gene during myoblast differentiation. Mol Endocrinol. 1993;7:291–302. doi: 10.1210/mend.7.2.8469241. [DOI] [PubMed] [Google Scholar]
- [63].Molkentin JD, Firulli AB, Black BL, Martin JF, Hustad CM, Copeland N, Jenkins N, Lyons G, Olson EN. MEF2B is a potent transactivator expressed in early myogenic lineages. Mol. Cell. Biol. 1996;16:3814–3824. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sambrook J, Russell DW. Rapid amplification of 5′ cDNA ends (Protocol 9, 8.54–8.60) Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- [65].Sambrook J, Russell DW. Rapid amplification of 5[prime] complementary DNA ends (5[prime] RACE) Nat Meth. 2005;2:629–630. doi: 10.1038/nmeth0805-629. [DOI] [PubMed] [Google Scholar]
- [66].Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–6. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- [68].Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- [69].Loots GG, Ovcharenko I. Dcode.org anthology of comparative genomic tools. Nucleic Acids Res. 2005;33:W56–64. doi: 10.1093/nar/gki355. Dcode.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–21. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Knuppel R, Dietze P, Lehnberg W, Frech K, Wingender E. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J Comput Biol. 1994;1:191–8. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- [72].Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucl. Acids Res. 2009 doi: 10.1093/nar/gkp950. gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–87. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.