Abstract
Damage to our genetic material is an ongoing threat to both our ability to faithfully transmit genetic information to our offspring as well as our own survival. To respond to these threats, eukaryotes have evolved the DNA Damage Response (DDR). The DDR is a complex signal transduction pathway that has the ability to sense DNA damage and transduce this information to the cell to influence cellular responses to DNA damage. Cells possess an arsenal of enzymatic tools capable of remodeling and repairing DNA, however, their activities must be tightly regulated in a temporal, spatial and DNA lesion-appropriate fashion to optimize repair and prevent unnecessary and potentially deleterious alterations in the structure of DNA during normal cellular processes. This review will focus on how the DDR controls DNA repair and the phenotypic consequences of defects in these critical regulatory functions in mammals.
Since the discovery of the DNA structure more than 50 years ago, the remarkable mechanisms that preserve the genetic information encoded by DNA and guarantee its faithful transmission across generations have been the subject of extensive investigation. To maintain genomic integrity, DNA must be protected from damage induced by environmental agents or generated spontaneously during DNA metabolism. Spontaneous DNA alterations can be due to dNTP misincorporation during DNA replication, interconversion between DNA bases caused by deamination, loss of DNA bases following DNA depurination and modification of DNA bases by alkylation (Table 1) (Lindahl and Barnes, 2000). Additionally, oxidized DNA bases and DNA breaks can be generated by reactive oxygen species derived from normal cellular metabolism. Altogether, it has been estimated that every cell could experience up to 105 spontaneous DNA lesions per day (Hoeijmakers, 2009).
Table 1. DNA lesions generated by endogenous and exogenous DNA damage.
Type and number of DNA lesions are indicated. The number of lesion/cell in (B) has been estimated as described below the table.
| A | ||
|---|---|---|
| Endogenous DNA Damage | DNA lesions generated | Number lesions/cell/day |
| Depurination | AP site | 10,000(a) |
| Cytosine deamination | Base transition | 100–500(a) |
| SAM-induced methylation | 3-meA | 600(a) |
| 7-meG | 4,000(a) | |
| O6-meG | 10–30(b) | |
| Oxidation | 8-oxo-dG | 400–1500(c) |
| B | |||
|---|---|---|---|
| Exogenous DNA Damage | Dose exposure (mSv) | DNA lesions generated | Estimated number lesions/cell |
| Peak hour sunlight | - | pyrimidine dimers, (6-4) photoproducts | 100,000/day(d) |
| Cigarette smoke | - | aromatic DNA adducts | 45–1029(e) |
| Chest X-rays | 0.02 (f-h) | DSBs | 0.0008* |
| Dental X-rays | 0.005(f-h) | DSBs | 0.0002* |
| Mammography | 0.4(f-h) | DSBs | 0.016* |
| Body CT | 7(f) | DSBs | 0.28* |
| Head CT | 2(f-g) | DSBs | 0.08* |
| Coronary angioplasty | 22(h) | DSBs | 0.88* |
| Tumor PET scan (18F) | 10(h) | DSBs | 0.4* |
| 131I treatment | 70–150(h) | DSBs | 2.8–6* |
| External beam therapy | 1800–2000(i) | DSBs | 72–80 |
| Airline travel | 0.005/hour (f) | DSBs | 0.0002/hour* |
| Space mission (60 days) | 50(j) | DSBs | 2* |
| Chernobyl accident | 300(k) | DSBs | 12* |
| Hiroshima and Nagasaki atomic bombs | 5–4000(l) | DSBs | 0.2–160* |
Based on the effective dose received by the whole body. Dose absorbed by the specific tissue irradiated may be higher. The number of DSBs has been calculated assuming that mammalian cells irradiated with 1 Sv (corresponding to 1 Gy for X and γ-rays) experience approximately 40 DSBs (Elkind and Redpath, 1977). Approx 1000 SSBs/cell are generated following 1 Gy irradiation (Elkind and Redpath, 1977).
Barnes and Lindahl, 2004
DNA adducts detected in the lung of smokers following 1–2 cigarette packs per day for approx 40 years (Phillips et al, 1988). Higher number of cigarettes consumed correlates with higher number of aromatic DNA adducts. Up to 6000 adducts per cell could be present in smokers, if all the types of DNA adducts generated by cigarette smoke carcinogens are taken into account. This does not include oxidative damage.
Typical single dose administered per day in the treatment of cancer. The number of DSBs has been calculated as described above.
NCRP Report 138, 2001
Average dose for people living near the Chernobyl plant. http://www.merck.com/mmhe/sec24/ch292/ch292a.html
Life Span Study (LSS) mortality data set, 2003. The average dose for the survivors of this study is 200 mSv.
Environmental DNA damage can be produced by physical or chemical sources. Examples of physical genotoxic agents are ionizing radiation (IR) and ultraviolet (UV) light from sunlight which can also induce up to 105 DNA lesions (pyrimidine dimers and (6-4) photoproducts) per cell per day (Table 1) (Hoeijmakers, 2009). IR (from e.g. cosmic radiation and medical treatments employing X-rays or radiotherapy) can induce oxidation of DNA bases and generate single-strand and double-strand DNA breaks (SSBs and DSBs, respectively) (Table 1). Chemical agents used in cancer chemotherapy can cause a variety of DNA lesions: alkylating agents, such as methyl-methane sulfonate (MMS) and temozolomide, attach alkyl groups to DNA bases, while crosslinking agents, such as mitomycin C (MMC), cisplatin, psoralen and nitrogen mustard, introduce covalent links between bases of the same DNA strand (intrastrand crosslinks) or of different DNA strands (interstrand crosslinks or ICLs). Other chemical agents, such as the topoisomerase inhibitors camptothecin (CPT) and etoposide, which inhibit topoisomerase I or II, respectively, induce the formation of SSBs or DSBs by trapping topoisomerase-DNA covalent complexes. Cigarette smoking, one of the most common mechanisms of self-inflicted DNA damage, causes a wide variety of adducts and oxidative damage in lung and other tissues. The measurement of lesions in smokers in Table 1 is likely to be a vast underestimate of the total damage produced per day since smoke-induced DNA adducts present in normal tissues adjacent to tumors are measured as a single snapshot at the time of tumor removal, presumably hours or days after the last cigarette exposure.
To counteract DNA damage, repair mechanisms specific for many types of lesion have evolved. Mispaired DNA bases are replaced with correct bases by mismatch repair (MMR) and small chemical alterations of DNA bases are repaired by base excision repair (BER) through excision of the damaged base (Jiricny, 2006; Lindahl and Barnes, 2000). More complex lesions, such as pyrimidine dimers and intrastrand crosslinks, are corrected by nucleotide excision repair (NER) through the removal of an oligonucleotide of approximately 30 bp containing the damaged bases, while ICLs are excised by interstrand crosslink repair with the assistance of proteins involved in the genetic syndrome Fanconi anemia (Hoeijmakers, 2009; Moldovan and D’Andrea, 2009). SSBs are repaired by single-strand break repair (SSBR), whereas DSBs are processed either by non-homologous end joining (NHEJ) or homologous recombination (HR) (Caldecott, 2008; West, 2003). While NHEJ promotes the potentially inaccurate religation of DSBs, HR precisely restores the genomic sequence of the broken DNA ends by utilizing sister chromatids as template for repair.
DNA repair is carried out by a plethora of enzymatic activities that chemically modify DNA to repair DNA damage including nucleases, helicases, polymerases, topoisomerases, recombinases, ligases, glycosylases, demethylases, kinases and phosphatases. These repair tools must be precisely regulated because each in its own right can wreck havoc on the integrity of DNA if misused or allowed to access DNA at the inappropriate time or place. Thus, eukaryotic cells have developed strategies to recruit and activate the right factors in the right place at the right time. Here we describe the cellular mechanisms that regulate the recruitment of DNA repair factors to sites of DNA damage, activate those factors and coordinate the choice of the pathways to employ for efficient DNA repair. Moreover, we describe the pathological consequences that result from a defective response to DNA damage in humans.
Signal Transduction: Sending an SOS to Repair
The DNA damage response (DDR) is a signal transduction pathway that senses DNA damage and replication stress and sets in motion a choreographed response to protect the cell and ameliorate the threat to the organism (Harper and Elledge, 2007; Jackson and Bartek, 2009). The DDR is primarily mediated by proteins of the phosphatidylinositol 3-kinase-like protein kinases (PIKKs) family — ATM, ATR and DNA-PK — and by members of the poly(ADP)ribose polymerase (PARP) family. ATM and DNA-PK are activated by DNA damaging agents (e.g. ionizing radiation) that create DSBs (Figure 1A) (Harper and Elledge, 2007; Meek et al., 2008). Unlike ATM, which has hundreds of substrates, DNA-PK primarily regulates a smaller group of proteins involved in DSB end joining. ATR, in complex with its partner protein ATRIP, is activated following recruitment to RPA-coated ssDNA regions generated at stalled replication forks (Figure 1B) and DSBs (Cimprich and Cortez, 2008). The PARP family has 16 members, but only PARP1 and PARP2 have been implicated in the DDR (Schreiber et al., 2006). PARP1 and PARP2 are activated by SSBs and DSBs and catalyze the addition of poly(ADP-ribose) chains on proteins to recruit DDR factors to chromatin at breaks (Figure 1A) (Schreiber et al., 2006).
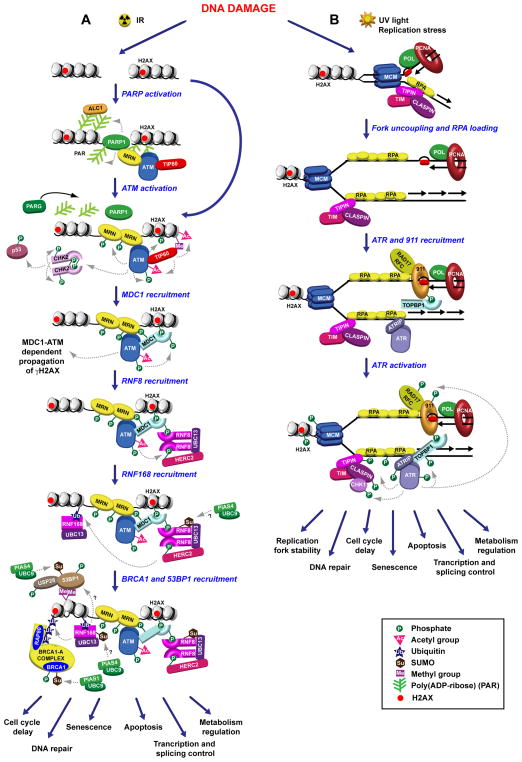
Figure 1. Schematic model for ATM and ATR activation in response to DNA damage.
(A) Formation of DSBs following IR treatment activates PARP1, which mediates the initial recruitment of the MRN/ATM complex at DSBs. Activation of the ATM kinase activity by MRN and TIP60 leads to the phosphorylation of CHK2 and p53, in addition to a wide number of other DDR factors, and the induction of the γH2AX-dependent signaling cascade, which results in the recruitment of MDC1, RNF8, RNF168, BRCA1 and 53BP1 to DSBs, as described in greater details in the main text. (B) DNA lesions induced by UV light or replication stress (denoted by red rectangular shapes) lead to replication fork stalling and accumulation of RPA-coated ssDNA regions, which recruit the ATR/ATRIP and the RAD17/RFC2-5 complexes. Loading of the 9-1-1 complex by RAD17/RFC2-5 and stimulation of the ATR kinase activity by the 9-1-1 associated protein TOPBP1 result in the activation of the ATR signaling cascade and CHK1 phosphorylation. Post-translational modifications of the DDR factors here depicted are represented by different colored shapes, as indicated by the legend at the bottom of the figure.
Much of the current understanding of the DDR is based on the study of the ATM and ATR kinases. Following the recognition of DNA lesions by sensor proteins, ATM and ATR initially phosphorylate mediator proteins, which can amplify the DNA damage response by acting as recruiters of ATM/ATR substrates (Zhou and Elledge, 2000). Effector proteins of the DNA damage response are either directly phosphorylated by ATM/ATR or by the CHK1 and CHK2 kinases, as well as other kinases such as CK2, p38 and MK2 (Harper and Elledge, 2007). The stability of ATM and ATR — and other PIKKs — is dependent on the TEL2-TTI1-TTI2 (Triple T) complex, which has been reported to associate with the heat shock protein HSP90 and possibly promote the maturation of newly synthesized PIKKs (Hurov et al., 2010; Takai et al., 2007; Takai et al., 2010).
The DDR regulates physiological processes that involve multiple layers of decisions, such as the determination to undergo apoptosis, enter terminal differentiation through senescence, activation of heightened immune surveillance, DNA damage prophylaxis through tanning as well as DNA repair itself (Cui et al., 2007; Gasser and Raulet, 2006; Zhou and Elledge, 2000). ATM and ATR are required for NHEJ, HR, ICL repair, and NER, as well as replication fork stability during unperturbed DNA replication and in response to replication blocks. While primarily mediated through relatively fast post-translational modifications — such as phosphorylation and inhibition of the cell cycle phosphatase CDC25 required for CDK activation — a significant portion of the decision processes are mediated through slower transcriptional responses that allow integration of information over time. The most extensively studied component of this response is p53, which is regulated by ATM and CHK2 in response to DSBs (Figure 1A) (Zhou and Elledge, 2000). P53 induces cell cycle arrest, apoptosis or senescence in response to DNA damage by transcriptionally regulating, among others, the CDK inhibitor p21 and the pro-apoptotic BAX and PUMA proteins (Riley et al., 2008). Moreover, p53 directly activates repair pathways such as NER through regulation of the NER factors XPC and DDB2 and induces dNTP synthesis as described below (Ford, 2005). Importantly, following DSB formation, p53 is activated by ATM in a cyclically periodic manner through a transcriptional circuit involving the WIP1 phosphatase and the MDM2 E3 ubiquitin ligase, both p53 targets, which turn off ATM and p53, respectively (Batchelor et al., 2009). This provides the cell with a time measurement mechanism that activates p53 transcriptional pulses in an oscillating fashion depending on whether the initiating damage has been repaired. This raises the interesting possibility that each succeeding pulse occurs in a different proteome environment and could impart distinct information to the cell on the persistence of damage, directing the cell to make different decisions such as apoptosis or senescence.
It is now clear that ATM and ATR coordinate a much wider variety of cellular activities than initially anticipated, from DNA replication and repair to transcription, metabolic signaling and RNA splicing (Bennetzen et al., 2010; Matsuoka et al., 2007; Paulsen et al., 2009). Defective regulation of any of these activities results in genomic instability after DNA damage. In the next sections we will focus our attention on the mechanisms employed by the DNA damage response to regulate DNA repair in order to preserve genomic integrity.
Spatiotemporal Regulation of DNA Repair: Sensing the damage
Localization of DDR factors to sites of DNA damage is initiated by sensor proteins that directly recognize specific DNA lesions and activate the DDR (Zhou and Elledge, 2000). Interestingly, experiments performed both in yeast and mammalian cells have demonstrated that forced tethering of sensor proteins to chromatin is sufficient to elicit the DDR cascade even in the absence of DNA damage (Bonilla et al., 2008; Soutoglou and Misteli, 2008). The next level of regulation of DNA repair resides in the DDR-regulated recruitment of factors to sites of DNA damage, which can be visualized as discrete nuclear foci by microscopy. DNA damage-induced foci are highly dynamic structures subjected to precise spatiotemporal regulation (Bekker-Jensen et al., 2006). The precise order and timing of recruitment is thought to provide a kinetic choice of repair options, presumably in an optimized order. The assembly of the DDR cascade is dependent on a broad spectrum of post-translational modifications — phosphorylation, ubiquitination, sumoylation, poly(ADP-ribosylation), acetylation, methylation — induced by the activation of the DNA damage response (Bergink and Jentsch, 2009; Harper and Elledge, 2007; Kleine and Luscher, 2009; Misteli and Soutoglou, 2009). These post-translational modifications are specifically recognized by a wide variety of protein domains, many of which mediate the recruitment to DNA damage sites. In the following sections we will highlight the different mechanisms by which different lesions are sensed, the DDR factors are recruited to sites of DNA damage and the consequences of DDR signaling on repair.
Single-Strand Break Repair
SSBs generated by IR and reactive oxidative species (ROS) or arising indirectly during BER of abasic sites and altered DNA bases, such as 8oxoG and 3meA, activate PARP family members (Caldecott, 2008). PARP1 and PARP2 act as molecular sensors of SSBs and DSBs, which are recognized by two PARP1 zinc finger motifs. Activation of PARP1 and PARP2 and subsequent synthesis of poly(ADP-ribose) (PAR) chains occurs within seconds at damage sites and is one of the earliest events of the DNA damage response PAR chains are rapidly disassembled by the PAR hydrolyzing enzyme PARG to provide a quick transient response lasting minutes (Schreiber et al., 2006). Upon DNA binding, PARP1/2 assembles PAR moieties from NAD+ on target proteins including histones H1 and H2B, and PARP1 itself (Schreiber et al., 2006). Histone PARylation is thought to contribute to chromatin reorganization and recruitment of DNA repair and chromatin modifying complexes, such as polycomb and histone deacetylases (HDACs) complexes, at DNA damage sites (Polo et al., 2010; Schreiber et al., 2006; Chou et al., 2010).
PAR Recruitment
PAR structures act as platforms upon which to recruit factors to promote DNA repair. Three PAR binding motifs have been described: the macrodomain, PAR-binding zinc finger (PBZ) and an 8 amino acid basic residue-rich cluster (Kleine and Luscher, 2009). Ten human proteins contain macrodomains, including PARP9, PARP14, PARP15, the histone variant macroH2A1.1 and the chromatin remodelling factor ALC1 (Kleine and Luscher, 2009; Schreiber et al., 2006). Recent studies have shown that macroH2A1.1 and ALC1 are recruited in a PAR-dependent manner to sites of DNA damage, where they contribute to the reorganization of chromatin structure (Ahel et al., 2009; Gottschalk et al., 2009; Timinszky et al., 2009).
Several DDR factors contain the acid basic residue-rich cluster, including p53, XRCC1, LIG3, MRE11 and ATM, whereas PBZ motifs have recently been identified in the nucleases APLF and SNM1 and in the cell cycle checkpoint protein CHFR (Ahel et al., 2008; Gagne et al., 2008). XRCC1 and LIG3 are recruited to SSBs in a PARP1-dependent manner and promote SSB repair following DNA end processing by XRCC1 interacting proteins, such as DNA polymerase β, PNK and the nucleases APE1, APTX and APLF (Caldecott, 2008). APLF is dependent on the PBZ motif for its recruitment to DNA damage sites (Bekker-Jensen et al., 2007; Kanno et al., 2007; Rulten et al., 2008).
Double-Strand Break Repair
DSBs are life threatening lesions whose repair is promoted by an intricate network of multiple DNA repair pathways. At least four independent pathways can repair DSBs — HR, NHEJ, alternative-NHEJ (alt-NHEJ) and single-strand annealing (SSA) (Figure 2). A main factor influencing the pathway choice is the extent of DNA end processing. Classical NHEJ does not require DNA end resection, whereas alt-NHEJ (also known as microhomology-mediated end-joining or MMEJ), HR and SSA are dependent on DSB resection, which is limited for alt-NHEJ (5–25 nt) and more extensive for HR and SSA (Hartlerode and Scully, 2009). In addition at least four partially independent sensors can detect DSBs, PARP, Ku70/Ku80, MRN and, with DSB processing, RPA.
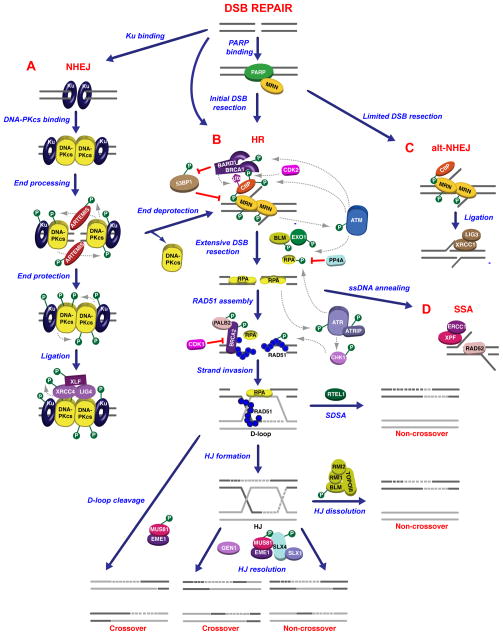
Figure 2. Alternative DNA repair pathways involved in the repair of double-strand breaks.
(A) Rapid association of Ku to DSBs promotes NHEJ by recruiting DNA-PKcs. Sequential phosphorylation events on multiple DNA-PKcs amino acid clusters favors the initial processing of DNA ends by ARTEMIS, followed by DNA-PKcs-dependent protection of DNA ends required for DNA ligation. (B) Alternatively to NHEJ, MRN, which is initially recruited to DSBs by PARP in competition with Ku, mediates the initial stages of DSB resection together with CtIP and BRCA1 to promote homologous recombination in S and G2. 53BP1 has an inhibitory role on DSB resection and is negatively regulated by BRCA1 by unknown mechanisms. The MRN/CtIP/BRCA1 complex can also promote DSB resection following deprotection of DNA ends when NHEJ fails. Extensive DSB resection and formation of RPA-coated 3′-ssDNA ends is induced by EXO1 and BLM. ATM plays a central role in the regulation of DSB resection as described in the main text. Displacement of RPA from the 3′-ssDNA ends and assembly of RAD51 filaments mediated by BRCA2 leads to strand invasion into homologous DNA sequences. Recruitment of RAD51 to ssDNA ends is regulated by the ATR pathway, which is activated following DSB resection. D-loop structures formed after strand invasion can be cleaved by MUS81/EME1 or displaced by RTEL1 during SDSA to generate crossover or non-crossover events, respectively. Non-crossovers are generated also by dissolution of Holliday junctions (HJs) by the BLM/TOPOIII complex, whereas HJ resolution by the nucleases GEN1 and SLX1/SLX4, which associates with MUS81/EME1, can generate both crossover and non-crossover events. (C) Limited DSB resection carried out by CtIP and MRN in G1 results in alternative NHEJ. (D) Following DSB resection, 3′-ssDNA ends with homologous sequences can be directly annealed by RAD52. Post-translational modifications are indicated as in Figure 1.
DNA end joining promoted by Ku70/Ku80 and PARP
Double strand DNA breaks are rapidly bound by the Ku heterodimer (Ku70 and Ku80), which has a toroidal structure with a hole through which it loads onto DSB ends (Figure 2A) (Mahaney et al., 2009). It also possesses a DNA end processing activity (Roberts et al., 2010). Ku localizes to DSBs within seconds, where it loads and activates the catalytic subunit of DNA-PK (DNA-PKcs) to initiate NHEJ (Mahaney et al., 2009).
DNA-PKcs
During NHEJ, DNA-PKcs plays a critical role in stabilizing DSB ends and preventing end resection through a series of phosphorylation reactions (Figure 2A) (Meek et al., 2008). Following DSB binding, DNA-PKcs autophosphorylation on the six residue ABCDE cluster (also known as the T2609 cluster) results in destabilization of the DNA-PKcs interaction with the DNA ends, thus providing access to end processing enzymes, such as ARTEMIS (Meek et al., 2008). Excessive end processing is then prevented by DNA-PKcs autophosphorylation on the five residue PQR cluster (also known as the S2056 cluster), which helps protect the DNA ends (Meek et al., 2008). Interestingly, ABCDE phosphorylation, which can also be induced by ATM, has been also shown to facilitate the access of DNA ends to DSB resecting enzymes in order to promote HR when NHEJ fails (Shrivastav et al., 2008). Conversely, PQR phosphorylation has an inhibitory effect on HR by preventing end resection (Meek et al., 2008). After DNA-PKcs is loaded, XRCC4/LIG4 is recruited, which promotes the religation of the broken ends with the help of the stimulatory factor XLF (Figure 2A) (Mahaney et al., 2009). DNA termini that contain non-ligatable end groups are processed by the ARTEMIS and APLF nucleases and the PNK kinase/phosphatase prior to DNA ligation (Mahaney et al., 2009). All three factors are phosphorylated in an ATM-dependent manner and ARTEMIS is a substrate for DNA-PKcs (Macrae et al., 2008; Mahaney et al., 2009; Matsuoka et al., 2007). ATM has been shown to play a role in 10% of NHEJ through ARTEMIS (Jeggo and Lobrich, 2005). This may be an underestimate because ATM and ATR often play redundant roles. Supporting this, telomeres deprotected by loss of TRF2 and POT1 resemble a DSB and undergo end-to-end fusion via NHEJ (Denchi and de Lange, 2007). This process requires either ATM or ATR as the double mutant abrogates end fusions (Denchi and de Lange, 2007).
PARP
As noted above, PARP1/2 also senses DSBs. PARP acts to promote alt-NHEJ that functions as backup to the classical pathway of NHEJ described above (Figure 2C) (Wang et al., 2006). PARP1 also competes with Ku binding to DNA ends to promote HR (Figure 2B) (Hochegger et al., 2006). During DSB repair, PARP1 is thought to mediate the initial accumulation of the MRN complex to DSBs in a γH2AX and MDC1 independent manner (Haince et al., 2008). Recruitment of ATM by MRN and PARP1 could then contribute to the activation of the γH2AX cascade and stabilization of DDR factors at sites of damage as discussed below (Figure 1A) (Haince et al., 2007). Indeed, PARP1 plays an initial role in the DDR by facilitating ATM activation, as indicated by the delayed phosphorylation of ATM substrates observed in the absence of PARP1 following treatment with DNA damaging agents (Haince et al., 2007). However, PARP1/2 and ATM also have independent functions, as shown by the synthetic lethality of PARP1 (or PARP2) deletion with ATM deficiency in mice (Huber et al., 2004).
Homologous Recombination Repair through MRN-ATM and RPA-ATR
DSBs can also be recognized by the MRE11-RAD50-NBS1 (MRN) complex, which promotes the activation of ATM and the preparation of DNA for HR (Figures 1 and 2) (Williams et al., 2007). RAD50, a member of the SMC family, contains ATPase domains that interact with MRE11 and associates with the DNA ends of the DSB (Williams et al., 2007). In addition to stabilizing DNA ends, MRE11 has endonuclease and exonuclease activities important for the initial steps of DNA end resection that is essential for HR, as described below (Williams et al., 2007). The third subunit of the MRN complex, NBS1, interacts with MRE11 and contains additional protein-protein interaction domains important for MRN function in the DNA damage response. NBS1 associates with ATM via its C-terminal region, which promotes the recruitment of ATM to DSBs, where ATM is activated by the MRN complex by yet to be defined mechanisms possibly involving the formation of ssDNA oligos during end resection (Figure 1A) (Jazayeri et al., 2008; Kanaar and Wyman, 2008; Lee and Paull, 2005).
Resection Control
DNA end resection is regulated by ATM through CtIP, which interacts with BRCA1 and MRN in the BRCA1-C complex (Figure 2B) (Huen et al., 2010b). DSB resection is primarily induced in the S and G2 phases of the cell cycle, when sister chromatids can be used for HR (You and Bailis, 2010). Limited DSB resection is carried out by CtIP in G1 in a BRCA1 independent manner to promote alt-NHEJ, which is mediated by the annealing of ssDNA microhomology regions, followed by LIG3 dependent DNA end ligation (Figure 2C) (You and Bailis, 2010; Yun and Hiom, 2009). In S and G2, CtIP associates with BRCA1, which ubiquitinates CtIP and facilitates its association with damage sites (Huen et al., 2010b). CtIP recruitment is also dependent on MRN and ATM kinase activity, helping explain ATM’s role in DSB resection (You and Bailis, 2010). Two and seven ATM phosphorylation sites have been identified in CtIP and BRCA1, respectively, but their precise function remains to be elucidated (Ouchi, 2006; You and Bailis, 2010). In addition, EXO1, which is involved in the processive stage of DSB resection together with BLM following the initial resection carried out by CtIP, is also stimulated by ATM phosphorylation (Figure 2B) (Bolderson et al., 2010). Moreover, ARTEMIS, which is also regulated by ATM, has been suggested to play a role in DSB resection (Beucher et al., 2009).
Pathway Choice
The orderly progression of choices between alternative DNA repair pathways could be facilitated by negative regulation of one pathway by another. Indeed, DSB resection promoted by CtIP and ATM can be inhibited by 53BP1 (Figure 2B) (Bunting et al., 2010). 53BP1 has been suggested to promote NHEJ by increasing the stability and mobility of DSBs to find each other for productive ligation (Difilippantonio et al., 2008; Dimitrova et al., 2008). Loss of 53BP1 partially rescues the HR defect of BRCA1 mutant cells, suggesting that BRCA1 might somehow overcome 53BP1 function at DSBs in order to promote DSB resection (Bouwman et al., 2010; Bunting et al., 2010). Defective DSB resection in BRCA1 mutant cells results in NHEJ-dependent chromosomal rearrangements, whose formation could be prevented by 53BP1 loss (Bunting et al., 2010). Interestingly, DSB resection induced by 53BP1 deletion was shown to increase alt-NHEJ and decrease classical NHEJ during immunoglobulin maturation in G1 phase B cells (Bothmer et al., 2010). It is known that abnormal activity of alt-NHEJ in the absence of functional NHEJ induces chromosomal translocations in mammalian cells (Simsek and Jasin, 2010). Altogether, these observations indicate that alterations of the correct balance between DSB repair pathways can lead to genomic instability. Another case in point, the chromosomal instability defects and DNA damage sensitivity of Fanconi anemia (FA) mutant cells have recently been shown to be due to aberrant NHEJ, indicating that FA proteins might promote HR and suppress NHEJ (Adamo et al., 2010; Pace et al., 2010). Thus, in the absence of the proper repair pathway choice, incorrect pathway choices can be deleterious.
The RPA Platform
DSB resection and formation of 3′-ssDNA ends leads to RPA accumulation (Figure 2B). RPA is an essential heterotrimeric complex (RPA1, RPA2, RPA3) that stabilizes ssDNA regions generated during DNA replication and repair (Wold, 1997). RPA-ssDNA complexes play a critical role in activation of the ATR pathway as described in greater detail below. In the presence of repetitive DNA sequences that are repaired by SSA, annealing of the resected 3′-ssDNA could be catalyzed by RAD52, followed by removal of DNA flaps by XPF/ERCC1 (Figure 2D) (Hartlerode and Scully, 2009; Motycka et al., 2004). Alternatively, assembly of RAD51 filaments on RPA-coated ssDNA mediated by BRCA2 can lead to HR (Figure 2B) (West, 2003). The interaction between RAD51 and BRCA2 C-terminus, which is important for HR, is thought to be limited to S and G2 phases of the cell cycle by CDK dependent phosphorylation of BRCA2 (Esashi et al., 2005). Further regulation of HR is provided by RAD51 phosphorylation mediated by CHK1, which is required for RAD51 recruitment to damage sites (Sorensen et al., 2005). BRCA2 is also phosphorylated by ATM/ATR (Matsuoka et al, 2007). Moreover, RPA2 undergoes ATM/ATR-mediated hyperphosphorylation followed by PP4-dependent dephosphorylation, which was shown to be important for HR (Lee et al., 2010a). Furthermore, sumoylation of RPA1 was recently suggested to promote HR by facilitating the recruitment of RAD51 (Dou et al., 2010).
Crossover Regulation
Following RAD51-dependent strand invasion into homologous sequences of the sister chromatid and formation of D-loop structures, the 3′-invading strand could be extended by DNA polymerases and then reanneal to the processed second end of the break (West, 2003). This pathway, which is known as synthesis dependent strand annealing (SDSA), is thought to be promoted by the RTEL helicase after displacement of the RAD51 filament and D-loop dissociation (Figure 2B) (Barber et al., 2008). Alternative to this pathway, double Holliday junctions (dHJ) could be formed after ligation of the invading strand with the second end captured by D-loop branch migration (West, 2003). HJ intermediates could be dissolved by the BLM/TOPOIII complex or cleaved by the endonucleases GEN1, MUS81/EME1 or SLX1/SLX4 to generate either crossover or non-crossover of the markers flanking the dHJ (Figure 2B) (Andersen et al., 2009; Ciccia et al., 2008; Fekairi et al., 2009; Ip et al., 2008; Munoz et al., 2009; Svendsen et al., 2009). Crossover events, which could be generated by GEN1, MUS81/EME1 or SLX1/SLX4, are highly regulated, as they can lead to loss of heterozygosity and genomic rearrangements in mitotic cells. Indeed, the high increase in the number of crossover events, which can be visualized as sister chromatid exchanges or SCEs, displayed by BLM defective cells causes genomic instability (Chu and Hickson, 2009). It is known that BLM and SLX4 are ATM/ATR substrates and yeast MUS81 is inhibited by phosphorylation by the yeast CHK2 ortholog (Bachrati and Hickson, 2008; Kai et al., 2005; Svendsen et al., 2009). How phosphorylation of BLM, SLX4 and MUS81 might affect their activity on HJ intermediates has not been determined. Nonetheless, the importance of both ATM and ATR in HR is indicated by the strong recombination defects displayed by cells with ATM or ATR deficiency (Beucher et al., 2009; Wang et al., 2004).
γH2AX – Phosphorylation-dependent Recruitment and Modification Cascades
ATM and ATR promote DSB repair in part through phosphorylation-dependent recruitment of DDR factors to sites of DNA damage. A critical aspect of this process involves the phosphorylation of Ser139 on a specialized histone H2AX called γH2AX (Figure 1A) (Rogakou et al., 1998). H2AX phosphorylation spreads for distances up to 1–2 megabases around DSBs in an ATM and MDC1 dependent manner and initiates a cascade of factor assembly (Harper and Elledge, 2007). MDC1 directly binds the phospho-Ser139 of H2AX through its C-terminal BRCT-repeats (Figure 1A) (Stucki, 2009). H2AX Tyr142 is constitutively phosphorylated by the kinase WSTF, a member of the BAZ/WAL family of chromatin remodelling enzymes, and blocks MDC1 recruitment (Xiao et al., 2009). Following DNA damage, Tyr142 is dephosphorylated by the Tyr phosphatases EYA1 and EYA3 (Cook et al., 2009; Krishnan et al., 2009). Interestingly, MDC1 binding to γH2AX was shown to depend on Tyr142 dephosphorylation by EYA1/3, whereas the proapoptotic kinase JNK1 was reported to associate with H2AX phosphorylated on both Ser139 and Tyr142 (Cook et al., 2009). This observation has led to the proposal that Tyr142 phosphorylation of γH2AX might provide a molecular switch between JNK mediated apoptosis and MDC1 dependent DSB repair (Stucki, 2009). In particular, MDC1 has been reported to facilitate both NHEJ and HR in H2AX dependent manner (Hartlerode and Scully, 2009).
The MDC1 Platform
MDC1 associates with the FHA and BRCT motifs of NBS1 through multiple SDTD sites that are constitutively phosphorylated by CK2 (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). Moreover, MDC1 binds ATM through its FHA domain to further propagate γH2AX spreading (Figure 1A) (Lou et al., 2006). Formation of extensive γH2AX regions is important for sustaining the DNA damage response as H2AX is not required for the initial localization of NBS1, BRCA1 and 53BP1 at DSBs via PARP, but rather for the maintenance of these DDR factors at sites of damage (Celeste et al., 2002). Stabilization of DDR factor recruitment to γH2AX nucleosomes is achieved through the recruitment of an intricate network of chromatin modifying enzymes regulating ubiquitination, sumoylation, acetylation and methylation, as described below. In addition, experiments in yeast and mammalian cells have shown that chromatin remodelling enzymes, such as the SNF2 family protein INO80 and SWI/SNF, are recruited to DSBs in a γH2AX dependent manner (Lee et al., 2010b; van Attikum and Gasser, 2009). INO80 is thought to promote nucleosome eviction to facilitate DSB resection and HR. Similarly, SWI/SNF is known to stimulate chromatin relaxation at DSBs. Accumulation of SWI/SNF at DNA damage sites is facilitated by its interaction with the BRCT domain containing protein MCPH1/BRIT1, which directly associates with γH2AX in an MDC1-independent manner (Lin et al., 2010).
Ubiquitin-mediated recruitment
DDR-dependent MDC1 phosphorylation and recruitment to γH2AX initiates a ubiquitination cascade at sites of DNA damage that primarily involve protein monoubiquitination or lysine-63 (K63)-linked polyubiquitination (Messick and Greenberg, 2009). The ubiquitin ligase RNF8 associates to MDC1 phospho-TQ sites through an N-terminal FHA domain and activates a DDR induced ubiquitination cascade by K63-linked ubiquitination of H2A and γH2AX (Figure 1A) (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). The E3 RNF168 then binds these chains through its motifs interacting with ubiquitin (MIU) to stimulate K63-ubiquitination (Doil et al., 2009; Stewart et al., 2009). A third E3 ubiquitin ligase, HERC2, interacts with the FHA domain of RNF8 in a phosphorylation dependent manner and facilitates the assembly of the RNF8/UBC13 complex, further stimulating K63-linked ubiquitin ligase activity (Bekker-Jensen et al., 2010). The K63-ubiquitin chains generated by RNF8 and RNF168 are recognized by the ubiquitin interacting motifs (UIM) of RAP80, which recruits the BRCA1-A complex, itself an E3 ligase, through the interaction with the scaffold protein ABRA1 (Figure 1A) (Huen et al., 2007; Kolas et al., 2007; Wang and Elledge, 2007). The BRCA1-A complex additionally includes the ubiquitin conjugating enzyme variant (UEV) motif containing protein BRE, the ubiquitin protease BRCC36 and the adaptor protein NBA1 and has structural similarities to the proteosome lid (Feng et al., 2009; Shao et al., 2009; Wang et al., 2009). Ubiquitin binding activity has been shown for several subunits of the BRCA1-A complex, including ABRA1, BRE, BRCC36, NBA1 and RAP80 (Wang et al., 2009). Interestingly, recent observations have shown that the deubiquitinating enzyme OTUB1 suppresses RNF168-dependent ubiquitination by direct inhibition of UBC13 (Nakada et al., 2010).
SUMO-mediated Recruitment
It has recently been reported that the accumulation of the BRCA1-A complex at DSBs is also dependent on sumoylation (Galanty et al., 2009; Morris et al., 2009). Indeed, the SUMO ligases PIAS1 and PIAS4 have been shown to localize at sites of DNA damage and PIAS4 was reported to stimulate the ubiquitin ligase activity of the RNF8/UBC13 complex, thus promoting the recruitment of RNF168 and BRCA1 to sites of DNA damage (Figure 1A). PIAS1 has instead been proposed to directly sumoylate BRCA1 and stimulate its ubiquitin ligase activity (Galanty et al., 2009; Morris et al., 2009). Therefore, PIAS1 and PIAS4 might function as regulators of the ubiquitin ligase activity of RNF8 and BRCA1, respectively. In addition, 53BP1 recruitment to DSBs depends on the SUMO ligase PIAS4, which is also thought to directly sumoylate 53BP1 (Figure 1A) (Galanty et al., 2009). Given that 53BP1 does not display ubiquitin binding activity, it has been proposed that RNF8/RNF168 ubiquitination might induce chromatin relaxation and subsequent exposure of dimethyl H4K20 — and possibly dimethyl H3K79 — which are recognized by 53BP1 tudor domains (FitzGerald et al., 2009). H3K79 and H4K20 dimethylation levels do not appear to change after DNA damage, indicating that de-novo histone methylation is not promoting 53BP1 recruitment (FitzGerald et al., 2009). 53BP1 in known to associate with the deubiquitinating enzyme USP28 and PTIP, a six-BRCT motif-containing protein, in a phospho-dependent manner (Mohammad and Yaffe, 2009; Zhang et al., 2006). Despite the interaction with 53BP1, PTIP appears to be recruited to damage sites in a 53BP1-independent but RNF8/UBC13 dependent manner (Mohammad and Yaffe, 2009).
All of the complexities in modifications that are responsible for recruiting these many factors to sites of DNA damage or replication stress pose a significant conundrum for the field. Why do we need so many different modifications to build these structures? Is this an evolutionary accident or is there method in this madness? One possibility is that each layer of modification, i.e. phosphorylation, ubiquitination, sumoylation, acetylation recruits a different constellation of factors with distinct repair capabilities. Since the focus of proteins is built in layers, there could be a kinetic ordering of repair choices evident in each layer. Furthermore, since multiple E3 ligases (RNF8, RNF168, HERC2, BRCA1/BARD1) and deubiquitinating enzymes (USP28, BRCC36) are simultaneously recruited and phosphorylated and modified themselves, significant regulation of modifications could occur within these structures, thereby directing repair choices as events unfold. For example, if a DSB is clean it can be directly religated. If it is not, nucleolytic restructuring must occur. If the break is not held together, factors that promote end searching may be required to bring the ends into proximity for repair as proposed for 53BP1. If this attempt fails, unwinding or resection of the ends might occur to allow a search for microhomologies for alt-NHEJ, or more extensive homologies as for classical RAD51-mediated HR. These choices require different, often competing repair activities that must be temporally and structurally coordinated in a manner that optimally deals with the eventualities that occur with different repair events. Multiple modification layers may allow exquisite control of these choices. Clearly, it will be critical to dissect out the contribution of each individual focus component to different repair pathways in order to understand how these dynamic structures influence repair choices.
DNA Replication Stress, Fork Stalling and ICLs
Perhaps the most dangerous lesion facing cells is the stalling of a replication fork. Failure to properly overcome such lesions leads to an inability to complete chromosome duplication and can lead to mitotic catastrophe, complex chromosomal rearrangements and cell death. Cells have evolved multiple mechanisms to sense and respond precisely to these catastrophic types of lesions.
RPA: Sensor of DNA Replication Stress
Bulky DNA lesions can lead to arrest of leading strand synthesis at the replication fork and formation of extensive RPA-coated ssDNA regions due to the uncoupling between the MCM helicase and the DNA polymerase (Figure 1B) (Byun et al., 2005). RPA1 polymerizes on this ssDNA to generate a platform that activates the central signaling pathway orchestrating DNA replication responses, the ATR pathway. RPA-ssDNA complexes recruits the ATR/ATRIP complex through direct interaction with ATRIP to localize it to the fork (Figure 1B) (Zou and Elledge, 2003). Furthermore, it stimulates binding and activation of the RAD17-RFC2-5 clamp loader, which loads the PCNA-related RAD9-HUS1-RAD1 (9-1-1) heterotrimer bound to the ATR-activating TOPBP1 protein, and stimulates ATR kinase activity (Cimprich and Cortez, 2008; Ellison and Stillman, 2003; Kumagai et al., 2006; Mordes et al., 2008; Zou et al., 2003). The 9-1-1 complex is loaded onto 5′- or 3′-DNA ends adjacent to RPA-coated ssDNA regions (Cimprich and Cortez, 2008). It is the colocalization of these two RPA-dependent complexes that sets in motion the ATR signaling cascade, which results in the activation of CHK1 and CHK2 kinase signaling and phosphorylation of many chromatin bound factors to promote fork stability and restart of stalled or collapsed replication forks in order to complete chromosome replication (Cimprich and Cortez, 2008).
Restart of Stalled or Collapsed Replication Forks
Replication forks are fragile DNA structures that must be stabilized when fork progression is arrested by DNA lesions. Fork stability is promoted by the TIM/TIPIN complex and CLASPIN, both ATR targets (Errico and Costanzo, 2010). TIPIN, and its partner protein TIMELESS, associate with RPA2 to stabilize stalled forks and promote the accumulation of CHK1 and its regulatory protein CLASPIN to RPA-ssDNA regions where CHK1 can be activated by ATR (Figure 1B) (Kemp et al., 2010). Restart of stalled replication forks is dependent on several DNA helicases or translocases, including BLM, WRN, FANCM, HLTF and SMARCAL1, many of which are recruited to forks by RPA or interact with RPA at forks (Bachrati and Hickson, 2008; Driscoll and Cimprich, 2009; Luke-Glaser et al., 2010; Unk et al., 2010). In particular, SMARCAL1 interacts with RPA2 through an RPA2 interaction motif common to TIPIN, RAD52, XPA and UNG2 (Bansbach et al., 2009; Ciccia et al., 2009; Postow et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009). SMARCAL1, BLM, WRN and FANCM undergo ATM/ATR dependent phosphorylation after DNA damage (Bachrati and Hickson, 2008; Bansbach et al., 2009; Postow et al., 2009; Whitby, 2010; Yuan et al., 2009). BLM phosphorylation by ATR was shown to be important for recovery after replication stress and FANCM phosphorylation results in its tighter association with chromatin (Bachrati and Hickson, 2008; Kim et al., 2008). BLM, WRN, FANCM and HLTF have been suggested to regress replication by favoring the annealing of the leading- and lagging-strands and generating a HJ-like structure also known as “chicken foot”, which could allow the restart of DNA synthesis by template switching and lesion bypass (Atkinson and McGlynn, 2009; Blastyak et al., 2010). SMARCAL1 might also promote replication fork regression given the similarity between the helicase domain of SMARCAL1 and yeast Rad5, which was shown to regress replication forks (Atkinson and McGlynn, 2009; Blastyak et al., 2007). Future studies will be needed to uncover the coordination between these enzymes at the replication fork.
Single-strand DNA breaks encountered by the replication fork can be converted into DSBs during DNA synthesis, thus inducing fork collapse. Replication fork collapse could also be induced by direct fork cleavage by the MUS81/EME1 endonuclease following replication arrest (Ciccia et al., 2008; Hanada et al., 2007). Different from DSBs formed in non-replicating regions, collapsed replication forks contain one-ended DSBs. Repair of one-ended DSBs is carried out by the break induced replication (BIR) pathway, which involves DSB resection, strand invasion and reassembly of a new replication fork at a RAD51-generated D-loop intermediates (Llorente et al., 2008). PARP1/2 could facilitate processing of DSBs generated after replication stress by recruiting MRN (Bryant et al., 2009). Incorrect regulation of BIR could lead to multiple rounds of strand invasion and DNA synthesis at non perfectly homologous DNA sequences, thus leading to chromosomal rearrangements (Llorente et al., 2008). BIR has been observed in yeast, however its role in replication fork recovery in higher eukaryotes has not yet been visualized.
PCNA and Post-Replication Repair of ssDNA Gaps
Replication restart of stalled replication forks could be accomplished by reinitiation of leading and lagging strand synthesis downstream of the DNA lesion, as demonstrated in bacteria and yeast (Figure 3A) (Branzei and Foiani, 2010). Replication restart would leave behind the replication forks ssDNA gaps, which in yeast have been shown to be subsequently repaired by either TLS or error-free PRR in a manner dependent on the RAD6/RAD18 ubiquitin ligase complex (Branzei and Foiani, 2010; Daigaku et al., 2010; Karras and Jentsch, 2010). Whereas TLS involves the direct bypass of the DNA lesion using the 3′-end of the DNA filament arrested, error-free PRR is thought to promote strand invasion and repair of the ssDNA gaps by template switch and homologous recombination (Budzowska and Kanaar, 2009). TLS or error-free PRR are thought to be dependent, respectively, on monoubiquitination or K63-linked polyubiquitination of PCNA (Figure 3A) (Ulrich and Walden, 2010). PCNA monoubiquitination is promoted by the RAD6/RAD18 complex, which is recruited by RPA to unreplicated ssDNA regions (Davies et al., 2008). Monoubiquitinated PCNA can be further subjected to K63-linked polyubiquitination by the ubiquitin ligases UBC13, HLTF and SHPRH (Unk et al., 2010). HLTF and SHPRH are the mammalian orthologs of yeast Rad5 (Unk et al., 2010).
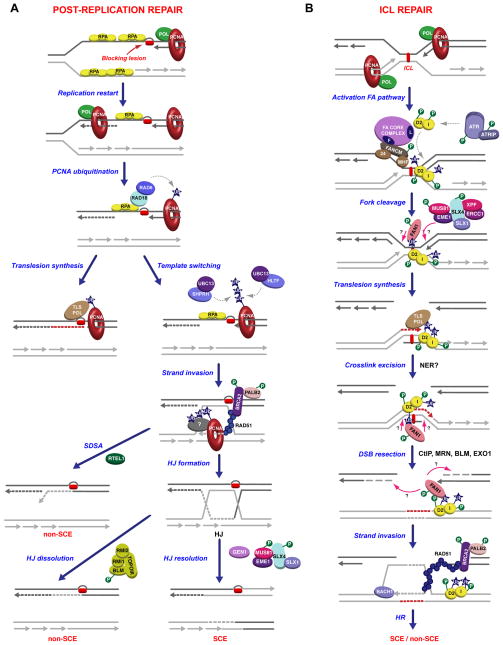
Figure 3. Repair of DNA lesions encountered during DNA replication.
(A) Post-replication repair of ssDNA gaps. Leading strand synthesis arrested at DNA lesions (red rectangular shapes) can be reprimed downstream of lesions, leaving ssDNA gaps behind the replication fork. Repair of ssDNA gaps is mediated by RAD6 and RAD18, which are recruited by RPA to ssDNA gaps, where they monoubiquitinate PCNA. Monoubiquitinated PCNA associates with translesion polymerases, which promote lesion bypass. Alternatively, polyubiquitination of PCNA by SHPRH, HLTF and UBC13 induces template switching and strand invasion into homologous sequences of the sister chromatid. Template switching could possibly involve proteins interacting with polyubiquitinated PCNA. Resolution of Holliday junctions formed after strand invasion can then result in sister chromatid exchanges (SCEs), whereas HJ dissolution and SDSA do not generate SCEs. (B) Repair of interstrand crosslinks. Converging replication forks blocked by interstrand crosslinks (red rectangle) activate the FA pathway. The FA core complex associated to blocked replication forks through the FANCM complex promotes the monoubiquitination of the FANCD2/FANCI (ID) complex. Phosphorylation of FANCI by ATR regulates the ubiquitination of the ID complex and its subsequent relocalization to blocked replication forks. Monoubiquitinated ID complex promotes fork cleavage, probably through the interaction with FAN1 and possibly other nucleases, translesion synthesis and crosslink excision. DSB resection, which could be dependent on FAN1, in addition to CtIP/MRN, BLM and EXO1, leads to strand invasion and homologous recombination with or without formation of SCEs as described in (A). Post-translational modifications are indicated as in Figure 1.
The PCNA Platform
PCNA is a homotrimeric DNA sliding clamp that functions as processivity factor for DNA polymerases (Moldovan et al., 2007). PCNA, which is loaded by the clamp loader RFC at 3′-primer-template junctions, tethers DNA polymerases to DNA and serves as loading platform for proteins that operate in conjunction with DNA synthesis. PCNA accumulates rapidly at sites of DNA damage, where it recruits DDR factors containing PCNA interaction motifs (Moldovan et al., 2007). How PCNA accumulates on DNA and whether that is regulated by the DDR is not yet clear although RFC is phosphorylated in response to DNA damage (Matsuoka et al., 2007). Three PCNA interaction motifs are known: the PCNA-interacting protein (PIP)-box, the AlkB homologue 2 PCNA-interacting motif (APIM) and the KA-box (Gilljam et al., 2009; Moldovan et al., 2007; Xu et al., 2001). More than 400 human proteins with putative PIP-box or APIM motif have been identified based on bioinformatic analyses (http://tare.medisin.ntnu.no/pcna/index.php). Approximately 30 PIP-box containing proteins have currently been shown to directly interact with PCNA, including TLS polymerases and NER, BER, MMR factors (Moldovan et al., 2007). Following DNA damage, the accumulation of PIP-box containing proteins, such as TLS polymerases, was shown to be dependent on a functional PIP-box (Moldovan et al., 2007). Given the intense competition among PIP-box containing proteins for the same binding pocket of PCNA (the interdomain connecting loop that connects the PCNA monomers), the PCNA binding affinity of specific DDR factors after DNA damage is regulated by DDR induced PCNA post-translational modifications, including RAD6/RAD18-dependent PCNA ubiquitination. Indeed, TLS polymerases associate with monoubiquitinated PCNA through UBZ or UBM ubiquitin binding motifs, which then provide an additional PCNA interaction surface required to target TLS polymerases to sites of DNA damage (Bienko et al., 2005; Ulrich and Walden, 2010). Monoubiquitination of the TLS polymerase eta (Polη) has been proposed to promote the intramolecular interaction between ubiquitin and the UBZ domain of Polη, thus impairing its association with monoubiquitinated PCNA (Bienko et al., 2010). Whether PCNA K63-linked polyubiquitin chains might serve as a signal to recruit factors involved in error-free PRR is currently unknown.
A key question in this field for both RPA and PCNA, which have many, many interacting factors, is how does the cell decide which factor to recruit? It is possible that everything is recruited simultaneously as a repair toolkit, and then the proper tool is selected based on geometrical constraints at the damage site. One parameter that could influence the selection of the proper tool might be the amount of ssDNA present or whether the recruitment is occurring in the context of a replication fork. This remains a significant challenge to unravel in the future.
The Fanconi Anemia Pathway for Interstrand Crosslink Repair
ICLs that covalently connect the two strands of DNA are a formidable bidirectional barrier to replication fork progression and require no less than 4 incision events, TLS polymerases and recombination events to be circumvented (Figure 3B). Central components of the ICL repair pathway are 13 genes mutated in the genetic syndrome Fanconi anemia (FA) (Moldovan and D’Andrea, 2009). Eight of the FA proteins (FANCA, B, C, E, F, G, L and M) form the FA core complex, an E3 ubiquitin ligase, with the addition of 4 associated factors (FAAP24, FAAP100 and the heterodimer MHF) (Thompson and Jones, 2010). The histone-fold heterodimer MHF has recently been shown to stimulate FANCM fork reversal activity, whereas FAAP24 could target the FANCM complex to ssDNA regions (Ciccia et al., 2007; Singh et al., 2010; Yan et al., 2010). FANCM has DNA binding activity and has been implicated in targeting the core complex to DNA (Figure 3B) (Kim et al., 2008). Moreover, FANCM has been suggested to associate with the BLM complex and also contribute to the activation of the ATR pathway at blocked replication forks (Collis et al., 2008; Deans and West, 2009; Huang et al., 2010; Schwab et al., 2010).
The FANCI-FANCD2 (ID) Platform
The FANC ID complex lies at the heart of the FA pathway and becomes monoubiquitinated on both subunits by the FANCL ubiquitin ligase within the FA core complex (Moldovan and D’Andrea, 2009). Once monoubiquitinated, the ID complex accumulates at sites of crosslinks and colocalizes with 3 additional FA proteins, BRCA2/FANCD1, PALB2/FANCN and BACH1/FANCJ (Figure 3B) (Moldovan and D’Andrea, 2009). The DDR tightly regulates the ID complex through ATR-dependent phosphorylation of both FANCI and D2 and several other components of the FA core complex like FANCM (Smogorzewska et al., 2007; Sobeck et al., 2009). Phosphorylation of FANCI was recently shown to play a critical role in promoting the monoubiquitination of FANCD2 and itself, thus acting as a molecular switch of the FA pathway (Ishiai et al., 2008). ID phosphorylation might facilitate the recruitment of the ID complex to the FA core complex for monoubiquitination. Monoubiquitinated ID complex is required for the incision and translesion synthesis steps of ICL repair (Figure 3B) (Knipscheer et al., 2009). The monoubiquitinated ID complex promotes the recruitment of DDR factors required for ICL repair, possibly the MUS81/EME1, XPF/ERCC1 nucleases and including the newly identified FAN1 endo- and exo-nuclease, which was shown to associate with the monoubiquitinated ID complex through a UBZ domain (Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010; Smogorzewska et al., 2010). Monoubiquitinated ID could also recruit the TLS polymerases REV1 and Polζ for the translesion step as they contain ubiquitin interaction domains (Moldovan and D’Andrea, 2009). Factors involved in HR are also likely to be recruited subsequent to these steps to repair the DSBs created in this process. Once the lesion has been repaired, the ID complex is deubiquitinated by the USP1-UAF1 complex, which also acts on monoubiquitinated PCNA when associated to the RFC subunit ELG1 (Cohn et al., 2007; Lee et al., 2010c). This is one of the clearest examples of how the DDR, through ATR/ATRIP localization and activation at the sites of replication stress, controls the enzymatic toolbox of repair at the right time and location to promote the appropriate repair event.
Additional DDR regulated pathways that promote DNA repair
dNTP Biosynthesis for DNA Replicational Repair Processes
Deoxyribonucleotides (dNTPs) are required for many aspects of DNA repair and their levels are tightly regulated. In yeast through humans, dNTP synthesis is increased in S phase to meet the demands of DNA replication. However, occasionally levels of dNTPs are insufficient, in particular for DNA repair outside of S phase. The enzyme ribonucleotide reductase (RNR), which controls the rate-limiting step in the synthesis of dNTPs, is one of the most highly regulated proteins in response to DNA damage. RNR is a tetramer containing a dimeric small and large subunit and is highly cell cycle regulated (Nordlund and Reichard, 2006). The transcription of many RNR subunits is highly inducible in response to DNA damage and replication stress in eukaryotes. Mutations that decrease the activity of yeast RNR or the failure to induce it result in sensitivity to DNA damage (Elledge and Davis, 1989). Mammals induce a specialized small subunit, p53R2, through the ATM/ATR-p53 pathway in response to DNA damage (Nordlund and Reichard, 2006). While RNR is primarily cytoplasmic, in response to DNA damage it acetylated by TIP60 and is localized to sites of DNA damage in the nucleus in a manner dependent on TIP60 (Niida et al., 2010). In theory, this produces dNTPs at the site of repair synthesis outside of S phase and may provide high local concentrations of dNTPs required for optimal function of TLS polymerases (Nordlund and Reichard, 2006).
DNA Repair of Transcribed Regions
Active transcription can provide obstacles to DSB repair because of possible collisions between the RNA polymerase and the HR machineries. Formation of DSBs in the highly repetitive nucleolar rDNA has been reported to induce transcriptional arrest and displacement of RNA polymerase I from rDNA in an ATM, NBS1 and MDC1 dependent manner (Kruhlak et al., 2007). The mechanism by which ATM regulates RNA polymerase transcription has not been defined. However, several nucleolar factors have been shown to be potential ATM/ATR substrates (Matsuoka et al., 2007). Recent observations have suggested that DSB formation leads to ATM-dependent repression of transcription in the vicinity of the DSB (Shanbhag et al., 2010). This could be reinforced by histone methyltranferase complexes, such as polycomb proteins, histone deacetylases (HDACs and sirtuins) and DNA methyltransferases, which have been shown to be recruited to sites of DNA damage (O’Hagan et al., 2008; Polo et al., 2010; Chou et al., 2010). Transcriptional repression could also be achieved by the RecQ helicase RECQ5, which prevents chromosomal rearrangements by both inhibiting RNA polymerase II and preventing excessive HR (Aygun et al., 2008; Aygun et al., 2009; Islam et al., 2010).
Bulky DNA lesions, such as UV-induced pyrimidine dimers, in transcribed genes can lead to RNA polymerase II stalling (Hanawalt and Spivak, 2008). Removal of stalled RNA polymerase and repair of these lesions is promoted by transcription coupled NER (TC-NER), which is initiated by the Cockayne syndrome proteins CSA and CSB. Once RNA polymerase has been removed, the repair of the DNA lesions is catalyzed by the Xeroderma pigmentosum (XP) proteins (Hanawalt and Spivak, 2008). TC-NER is distinct from global genome NER (GG-NER), which is initiated by XPC and operates throughout the genome (Hoeijmakers, 2009). Following UV radiation, XPC is phosphorylated by ATM/ATR and polyubiquitinated by the cullin ligase complex CUL4-DDB1-DDB2, however the significance of these modifications is still unknown (Bergink and Jentsch, 2009; Matsuoka et al., 2007).
DNA Repair of Heterochromatic Regions
Heterochromatic regions, such as centromeric regions and inactive X chromosome in females, pose a particular problem to DNA repair because of extreme chromatin compaction. Establishment and maintenance of heterochromatin is dependent on H3K9me3 marks, which are recognized by the chromodomain of the heterochromatin protein HP1 (Fischle, 2009). Recent studies have shown that the chromodomain of HP1 is phosphorylated by CK2 after DNA damage, thus releasing HP1 from H3K9me3 (Ayoub et al., 2009). Association of TIP60 with exposed H3K9me3 through its chromodomain has been suggested to stimulate its acetyltranferase activity (Figure 1A) (Sun et al., 2009). TIP60 acetylation of histones H3 and H4 could then induce chromatin relaxation and facilitate the DSB repair of heterochromatic regions (Sun et al., 2010). TIP60 could also promote chromatin remodelling at DSBs by acetylating γH2AX modified histones, which are then removed following ubiquitination by the UBC13 ubiquitin ligase (van Attikum and Gasser, 2009). TIP60 is also known to interact with ATM and stimulate its kinase activity by directly acetylating ATM (Sun et al., 2010). Activation of ATM leads to very rapid phosphorylation of the transcriptional repressor KAP1, which accumulates at heterochromatic DSBs in a 53BP1-dependent manner, where it contributes to chromatin relaxation (Noon et al., 2010). Moreover, 53BP1 interacts with the chromatin remodelling factor EXPAND1, which induces chromatin decondensation at sites of DNA damage (Huen et al., 2010a). Altogether, these observations point towards a critical role of the ATM signaling for the repair of DSBs in heterochromatin regions (Sun et al., 2010).
The DNA damage response and human disease
The central role of the DNA damage response in human physiology is indicated by the broad spectrum of defects displayed by individuals carrying mutations in DDR genes. DDR genetic syndromes primarily affect the homeostasis of the nervous, immune and reproductive systems and can also lead to premature aging or cancer predisposition (Table 2) (Jackson and Bartek, 2009). In this section we will describe the pathologies associated with DDR defects.
Table 2.
Human genetic diseases associated with DDR defects.Mutated genes, DDR defects and phenotypes of each genetic disorder are indicated.
| Syndrome | Mutated gene | DDR defect | Phenotype |
||||
|---|---|---|---|---|---|---|---|
| Neurological disorder | Immunodeficiency | Progeria | Cancer | Other | |||
| Cerebro-oculo-facio-skeletal syndrome (COFS) |
CSB, XPD XPG, ERCC1 |
TC-NER | Brain calcification Hypomyelination Microcephaly Neurodegeneration |
Cataracts Hearing loss Optic atrophy Osteoporosis |
Facial dysmorphism Joint contractures Photosensitivity Growth defects |
||
| Cockayne syndrome (CS) |
CSA, CSB XPB, XPD, XPG |
TC-NER | Microcephaly Neurodegeneration Neuronal demyelination |
Cachexia Cataract Hearing loss Retinopathy |
Photosensitivity Growth defects |
||
| Trichothiodystrophy (TTD) |
XPB, XPD TTDA |
TC-NER | Hypomyelination Neurodegeneration |
Cachexia Cataracts Osteoporosis |
Brittle hair, nails Photosensitivity Scaly skin |
||
| Xeroderma pigmentosum (XP) |
XPA-G POLH |
NER | Microcephaly Neurodegeneration |
Squamous & Basal cell carcinoma Melanoma |
Photosensitivity Scaly skin |
||
| XPF-ERCC1 (XFE) syndrome | XPF | NER ICL repair |
Microcephaly | Cachexia Osteoporosis Scoliosis |
Photosensitivity Liver and renal dysfunction |
||
| Fanconi anemia (FA) |
FANCA-C FANCD1, D2, FANCE, FANCG FANCI, J, L-N |
ICL repair HR |
Microcephaly | Pancytopenia | Bone marrow failure | AML Myelodysplasia Squamous cell carcinoma |
Abnormal Skin pigmentation Infertility Limb deformities Renal dysfunction |
| Fanconi anemia-like disorder | RAD51C | ICL repair HR |
Growth defects | Hypogonadism Limb deformities Renal dysfunction |
|||
| Familial breast cancer | ATM, BRCA1, BRCA2, BRIP1, CHK2, NBS1, PALB2, RAD50, RAD51C | HR Damage signaling |
Breast and Ovarian cancer (BRCA1 BRCA2 RAD51C) | ||||
| Bloom syndrome (BS) | BLM | HR | Microcephaly Mild mental retardation |
Immunoglobulin deficiency | Carcinomas Leukemias Lymphomas |
Abnormal Skin pigmentation Facial dysmorphism Infertility Growth defects |
|
| Rothmund Thomson syndrome (RTS) | RECQL4 | BER HR? |
Cataracts Grey hair |
Osteosarcoma Skin cancers |
Skin and skeletal abnormalities Growth defects |
||
| Werner syndrome (WS) | WRN | HR BER Telomere maintenance |
Atherosclerosis Cataracts Grey hair Osteoporosis |
Sarcomas | Type II diabetes Growth defects |
||
| Dyskeratosis congenita (DKC) |
DKC1 TERC |
Telomere maintenance | Microcephaly Mental retardation |
Pancytopenia | Bone marrow failure Osteoporosis |
Carcinomas | Abnormal skin pigmentation Nail dystrophy Growth defects |
| Ataxia with oculomotor apraxia 1 (AOA1) | APTX | SSB repair | Ataxia Neurodegeneration Oculomotor apraxia |
Hypercolesterolemia | |||
| Ataxia with oculomotor apraxia 2 (AOA2) | SETX | SSB repair? | Ataxia Neurodegeneration Oculomotor apraxia |
Hypercolesterolemia | |||
| Spinocerebellar ataxia with axonal neuropathy (SCAN1) | TDP1 | SSB repair | Ataxia Neurodegeneration Muscle weakness |
Hypercolesterolemia | |||
| Ligase I syndrome | LIG1 | SSB repair NER |
Immunoglobulin deficiency | Growth defects Photosensitivity |
|||
| MYH-associated polyposis (MAP) | MYH | BER, Oxidative damage repair | Colorectal cancer | ||||
| Hereditary non-polyposis colorectal cancer (HNPCC) | MSH2, MSH6, MLH1, PMS2 | MMR | Colorectal cancer Carcinomas |
||||
| Immunodeficiency with microcephaly | XLF | NHEJ | Microcephaly | Hypogammaglobulinemia Lymphopenia |
Growth defects | ||
| Ligase IV syndrome | LIG4 | NHEJ | Microcephaly | Hypogammaglobulinemia Lymphopenia |
ALL Lymphomas | Growth defects | |
| Radiosensitive severe combined immunodeficiency (RS-SCID) | ARTEMIS | NHEJ | Agammaglobulinemia Lymphopenia |
Lymphomas | Growth defects | ||
| Severe combined immunodeficiency (SCID) | RAG1, RAG2 | NHEJ | Agammaglobulinemia Lymphopenia |
Growth defects | |||
| Microcephaly, intractable seizures and developmental delay syndrome (MCSZ) | PNKP | NHEJ SSB repair |
Microcephaly | Seizures Growth defects |
|||
| Hyper-IgM syndrome | AID, UNG | CSR | Increased IgM levels Lymphoid hyperplasia |
||||
| Aicardi Goutieres syndrome (AGS) |
RNASEH2 TREX1 |
Damage signaling Immunological response |
Cerebral atrophy Intracranial calcifications Microcephaly Neurodegeneration |
||||
| Ataxia telangiectasia (AT) | ATM | Damage signaling DSB repair Oxidative stress |
Ataxia Cerebellar degeneration Oculomotor apraxia |
Immunodeficiency | Lymphomas Leukemias Breast cancer |
Dilated blood vessel Infertility Metabolic defects Growth defects |
|
| Ataxia telangiectasia-like disorder (ATLD) | MRE11 | Damage signaling DSB repair Oxidative stress |
Ataxia Cerebellar degeneration Oculomotor apraxia |
Immunodeficiency | |||
| Li-Fraumeni syndrome (LFS) | TP53 | DNA damage signaling DSB repair |
Brain and Breast cancer Sarcomas |
||||
| Nijmegen breakage syndrome (NBS) | NBS1 | Damage signaling DSB repair Repl. fork repair |
Microcephaly | Immunodeficiency | B-cell lymphoma | Facial dysmorphism Growth defects |
|
| Nijmegen breakage syndrome-like disorder (NBSLD) | RAD50 | Damage signaling DSB repair Repl. fork repair |
Microcephaly | Facial dysmorphism Growth defects |
|||
| Riddle syndrome | RNF168 | Damage signaling DSB repair |
Immunodeficiency | Facial dysmorphism Growth defects |
|||
| Seckel syndrome (SS) |
ATR, PCTN SCKL2, SCKL3 |
Damage signaling DSB repair Repl. fork repair |
Microcephaly Mental retardation |
AML? | Facial dysmorphism Growth defects |
||
| Primary microcephaly 1 | MCPH1 | Damage signaling DSB repair Repl. fork repair |
Microcephaly Mental retardation |
||||
| Schimke immuno-osseous dysplasia (SIOD) | SMARCAL1 | Repl. fork repair | Microcephaly | T-cell deficiency | Nephritis Skeletal dysplasia Growth defects |
||
| Roberts syndrome (RBS) | ESCO2 | Cohesion Repl. fork repair |
Cleft palate Phocomelia |
||||
| Hutchinson-Gilford progeria syndrome (HGPS) | LMNA | Nuclear organization Damage signaling DSB repair |
Alopecia Atherosclerosis |
Adipose tissue deficiency | |||
| Restrictive dermopathy (RD) |
LMNA ZMPSTE24 |
Nuclear organization Damage signaling DSB repair |
Facial dysmorphism Tight skin Growth defects |
||||
| Amyotrophic lateral sclerosis (ALS) |
SOD1 SETX |
Oxidative stress SSB repair? |
Degeneration motor neurons | Muscular atrophy | |||
| Charcot-Marie-Thooth syndrome (CMT) | PMP22, GJB1, EGR2, SH3TC2, MTMR2, MTMR13 | Damage processing? Oxidative stress? |
Motor and sensory neuropathy Neuro-demyelination |
Muscular atrophy | |||
| Spino-cerebellar ataxia-epilepsy syndrome (SCAE) |
POLG TWINKLE |
mtDNA maintenance Oxidative stress? |
Ataxia, Dysarthria Neuropathy |
Epileptic seizures | |||
| Progressive external ophtalmoplegia (PEO) |
POLG, POLG2 TWINKLE, RRM2B |
mtDNA maintenance Oxidative stress? |
Eye, limb and facial muscle weakness | ||||
Neurological Defects
The nervous system relies heavily on an intact DNA damage response for functionality. Given that neurons display limited capacity of replacement, they must overcome DNA damage lesions often on a lifetime basis. Neurons exhibit high oxygen consumption by mitochondrial respiration, which can result in oxidative stress and subsequent DNA damage, such as DNA base lesions and DNA breaks (Jackson and Bartek, 2009). Consistent with this observation, defects in the regulation of oxidative stress and repair of DNA lesions often result in neuronal death and neurodegeneration. Several DDR syndromes affect primarily the cerebellum, which is responsible for motor coordination (Table 2) (Katyal and McKinnon, 2008). The cerebellum is composed by three classes of neurons — granule cells, Purkinjie cells and interneurons — which account for approximately 50% of the neurons of the whole brain (Katyal and McKinnon, 2008). Degeneration of cerebellar neurons often result in ataxia (impaired motor coordination), oculomotor apraxia (eye movement defect) and dysarthria (speech disorder). One of the most extensively studied neurodegenerative diseases is ataxia telangiectasia (A-T), which is caused by mutations in the ATM gene (Biton et al., 2008). A-T patients develop profound ataxia due to the progressive loss of granule and Purkinjie cells and are confined to a wheelchair before 10 years of life. Similar symptoms, although characterized by later onset and slower progression, are developed by patients with the ataxia telangiectasia like syndrome (A-TLD), which is caused by mutations in MRE11. The similarity between ATM and MRE11 deficient syndromes is consistent with the previously described role of MRE11 in ATM activation. The mechanism by which ATM deficiency causes cerebellar degeneration is still object of extensive debate. ATM has been shown to regulate oxidative stress, as indicated by the increased ROS levels in the absence of ATM (Biton et al., 2008). Neurodegeneration could then be caused by excessive ROS-induced DNA damage in ATM deficient cerebellar neurons. Interestingly, treatment with antioxidant agents was reported to enhance the survival of ATM−/− Purkinjie cells in vitro (Biton et al., 2008). Moreover, A-T patients subjected to steroid therapy showed improved cerebellar functions associated with reduction of ROS levels (Russo et al., 2009).
Defects in repair of SSBs, one of the primary ROS-induced lesions, have also been associated with cerebellar degeneration and ataxia (Table 2). The neurodegenerative disorders ataxia with axonal neuropathy (SCAN1) and ataxia with oculomotor apraxia 1 (AOA1) are caused by mutations of the DNA end processing enzymes TDP1 and APTX, respectively (Caldecott, 2008). TDP1 processes 3′-ends linked to TOP1, which are generated by abortive release of the topoisomerase from DNA, and other non-ligatable 3′-ends induced by ROS and other DNA damaging agents, whereas APTX is a 5′-end processing enzyme that removes abortive ligation intermediates from ssDNA and dsDNA ends (Rass et al., 2007). The helicase SETX, which is mutated in ataxia with oculomotor apraxia 2 (AOA2), has also been linked to the repair of SSBs generated by oxidative damage, but its precise role is still unknown (Rass et al., 2007).
Accumulation of mutations in mitochondrial DNA (mtDNA), which can lead to defects in oxygen metabolism and increased ROS levels, has also been associated with neurodegenerative disorders (Table 2). Indeed, mutations in the mitochondrial polymerase Polγ and its DNA helicase TWINKLE have been identified in the spino-cerebellar ataxia-epilepsy syndrome (SCAE), which is characterized by neuropathy, dysarthria and epileptic seizures (Copeland, 2008). Defects in Polγ, TWINKLE and the ribonucleotide reductase subunit RRM2B have also been linked to progressive external ophtalmoplegia (PEO), a disorder leading to degeneration of the external eye muscle (Copeland, 2008). Mutations and deletions of mtDNA have also been found in Parkinson’s, Alzheimer’s and Huntington’s diseases and amyotrophic lateral sclerosis (ALS) and they have been correlated with an increase in oxidative damage in the brain (Druzhyna et al., 2008). ALS, which causes progressive degeneration of motor neurons resulting in paralisis, can be induced by mutations of the ROS detoxifying enzyme SOD1 and the helicase SETX (Chen et al., 2004; Valdmanis and Rouleau, 2008). Recently, genes causing another motor neuron disorder, the Charcot-Marie-Thooth (CMT) syndrome, which leads to myelination defects and motor and sensory neuropathy, have been shown to prevent accumulation of DNA damage (Paulsen et al., 2009). Whether CMT genes might prevent oxidative damage or might regulate other DDR pathways has not been yet defined.
In addition to maintaining the homeostasis of the nervous system, the DNA damage response plays a critical role during brain development. Indeed, a wide number of DDR syndromes display microcephaly, a reduced head circumference resulting from defective proliferation of neuroprogenitor cells during fetal development (Table 2) (O’Driscoll and Jeggo, 2008). Microcephaly is one of the typical phenotypes of Seckel syndrome (SS) patients, in addition to dwarfism and “bird-like” facial dysmorphism (Kerzendorfer and O’Driscoll, 2009). Mutations of four different loci — SCKL1 to SCKL4 — have been found in Seckel syndrome patients. SCKL1 Seckel syndrome is caused by a hypomorphic ATR mutation that decreases ATR protein levels due to aberrant splicing of the ATR transcript, whereas the centrosomal protein PCNT, which has been shown to function in the ATR pathway, is mutated in SCKL4 patients (Kerzendorfer and O’Driscoll, 2009). PCNT mutations have recently been found in the microcephalic osteodysplastic primordial dwarfism type II (MOPDII), a disorder similar to Seckel syndrome (Kerzendorfer and O’Driscoll, 2009). Given that Seckel syndrome appears to be a disorder of defective ATR pathway, the uncharacterized genes responsible for SCKL2 and SCKL3 are probably novel ATR pathway components. Another protein shown to mediate the activation of the ATR pathway, MCPH1/BRIT1, has been shown to be defective in patients with primary microcephaly (O’Driscoll and Jeggo, 2008). Moreover, defects of the ATR pathway have been proposed to cause the microcephalic phenotype of the Blepharophimosis-ptosis-epicanthus inversus syndrome, Miller-Dieker lissencephaly syndrome and the Williams-Beuren syndrome, which are haploinsufficient for ATR, RPA1 and RFC2, respectively (Kerzendorfer and O’Driscoll, 2009).
Microcephaly is also characteristic of the Nijmegen breakage syndrome (NBS) and the Nijmegen breakage syndrome-like disorder (NBSLD), which are caused by hypomorphic mutations of NSB1 and RAD50, respectively (Katyal and McKinnon, 2008; Waltes et al., 2009). Similar to Seckel syndrome, NBS and NBSLD patients display growth retardation and “bird-like” face. These phenotypes are remarkably different from the A-T like symptoms due to MRE11 hypomorphic mutations in A-TLD patients, as mentioned above. This might reflect the complex functions of the MRN complex in both ATM activation and regulation of ATR-mediated processes, such as replication fork stabilization and restart. Therefore, the different impact of the hypomorphic mutations in MRE11, NBS1 and RAD50 on the ATM or ATR pathways might give rise to the distinct phenotypes of A-TLD and NBS/NBSLD patients. Interestingly, it has been recently reported that A-TLD mutant mice display loss of ATM-dependent apoptosis in the nervous system after DNA damage, thus causing faulty incorporation of damaged cells in the brain, whereas NBS mutant neurons still exhibit ATM-dependent apoptosis, which could result in cell loss after damage and microcephaly (Shull et al., 2009). Therefore the level of ATM residual activity in the brain might determine the outcome between neurodegeneration and microcephaly.
Syndromes with defective HR, such as Bloom syndrome, which is caused by mutations in the RecQ helicase BLM, or impaired ICL repair, like Fanconi anemia and the XFE syndrome, which is due to XPF mutations, lead to microcephaly (O’Driscoll and Jeggo, 2008). Moreover, microcephaly could also be caused by disorders that exhibit defective replication fork restart after DNA damage, as the Schimke immunosseous dysplasia (SIOD), which is caused by mutations of the SMARCAL1 helicase, or by NHEJ defective disorders with LIG4, XLF or PNKP mutations (Driscoll and Cimprich, 2009; O’Driscoll and Jeggo, 2008; Shen et al., 2010). Defective neuronal development is also observed in Ku deficient mice (Gu et al., 2000). Altogether, these observations have led to the proposal that the developing brain, and expecially neuroprogenitor cells, might be particularly sensitive to the presence of DSBs, which could be formed following replication stress or oxidative damage (O’Driscoll and Jeggo, 2008). Defective DSB repair could then lead to microcephaly as a consequence of ATM-dependent apoptosis of damaged neural cells. Interestingly, prenatal exposure to ionizing radiation is known to cause microcephaly in humans, as indicated by the reduced brain size of the Hiroshima and Nagasaki atomic bomb survivors that were in utero at the time of the radiation exposure (Fernandez-Capetillo, 2010).
Infertility and Immunological Defects
The DNA damage response plays an essential role in the generation of gametes and the development of the immune system. The generation of gametes during meiosis requires the exchange of genetic material between homologous chromosomes, which involves the formation of DSBs by the nuclease SPO11 and their subsequent repair by homologous recombination (Neale and Keeney, 2006). Defective DSB repair during meiosis results in infertility. A large number of DDR deficient mice and various human DDR syndromes, such as A-T, BS and FA, display aberrant meiotic progression and infertility (Table 2) (Biton et al., 2008; Bohr, 2008; Matzuk and Lamb, 2008). Given the requirement of a functional DDR for meiotic progression, a significant proportion of human infertility could be due to DDR defects (Jackson and Bartek, 2009).
The development of a functional immune system necessitates the generation of a wide number of unique immunoglobulins and T-cell receptors. This is obtained by joining different combinations of variable (V), diversity (D) and junction (J) DNA sequences through V(D)J recombination (Gennery, 2006). V(D)J recombination is initiated by the nucleases RAG1 and RAG2, which introduce specific DSBs at the segments to be rearranged. The ends of the RAG1/RAG2 DSBs are generated as hairpin loops, that are opened by the nuclease ARTEMIS prior to end joining by NHEJ factors (Gennery, 2006). Mutations of RAG1 and RAG2 have been identified in patients with severe combined immunodeficiency (SCID) caused by profound lymphopenia with diminished or absent immunoglobulins (Table 2) (Sobacchi et al., 2006). Similarly, ARTEMIS mutations give rise to SCID, which is however accompanied by radiosensitivity (RS-SCID) due to the role of ARTEMIS in the repair of radiation induced DSBs (Gennery, 2006). Hypomorphic mutations of RAG1, RAG2 or ARTEMIS have been found in the Omenn’s syndrome, a less severe immunodeficiency disorder characterized by low levels of V(D)J recombination and clonal expansion of limited set of T lymphocytes (Sobacchi et al., 2006). Low immunoglobulin levels, lymphopenia and radiosensitivity are also characteristic of the Ligase IV and immunodeficiency with microcephaly syndromes, which are defective in the NHEJ factors LIG4 and XLF, respectively (Gennery, 2006). As mentioned above, LIG4 and XLF, unlike ARTEMIS, are also required for brain development, thus reflecting a more essential role of LIG4 and XLF in NHEJ. No mutations in Ku70, Ku80 and DNA-PKcs, which are all essential for V(D)J recombination as indicated by the SCID phenotype of knockout mouse models, have yet been reported in human immunodeficiency syndromes (Brugmans et al., 2007; Gennery, 2006).
Diversification of V(D)J variable domains is further achieved by class switch recombination (CSR) and somatic hypermutation (Stavnezer et al., 2008). CSR promotes the switch between the constant regions of immunoglobulins, whereas somatic hypermutation introduces point mutations in the V(D)J variable domains. Both CSR and somatic hypermutation are triggered by the AID enzyme, which deaminates cytosines to uracil (Stavnezer et al., 2008). In CSR DNA breaks are then generated by the combined action of the uracil DNA glycosylase UNG and the AP endonuclease APE1. Classical NHEJ or alt-NHEJ could then lead to the exchange of the antibody constant regions. Defects in AID and UNG have been shown to cause the Hyper-IgM syndrome, which is characterized by normal IgM levels, but little or none IgG, IgA and IgE isotypes (Stavnezer et al., 2008).
Immunodeficiency has also been reported for A-T patients, who display lymphopenia and decreased levels of IgG, IgA and IgE isotypes (Gennery, 2006). Indeed, it has been shown that ATM stabilizes DNA repair complexes during V(D)J recombination and facilitates CSR (Stavnezer et al., 2008). Defective CSR has also been reported for mice deficient for H2AX, MDC1, 53BP1 and NBS1 (Stavnezer et al., 2008). Given that other DDR syndromes with defects of the ATM pathway — A-TLD, NBS and the Riddle syndrome, which is due to RNF168 mutation — lead to immunodeficiency resulting from reduced efficiency of V(D)J recombination and/or CSR, the ATM pathway appears to play an important role in the development of the immune system in humans (Gennery, 2006; Stewart et al., 2007). Interestingly, the recently identified SIOD syndrome leads to deficiency of T- but not B-lymphocytes (Boerkoel et al., 2002). The specific role of the SIOD protein SMARCAL1 in T-cell development or maturation awaits further investigation.
Premature Aging and Stem Cell Exhaustion
Accumulation of DNA damage can lead to premature aging, as indicated by several DDR syndromes with progeroid phenotypes (Table 2). Deficiency in multiple DDR pathways, including NER, BER and DSB repair, have been linked to premature aging. The progeroid syndromes Cockayne syndrome (CS), Cerebro-oculo-facio-skeletal syndrome (COFS) and Trichotiodystrophy (TTD) are caused by defects in TC-NER, whereas the related cancer-prone syndrome Xeroderma pigmentosum (XP) — classified into 8 complementation groups, XPA to XPG and XPV — is associated with defects in GG-NER or TLS (Hoeijmakers, 2009). CS, which can be caused by mutations of the TC-NER factors CSA and CSB, the TFIIH helicases XPB and XPD and the NER endonucleases XPG, is characterized by hearing loss, cataracts, weight loss with muscle atrophy (cachexia) and retinal defects associated to microcephaly and neurodegeneration. Mutations in XPD, XPG and CSB can also be found in COFS, an early onset form of CS with severe symptoms apparent at birth (Garinis, 2008). Moreover, some XPB and XPD mutations are associated with TTD, which has similar symptoms to CS with the addition of brittle hair and nails, and with an XP and CS combined syndrome (XP-CS) (Schumacher et al., 2008).
Deficiency in the RecQ helicase WRN leads to the development of Werner syndrome (WS), one the best examples of progeroid syndromes for its remarkable resemblance to normal aging (Bohr, 2008). WS patients prematurely develop grey hair, osteoporosis, cataracts, type II diabetes and atherosclerosis and generally die in the fifth decade of life because of cancer or cardiovascular diseases. Premature aging features, such as grey hair and cataracts, have also been associated with the Rothmund-Thomson syndrome (RTS), which is caused by mutations of the RecQ helicase RECQL4 (Chu and Hickson, 2009). It has been proposed that WRN and RECQL4 might participate in the removal of oxidative lesions by BER (Bohr, 2008). Moreover, WRN can promote HR and telomere maintenance (Chu and Hickson, 2009). Indeed, telomere shortening is known to lead to DDR activation and induction of senescence (Sahin and Depinho, 2010).
The impact of telomere dysfunction on organismal aging is further demonstrated by the progeroid syndrome dyskeratosis congenita (DKC), which is due to mutations of the telomerase associated TERC RNA or its interacting protein DKC1 (Schumacher et al., 2008). DCK patients develop progressive bone marrow failure and pancytopenia, accompanied by growth defects, osteoporosis and abnormal skin pigmentation due to telomere shortening resulting in the deprotection of telomeres and the activation of the DDR. Bone marrow failure and pancytopenia are also found in Fanconi anemia patients, who additionally display skeletal abnormalities, renal dysfunction, abnormal skin pigmentation, sensitivity to ICL agents and infertility, as mentioned above (Moldovan and D’Andrea, 2009). The bone marrow failure in DKC and FA patients is probably due to exhaustion of hematopoietic stem cells (HSC) caused by telomere shortening or DNA damage, respectively (Park and Gerson, 2005).
Depletion of HSCs has been observed in a variety of mouse models defective for DDR components, including ATM, BRCA2, ERCC1, FANCC, Ku80, LIG4, MSH2, XPD and mouse telomerase RNA (mTR) (Niedernhofer, 2008; Park and Gerson, 2005; Sharpless and DePinho, 2007). Spontaneous DNA damage accumulated in mice deficient for ATM, LIG4, XPD, mTR or Ku80 was shown to impair HSC self-renewal (Niedernhofer, 2008). In ATM deficient mice, HSC failure was reported to depend on increased oxidative damage, which induces the accumulation of the CDK inhibitor p16INK4A (Sharpless and DePinho, 2007). p16INK4A, which functions together with the tumor suppressor Rb to induce cell cycle arrest and cellular senescence, has been shown to accumulate with age in adult stem cells, thus reducing their regenerative capacity (Collado et al., 2007). Deficiency of p53, which also regulates cellular senescence, was shown to rescue many of the progeroid features displayed in mTR deficient mice due to defects of HSC and stem cells from the intestinal crypt and testis (Sharpless and DePinho, 2007). Similarly, the accelerated aging phenotypes of mice defective for BRCA1, Ku80 and ZMPSTE24, a nuclear membrane protease mutated in the Hutchinson-Gilford progeria syndrome, were rescued by TP53 deletion (Sahin and Depinho, 2010). Deficiency of p53 in these mouse models led to increased cancer formation, indicating that p53 rescues aging at the expenses of cancer. Interestingly, TP53 deletion was shown to exacerbate the progeroid phenotypes of a Seckel syndrome mouse model with ATR deficiency (Fernandez-Capetillo, 2010). Given that the progeroid features of Seckel syndrome mice might be due to replication stress during embryonic development, the increased proliferation rate induced by p53 deficiency could further increase the amount of replication stress and therefore accelerate the aging phenotype. Deletion of ATR in adult mice was also shown to lead to stem cell loss and premature aging (Ruzankina et al., 2007).
Analysis of several DDR mouse models, including mouse models for Seckel syndrome, CS and XFE progeria, have revealed a reduction of the growth hormone and insulin-like growth factor 1 (GH-IGF1) axis (Schumacher et al., 2008). Attenuation of the GH-IGF1 pathway is known to lead to extended lifespan and increased stress resistance and occurs during natural aging. The decreased function of the GH-IGF1 pathway after DNA damage might represent an attempt to reduce further damage accumulation by shifting the organismal resources from growth to tissue maintenance (Schumacher et al., 2008).
Cancer Development and Therapy
Cancer is an evolutionary disease fueled by genomic instability (Negrini et al., 2010). The majority of cancers display chromosomal instability (CIN), which is characterized by alterations of chromosomal numbers and/or structure. Other forms of genomic instability include accumulation of DNA base mutations and microsatellite instability (MIN), which results in contraction or expansion of the number of repetitive microsatellite sequences (Negrini et al., 2010). Maintenance of genomic integrity by the DNA damage response is critical to prevent tumorigenesis, as indicated by the cancer-prone phenotype of several DDR syndromes (Table 2). Hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch syndrome, is caused by heterozygous mutations of MMR genes, such as MLH1, MSH2, MSH6 and in fewer cases PMS2 (Spry et al., 2007). HNPCC is associated with MIN, which predisposes primarily to the development of colorectal cancer, with additional possible carcinomas of the endometrium, ovary, stomach and kidney. The risk of colorectal cancer is increased also in the MYH-associated polyposis (MAP), which is caused by defective repair of oxidative lesions due to mutations of the DNA glycosylase MYH (David et al., 2007). The mechanism by which deficiency of MMR factors and MYH preferentially affects intestinal cells has not been determined. Instead, deficiency of NER factors in the XP syndrome is known to greatly increase the risk of skin cancer and melanoma because of defective repair of UV lesions in skin cells following sun exposure (Hoeijmakers, 2009).
Familial breast cancer accounts for approximately 5–10% of breast cancer cases (Fackenthal and Olopade, 2007). The most prevalent mutations leading to hereditary breast and ovarian cancer affect the HR genes BRCA1 and BRCA2. Heterozygous individuals carrying mutations of the BRCA1 or BRCA2 genes have a 40–80% risk of developing breast cancer (Fackenthal and Olopade, 2007). Patients with BRCA2 mutations have increased incidence of male breast, pancreas and prostate cancer (Moynahan and Jasin, 2010). Tumors with BRCA1 or BRCA2 mutations are significantly associated with low level of 53BP1, indicating that 53BP1 mutation might confer a survival advantage in the absence of BRCA1 and BRCA2 (Bouwman et al., 2010). This is probably due to partial restoration of HR in BRCA1 mutant tumors, as previously described. Recently, mutations in three additional HR genes, BACH1, PALB2 or RAD51C, have been identified in approximately 3% of familiar breast cancer patients and have been associated with a 2-fold increased risk of breast cancer (Levy-Lahad, 2010; Walsh and King, 2007). Mutations of CHK2, ATM, NBS1, RAD50 have also been associated with a doubled risk of breast cancer, thus indicating the importance of the ATM pathway, together with HR, in preventing breast cancer formation (Walsh and King, 2007).
It has recently become apparent the strong connection between familial breast cancer and FA syndrome (Levy-Lahad, 2010). Indeed, BRCA2, BACH1 and PALB2 mutations predispose to breast cancer formation when monoallelic and lead to FA syndrome — complementation groups D1, J and N, respectively — when biallelic. Similarly, biallelic mutations of RAD51C have been identified in an FA-like disorder (Levy-Lahad, 2010). FA is also associated with increased cancer risk, in particular for the development of myelodisplasia (pre-leukemic syndrome) and acute myeloid leukemia (AML) (Moldovan and D’Andrea, 2009). Hematological neoplasia is also found in A-T patients (lymphomas and leukemias) and NBS patients (B-cell lymphomas) (Gennery, 2006; Spry et al., 2007). Some cases of hematological malignancies, such as EBV-associated lymphomas, have been associated with LIG4 syndrome and RS-SCID defective in ARTEMIS (Gennery, 2006). The preferential development of lymphoid tumors in A-T, NBS and NHEJ defective syndromes is due to their critical role during lymphocite development. Lymphoid tumors are often caused by chromosomal translocations, which can lead to aberrant expression of oncogenes, such as c-Myc, or generation of deregulated chimeric proteins with enhanced activity (Nussenzweig and Nussenzweig, 2010). Experiments in mice have shown that NHEJ defective mice in the absence of p53 develop B-cell lymphomas harboring translocations between the Ig locus and c-Myc, which are dependent on the unrepaired breaks created by RAG1/RAG2 during V(D)J recombination (Nussenzweig and Nussenzweig, 2010). Similarly, ATM deficiency was shown to lead to persistent RAG-induced breaks, which could join other DSBs, such as breaks induced by aberrant activity of AID, to generate translocations and promote tumorigenesis (Nussenzweig and Nussenzweig, 2010).
A broad spectrum of malignancies is displayed by the DDR disorders Bloom syndrome (BS) and Li-Fraumeni syndrome (LFS). The high level of chromosomal instability of BS patient cells due to hyper-recombination predispose to lymphomas, leukemias and carcinomas (Chu and Hickson, 2009). LFS, which is primarily caused by germline TP53 mutations, predisposes to breast cancer, brain tumors, leukemia, sarcomas, melanomas and gastrointestinal cancers (D’Orazio, 2010). Sporadic cancers arise from the accumulation of stochastic DNA lesions that increase the fitness of cancer cells. It is known that early stages of human tumors have elevated levels of DNA damage (Bartek et al., 2007). DNA damage could be generated by exposure to carcinogens, as confirmed by the complex mutational signature identified in the lung cancer genome of a smoker and in the genome of a malignant melanoma (Pleasance et al., 2010a; Pleasance et al., 2010b). Moreover, DNA damage can be induced by telomeric shortening, increased oxidative damage or replication stress induced by oncogene activation (Bartek et al., 2007; Luo et al., 2009). Activation of the DDR could normally prevent tumorigenesis by inducing cellular senescence or apoptosis of early tumor cells (Bartek et al., 2007). However, mutation of DDR genes, such as TP53 or ATM, could predispose to cancer formation by facilitating senescence and apoptosis bypass and cellular proliferation despite accumulation of DNA damage (Luo et al., 2009). Activation of p53 occurs in response to multiple stimuli. However, it is likely to be its role in responding to DNA damage that provides the majority of its tumor suppressive function as demonstrated by the observation that more than 50% of sporadic human cancers harbor somatic TP53 mutations and another 15% have ATM mutations in a mutually exclusive manner, indicating that they function in the same pathway (Ding et al., 2008).
In an attempt to develop better cancer therapeutic approaches, the concept of targeting non-oncogene addiction (NOA) for cancer therapy has recently been proposed (Luo et al., 2009). This idea is based on the observation that the cancer cells rely on many non-oncogenic pathways, which are not essential for the survival of normal cells. Therefore, targeting NOA pathways would provide more selective cancer treatments. In particular, given the elevated levels of DNA damage in cancer cells compared to normal cells, further increasing the amount of DNA damage by inhibiting DDR components, in combination with other chemotherapeutic drugs, could lead to cancer cell death. Indeed, recent studies have shown that ATM, DNA-PK and CHK1 inhibitors have preferential toxicity towards cancer cells following treatment with genotoxic agents (Bolderson et al., 2009). Interestingly, ATM and DNA-PK depletion have been shown to sensitize TP53 and ATM mutant tumor cells to genotoxic agents, respectively (Jiang et al., 2009). Moreover, PARP1 inhibitors have been successfully used to treat tumors that carry mutations in HR genes, such as BRCA1 and BRCA2 (Jackson and Bartek, 2009). This effect has been proposed to be caused by the accumulation of unrepaired DNA breaks in the absence of both PARP-dependent SSBR and HR. Recently, methotrexate, an inhibitor of DNA base synthesis, has been shown to selectively kill MMR deficient cancer cells by leading to accumulation of oxidative DNA lesions (Martin et al., 2010a). Interestingly, depletion of the BER polymerase Polβ and the mitochondrial polymerase Polγ was shown to be synthetic lethal in combination with MSH2 and MLH1 deficiency in tumor cells because of an increased number of oxidative lesions (Martin et al., 2010b).
In summary, the coordination of DNA repair processes play a critical role in allowing the proper development and survival of organisms. They are responsible for preventing numerous human diseases and conditions including cancer and aging. We envision that the further understanding of the molecular mechanisms through which the DNA damage response operate, in combination with the elucidation of the genetic interactions between different DDR pathways and between DDR pathways and other cellular pathways, will provide therapeutic opportunities for many human diseases.
Acknowledgments
We would like to thank Andrew Elia, Lawrence Loeb, Stuart Linn, David DeMarini and Stephen Hecht for helpful discussions. A.C. is a recipient of an EMBO long-term fellowship. This work has been supported by grants from the N.I.H. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2945–2950. [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Fugger K, Danielsen JR, Gromova I, Sehested M, Celis J, Bartek J, Lukas J, Mailand N. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem. 2007;282:19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12:80–86. 81–12. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028–1038. doi: 10.1016/j.dnarep.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Blastyak A, Hajdu I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15:6314–6320. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Brugmans L, Kanaar R, Essers J. Analysis of DNA double-strand break repair pathways in mice. Mutat Res. 2007;614:95–108. doi: 10.1016/j.mrfmmm.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals PARP-regulated recruitment of the repressive Polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1012946107. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D’Orazio J, Fung CY, Schanbacher CF, Granter SR, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- D’Orazio JA. Inherited cancer syndromes in children and young adults. J Pediatr Hematol Oncol. 2010;32:195–228. doi: 10.1097/MPH.0b013e3181ced34c. [DOI] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Dou H, Huang C, Singh M, Carpenter PB, Yeh ET. Regulation of DNA Repair through DeSUMOylation and SUMOylation of Replication Protein A Complex. Mol Cell. 2010;39:333–345. doi: 10.1016/j.molcel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Cimprich KA. HARPing on about the DNA damage response during replication. Genes Dev. 2009;23:2359–2365. doi: 10.1101/gad.1860609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–390. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind MM, Redpath JL. Molecular and cellular biology of radiation lethality. Plenum Press, New York. 1977;6:51–99. [Google Scholar]
- Elledge SJ, Davis RW. DNA damage induction of ribonucleotide reductase. Mol Cell Biol. 1989;9:4932–4940. doi: 10.1128/mcb.9.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep. 2010;11:270–278. doi: 10.1038/embor.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7:937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O. Intrauterine programming of ageing. EMBO Rep. 2010;11:32–36. doi: 10.1038/embor.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W. Tip60-ing the balance in DSB repair. Nat Cell Biol. 2009;11:1279–1281. doi: 10.1038/ncb1109-1279. [DOI] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- Ford JM. Regulation of DNA damage recognition and nucleotide excision repair: another role for p53. Mutat Res. 2005;577:195–202. doi: 10.1016/j.mrfmmm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA. Nucleotide excision repair deficiencies and the somatotropic axis in aging. Hormones (Athens) 2008;7:9–16. doi: 10.14310/horm.2002.1111032. [DOI] [PubMed] [Google Scholar]
- Gasser S, Raulet DH. The DNA damage response arouses the immune system. Cancer Res. 2006;66:3959–3962. doi: 10.1158/0008-5472.CAN-05-4603. [DOI] [PubMed] [Google Scholar]
- Gennery AR. Primary immunodeficiency syndromes associated with defective DNA double-strand break repair. Br Med Bull. 2006;77–78:71–85. doi: 10.1093/bmb/ldl006. [DOI] [PubMed] [Google Scholar]
- Gilljam KM, Feyzi E, Aas PA, Sousa MM, Muller R, Vagbo CB, Catterall TC, Liabakk NB, Slupphaug G, Drablos F, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YS, Sekiguchi J, Gao YJ, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt FW. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hall EJ, Giaccia AJ. Radiobiology for the radiologist. Lippincott Williams & Wilkins; 2006. pp. 206–216. [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Huang J, Leung JW, Sy SM, Leung KM, Ching YP, Tsao SW, Chen J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010a;37:854–864. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010b;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24 doi: 10.1101/gad.1934210. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Fox D, 3rd, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms--interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Lobrich M. Artemis links ATM to double strand break rejoining. Cell Cycle. 2005;4:359–362. doi: 10.4161/cc.4.3.1527. [DOI] [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kai M, Boddy MN, Russell P, Wang TS. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 2005;19:919–932. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Wyman C. DNA repair by the MRN complex: break it to make it. Cell. 2008;135:14–16. doi: 10.1016/j.cell.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Katyal S, McKinnon PJ. DNA strand breaks, neurodegeneration and aging in the brain. Mech Ageing Dev. 2008;129:483–491. doi: 10.1016/j.mad.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, et al. Tipin-RPA interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010 doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C, O’Driscoll M. Human DNA damage response and repair deficiency syndromes: linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst) 2009;8:1139–1152. doi: 10.1016/j.dnarep.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Luscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–19. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, Kim SJ, Tonks NK. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010a;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010b;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee KY, Yang K, Cohn MA, Sikdar N, D’Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem. 2010c;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E. Fanconi anemia and breast cancer susceptibility meet again. Nat Genet. 2010;42:368–369. doi: 10.1038/ng0510-368. [DOI] [PubMed] [Google Scholar]
- Lin SY, Liang Y, Li K. Multiple roles of BRIT1/MCPH1 in DNA damage response, DNA repair, and cancer suppression. Yonsei Med J. 2010;51:295–301. doi: 10.3349/ymj.2010.51.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci U S A. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Hewish M, Lord CJ, Ashworth A. Genomic instability and the selection of treatments for cancer. J Pathol. 2010a;220:281–289. doi: 10.1002/path.2631. [DOI] [PubMed] [Google Scholar]
- Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010b;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad DH, Yaffe MB. 14–3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279:13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, et al. Non-canonical inhibition of DNA damage dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair (Amst) 2008;7:523–529. doi: 10.1016/j.dnarep.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24:333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, Jeggo PA. The role of the DNA damage response pathways in brain development and microcephaly: insight from human disorders. DNA Repair (Amst) 2008;7:1039–1050. doi: 10.1016/j.dnarep.2008.03.018. [DOI] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi T. BRCA1 phosphorylation: biological consequences. Cancer Biol Ther. 2006;5:470–475. doi: 10.4161/cbt.5.5.2845. [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson M, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA Repair in the Absence of the Fanconi Anemia Pathway. Science. 2010 doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annu Rev Med. 2005;56:495–508. doi: 10.1146/annurev.med.56.082103.104546. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010 doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009 doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010 doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine-139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo I, Cosentino C, Del Giudice E, Broccoletti T, Amorosi S, Cirillo E, Aloj G, Fusco A, Costanzo V, Pignata C. In ataxia-teleangiectasia betamethasone response is inversely correlated to cerebellar atrophy and directly to antioxidative capacity. Eur J Neurol. 2009;16:755–759. doi: 10.1111/j.1468-1331.2009.02600.x. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–6. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Shull ER, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, Petrini JH, McKinnon PJ. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Landais I, de Graaf B, Hoatlin ME. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem. 2009;284:25560–25568. doi: 10.1074/jbc.M109.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–U121. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spry M, Scott T, Pierce H, D’Orazio JA. DNA repair pathways and hereditary cancer susceptibility syndromes. Front Biosci. 2007;12:4191–4207. doi: 10.2741/2380. [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ, Taylor AM. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci U S A. 2007;104:16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M. Histone H2A.X Tyr142 phosphorylation: a novel sWItCH for apoptosis? DNA Repair (Amst) 2009;8:873–876. doi: 10.1016/j.dnarep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010 doi: 10.1101/gad.1956410. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Jones NJ. Stabilizing and remodeling the blocked DNA replication fork: anchoring FANCM and the Fanconi anemia damage response. Mol Cell. 2010;37:749–751. doi: 10.1016/j.molcel.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Rouleau GA. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. doi: 10.1212/01.wnl.0000296811.19811.db. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA binding protein required for eukaryotic DNA metabolism. Ann Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang P, Liu L, Lee MY. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry. 2001;40:4512–4520. doi: 10.1021/bi010103+. [DOI] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]