Recent findings and technological advances provide insight into how a few transcription factors work together in complex ways to orchestrate red blood cell differentiation.
Abstract
A relatively small cadre of lineage-restricted transcription factors largely orchestrates erythropoiesis, but how these nuclear factors interact to regulate this complex biology is still largely unknown. However, recent technological advances, such as chromatin immunoprecipitation (ChIP) paired with massively parallel sequencing (ChIP-seq), gene expression profiling, and comprehensive bioinformatic analyses, offer new insights into the intricacies of red cell molecular circuits.
Red blood cells (RBCs) fulfill the essential functions of transporting oxygen to tissues and facilitating gas exchange in the lungs. They are continuously produced throughout life in a tightly controlled growth process termed erythropoiesis. Erythroid differentiation is accompanied by temporally regulated changes in cell surface protein expression, a reduction in cell size, progressive hemoglobinization, and nuclear condensation, which culminates in extrusion of the nucleus, RNA, and mitochondria (Richmond et al., 2005).
Erythropoiesis is largely mediated by a relatively small number of lineage-restricted transcription factors, including GATA-1, SCL/TAL1, LMO2, LDB1, and KLF1 (Cantor and Orkin, 2002). The importance of these transcription factors in erythropoiesis has been demonstrated unequivocally by cell-based ex vivo assays, as well as in knockout mouse models and rare patients with anemias. The critical transcription factors are present in diverse multiprotein complexes. However, how distinct multiprotein complexes activate or repress transcription, and thereby regulate the erythroid maturation program, remains incompletely understood. New techniques, including ChIP coupled with massively parallel sequencing (ChIP-seq), gene expression profiling, and bioinformatic analyses, provide new information about the regulatory networks that coordinate erythroid cell maturation and function. This minireview will summarize recent findings relevant to the understanding of gene expression regulation in red blood cells.
GATA-1
The transcription factor GATA-1 recognizes the DNA consensus sequence (A/T)GATA(A/G) through two Cys-X2-Cys-X17-Cys-X2-Cys zinc fingers that are characteristic of the GATA family (Wall et al., 1988; Evans and Felsenfeld, 1989). Annotation of GATA consensus sites, even those that are phylogenetically conserved, is a poor predictor of in vivo GATA-1 chromatin binding (Bresnick et al., 2005). Hence, several groups generated whole-genome occupancy maps for GATA-1 by using ChIP-seq in erythroid cell lines (Cheng et al., 2009; Fujiwara et al., 2009; Yu et al., 2009; Soler et al., 2010). Although three studies identified ∼4,000–6,000 in vivo binding sites for GATA-1 in mouse erythroleukemia (MEL) cells expressing a tagged form of GATA-1 (Yu et al., 2009; Soler et al., 2010) or human K562 erythroleukemia cells (Fujiwara et al., 2009), a fourth study identified >15,000 sites occupied by GATA-1 in G1E-ER4 cells, which were derived from GATA-1 knockout mouse embryonic stem cells and express an estrogen-inducible GATA-1 construct. Careful assessment of the data may help explain discrepancies in the number of GATA-1–occupied sites. These may have arisen from usage of different cell lines, employment of different peak calling algorithms, differences in the ChIP protocols, or simply differences in choice of statistical cut offs.
All studies demonstrated that a minority of GATA-1 binding sites (∼10–15%) are located at proximal promoter regions close to the transcription start site (TSS). The bulk of GATA-1 binding (∼85%) occurs at distal regulatory elements with equal distribution between intra- and intergenic regions (Fujiwara et al., 2009; Yu et al., 2009). High-level H3K4 monomethlyation (H3K4me1), a histone mark strongly enriched at functional enhancer regions (Heintzman et al., 2007), was observed at nearly all GATA-1–occupied DNA segments, further supporting the notion that GATA-1 principally binds enhancer regions (Cheng et al., 2009). To identify direct GATA-1 target genes, microarray gene expression profiling was performed (Yu et al., 2009) using G1E-ER4 cells (Weiss et al., 1997). G1E cells are arrested at the proerythroblast stage of differentiation, but undergo synchronous terminal maturation upon restoration of GATA-1 function (Weiss et al., 1997). Reexpression of GATA-1 triggers an extensive program of gene activation and repression (Weiss et al., 1997). Superimposition of GATA-1 whole-genome occupancy and gene expression data permitted identification of putative, direct GATA-1 targets. Although up to 5,000 genes were found to be differentially expressed upon GATA-1 activation (Cheng et al., 2009; Fujiwara et al., 2009; Yu et al., 2009), a surprisingly small fraction (∼300–700) of genes could be identified as direct GATA-1 target genes (Fujiwara et al., 2009; Yu et al., 2009). It should also be noted that within those genes identified as direct GATA-1 targets, 40–57% were up-regulated and 41–60% were down-regulated (Cheng et al., 2009; Fujiwara et al., 2009; Yu et al., 2009), demonstrating that GATA-1 activates or represses nearly equivalent numbers of genes. Bioinformatic analysis of transcription factor motifs further revealed that among activated genes, binding sites for SCL/TAL1 were highly enriched (Cheng et al., 2009; Fujiwara et al., 2009; Tripic et al., 2009; Yu et al., 2009; Kassouf et al., 2010). Based on this finding, one may infer that GATA-1 activates gene expression specifically in concert with SCL/TAL1 (Fig. 1). However, partners for GATA-1 in gene repression are less clear. GATA-1 is thought to facilitate gene repression via interaction with the NuRD complex; this may be mediated through a direct interaction between GATA-1 and FOG-1 (Hong et al., 2005; Rodriguez et al., 2005), as well as via the transcriptional repressor Gfi-1b in concert with the LSD1–CoREST corepressor complex (Fig. 1; Rodriguez et al., 2005; Saleque et al., 2007). Interestingly, the genome-wide occupancy maps revealed an additional level of complexity, as a subset of GATA-1–repressed genes was also found to carry the repressive H3K27me3 histone mark (Cheng et al., 2009; Yu et al., 2009). This mark is catalyzed by the polycomb repressive complex 2 (PRC2), a multiprotein complex containing EED, Ezh1/2, and Suz12 (Müller et al., 2002; Schuettengruber et al., 2007). Erythroid differentiation is impaired in mice with erythroid-specific loss of EED (Yu et al., 2009). Thus, the PRC2 complex participates in GATA-1–mediated gene repression during erythroid differentiation. Whether GATA-1 recruits PRC2 directly, or indirectly, will be of interest in future studies.
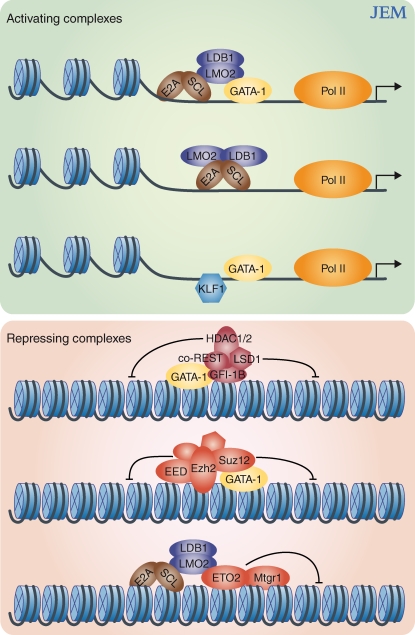
Figure 1.
Model of the multiprotein complexes orchestrating gene expression or repression in erythroid cells. Comparison of GATA-1, SCL/TAL1, and LDB1 whole-genome occupancy maps with gene expression profiling data suggests that the GATA-1/SCL/TAL1–LMO2–LDB1–E2A pentameric complex, as well as a GATA-1–independent SCL/TAL-1–containing complex, largely activate gene expression. GATA-1 may also activate gene expression in coordination with KLF1 (activating complexes, green box). GATA-1 might repress gene expression via a multi-step process. Interaction with the transcriptional repressor GFI-1B recruits the LSD1/coREST complex, which results in removal of the activating H3K4me2 mark. To permanently silence gene expression, GATA-1 can recruit the PRC2 complex (EED, Ezh2, and Suz12) resulting in H3K27 trimethylation and gene repression. The SCL/TAL1 complex can recruit the corepressors ETO2 and Mtgr1 resulting in SCL/TAL1 mediated gene silencing (repressing complexes, red box).
It should be recognized that these chromatin occupancy studies do not account for posttranslational modifications of GATA-1. For example, GATA-1 is acetylated (Boyes et al., 1998), and this modification appears to be important for erythroid differentiation (Hung et al., 1999; Lamonica et al., 2006). A study recently published in JEM revealed the importance of GATA-1 SUMOylation (Yu et al., 2010b). Genetic ablation of the SUMO-specific protease SENP1 resulted in severe anemia and embryonic lethality in mice at embryonic day 13.5. Accumulation of a SUMOylated form of GATA-1 was observed and coincided with down-regulation of GATA-1 target genes (Yu et al., 2010b). SUMOylation may modulate aspects of GATA-1 function beyond DNA binding, as suggested by Yu et al. (2010b), given that SUMOylation of FOG1 affects its interaction with other proteins (Snow et al., 2010). Further work is needed to interrogate protein–protein and protein–DNA interactions of SUMOylated GATA-1.
In recent years, microRNAs (miRNAs) have emerged as additional regulators of overall gene expression, representing yet another layer of control. Indeed, recent work demonstrates that the miR-144/451 locus is a direct target of GATA-1 and that mice lacking miR-144/451 or miR-451 alone show impaired erythropoiesis, particularly under conditions of stress (Dore et al., 2008; Rasmussen et al. 2010; Patrick et al., 2010; Yu et al., 2010a).
SCL/TAL1–LMO2–LDB1–E2A complex
The basic helix–loop–helix (bHLH) transcription factor SCL/TAL1 recognizes a short consensus DNA motif (CANNTG), the E-box. SCL/TAL1 expression largely parallels that of GATA-1, as it is expressed in erythroid cells, megakaryocytes, and mast cells (Cantor and Orkin, 2002). In erythroid cells, SCL/TAL1 forms a complex with the ubiquitous bHLH protein E2A, and also with the LIM domain containing cofactors LMO2 and LDB1 (Cantor and Orkin, 2002). These proteins interact with GATA-1 to form a pentameric complex (Fig. 1) that binds to composite E-box/GATA-1 DNA motifs spaced 9–11 nt apart (Wadman et al., 1997; Cohen-Kaminsky et al., 1998). LMO2, GATA-1, and SCL/TAL1 are all required for erythropoiesis in mice (Cantor and Orkin, 2002), and a conditional knockout mouse model of SCL/TAL1 is available (Mikkola et al., 2003). In this issue, Li et al. present the first conditional knockout of LDB. They find that embryos lacking LDB1 show defective primitive erythropoiesis and that Mx-Cre–driven deletion of LDB1 in adult mice results in a persistent drop in hematocrit and, ultimately, death, demonstrating that LDB1 is continuously required for definitive erythropoiesis.
For some time, only a handful of red cell–specific direct target genes of this complex had been identified. Two recent studies mapped whole-genome occupancy of this complex by performing ChIP-seq for endogenous SCL/TAL1 in primary mouse proerythroblasts (Kassouf et al., 2010) or for tagged LDB1 and SCL/TAL1 in MEL cells (Soler et al., 2010). A third group generated an occupancy map of SCL/TAL1 in G1E-ER4 cells, performing ChIP-on-chip analysis using a tiling array covering mouse chromosome 7 (Tripic et al., 2009). Approximately 3,000–4,000 and 5,000 genome-wide binding sites were identified for SCL/TAL1 and LDB1, respectively. Approximately 30% of all SCL/TAL1 binding sites were located at proximal promoter regions (in this study defined as ±3 kb of the TSS), whereas the bulk of SCL/TAL1 binding (∼70%) resided at distal regulatory elements with a distribution of 40 or 25% in intragenic or intergenic regions, respectively (Kassouf et al., 2010). To identify putative direct SCL/TAL1 target genes, microarray gene expression profiling was used to compare wild-type primary proerythroblasts with proerythroblasts derived from mice carrying a mutation in the DNA-binding domain of SCL/TAL1 (SCL/TAL1RER; Kassouf et al., 2008). 511 differentially expressed genes were identified, with 51% up-regulated and 49% down-regulated. The intersection of SCL/TAL1 occupancy and gene expression data resulted in an overlap of only 83 genes, which may be considered direct SCL/TAL1 targets. Strikingly, 75% of these genes were down-regulated as compared with wild-type cells, indicating that SCL/TAL1 largely activates gene expression. Analysis of motifs revealed enrichment of GATA binding sites close to SCL/TAL1 binding sites at genes activated by SCL/TAL1, in accordance with the reciprocal findings for GATA-1 (see above). Gene repression mediated by the SCL/TAL1 complex may be performed via recruitment of the corepressors ETO2 and Mtgr1 (Fig. 1; Fujiwara et al., 2009; Tripic et al., 2009; Soler et al., 2010). This conclusion is supported by cooccupancy of ETO2/Mtgr1 at a subset of SCL/TAL1 target genes (Soler et al., 2010), as well as de-repression of some SCL/TAL1 target genes upon depletion of ETO2 in erythroid cells (Tripic et al., 2009). The observation that SCL/TAL1 and LDB1 have been found binding far from their closest repressed gene prompted Soler et al. (2010) to perform chromosome conformation capture sequencing (3C-seq). Combination of the LDB1 ChIP-seq and 3C-seq data revealed direct binding of LDB1 to DNase-hypersensitive sites HS2, HS3, and HS4 of the β-globin locus control region (LCR) and long-range interactions with the β-globin promoter, despite the absence of a functional LDB1 binding site at the β-globin promoter (Soler et al., 2010). It would be of interest to study the nature of these long-range interactions in the absence of SCL/TAL1 or LDB1.
KLF1
KLF1 (formerly called EKLF), a zinc finger transcription factor with three highly similar C-terminal C2H2-type Kruppel zinc fingers, recognizes a subset of CACC box motifs (Miller and Bieker, 1993). Expression of KLF1 is remarkably restricted to erythroid cells and their precursors (Miller and Bieker, 1993). Although its essential role in erythropoiesis has been known for quite some time (Cantor and Orkin, 2002), few direct transcriptional targets have been identified.
Tallack et al. (2010) generated a whole-genome occupancy map for KLF1 in primary erythroid cells. In two independent ChIP-seq runs, KLF1 occupied between 940 and 1,400 binding sites in erythroid cells. 16% of these binding events occurred within 1 kb of the TSS, whereas the majority of sites were located at distances of >10 kb away from TSSs (Tallack et al., 2010). To identify new direct KLF1 target genes, the authors compared ChIP-seq data with gene expression profiles of wild-type and Klf1−/− fetal liver cells (Hodge et al., 2006). A total of 1,099 genes were differentially expressed in the absence of KLF1 in erythroid cells; 730 genes were down-regulated and 369 genes were up-regulated (Hodge et al., 2006). Only ∼19% of these genes were occupied by KLF1. The bulk of binding events occurred at genes that were down-regulated in the absence of KLF1, suggesting that KLF1 acts primarily as a transcriptional activator (Tallack et al., 2010). The authors investigated a potential functional DNA-dependent interaction between KLF-1 and GATA-1. Comparing KLF1 ChIP-seq data with results from GATA-1 whole genome occupancy maps, Tallack et al. determined the distances between the nearest GATA-1 peak and all KLF1 peaks. Approximately 48% of KLF1 peaks are located within 1 kb of GATA-1 peaks, strongly supporting an in vivo cooperation of the two factors (Fig. 1). Finally, the authors compared GATA-1/SCL (Cheng et al., 2009; Wilson et al., 2009) and GATA-1/KLF1–cooccupied regions and found minimal overlap (Tallack et al., 2010). This finding was surprising, given studies implicating GATA-1 in gene activation almost exclusively in complex with SCL/TAL1 (see above), but it suggests that GATA-1 may exist in two mutually exclusive activating complexes (Fig. 1).
Concluding remarks
In considering genome-wide occupancy data, one must be cognizant of potential methodological pitfalls. For example, although the widely applied “nearest-neighbor approach” (Kent et al., 2002; Pepke et al., 2009) provides a convenient way to assign transcription factor–binding peaks to nearby genes, it may oversimplify the situation, as it does not take into account long-range cis or trans interactions that frequently occur between promoter and enhancer elements. This limitation may account, in part, for the relatively small overlap between gene expression and transcription factor occupancy data.
Nevertheless, whole genome mapping of transcription factor occupancy is a relatively new technology that is providing prodigious datasets for computational and functional analyses. The integration of such data with profiling of mRNA and miRNA expression, coupled with sensitive proteomics, will lead to an enhanced view of the transcriptional networks orchestrating erythroid differentiation. The basic principles elucidated in these studies will inform transcriptional biology more broadly.
References
- Boyes J., Byfield P., Nakatani Y., Ogryzko V. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 396:594–598 10.1038/25166 [DOI] [PubMed] [Google Scholar]
- Bresnick E.H., Martowicz M.L., Pal S., Johnson K.D. 2005. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 205:1–9 10.1002/jcp.20393 [DOI] [PubMed] [Google Scholar]
- Cantor A.B., Orkin S.H. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 21:3368–3376 10.1038/sj.onc.1205326 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Wu W., Kumar S.A., Yu D., Deng W., Tripic T., King D.C., Chen K.B., Zhang Y., Drautz D., et al. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19:2172–2184 10.1101/gr.098921.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kaminsky S., Maouche-Chrétien L., Vitelli L., Vinit M.A., Blanchard I., Yamamoto M., Peschle C., Roméo P.H. 1998. Chromatin immunoselection defines a TAL-1 target gene. EMBO J. 17:5151–5160 10.1093/emboj/17.17.5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore L.C., Amigo J.D., Dos Santos C.O., Zhang Z., Gai X., Tobias J.W., Yu D., Klein A.M., Dorman C., Wu W., et al. 2008. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. USA. 105:3333–3338 10.1073/pnas.0712312105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Felsenfeld G. 1989. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 58:877–885 10.1016/0092-8674(89)90940-9 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., O’Geen H., Keles S., Blahnik K., Linnemann A.K., Kang Y.A., Choi K., Farnham P.J., Bresnick E.H. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 36:667–681 10.1016/j.molcel.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311–318 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Hodge D., Coghill E., Keys J., Maguire T., Hartmann B., McDowall A., Weiss M., Grimmond S., Perkins A. 2006. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 107:3359–3370 10.1182/blood-2005-07-2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., Nakazawa M., Chen Y.Y., Kori R., Vakoc C.R., Rakowski C., Blobel G.A. 2005. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24:2367–2378 10.1038/sj.emboj.7600703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.L., Lau J., Kim A.Y., Weiss M.J., Blobel G.A. 1999. CREB-Binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf M.T., Chagraoui H., Vyas P., Porcher C. 2008. Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood. 112:1056–1067 10.1182/blood-2007-12-128900 [DOI] [PubMed] [Google Scholar]
- Kassouf M.T., Hughes J.R., Taylor S., McGowan S.J., Soneji S., Green A.L., Vyas P., Porcher C. 2010. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 20:1064–1083 10.1101/gr.104935.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamonica J.M., Vakoc C.R., Blobel G.A. 2006. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 108:3736–3738 10.1182/blood-2006-07-032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H.K., Klintman J., Yang H., Hock H., Schlaeger T.M., Fujiwara Y., Orkin S.H. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 421:547–551 10.1038/nature01345 [DOI] [PubMed] [Google Scholar]
- Miller I.J., Bieker J.J. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 111:197–208 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- Patrick D.M., Zhang C.C., Tao Y., Yao H., Qi X., Schwartz R.J., Jun-Shen Huang L., Olson E.N. 2010. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 24:1614–1619 10.1101/gad.1942810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S., Wold B., Mortazavi A. 2009. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 6:S22–S32 10.1038/nmeth.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K.D., Simmini S., Abreu-Goodger C., Bartonicek N., Di Giacomo M., Bilbao-Cortes D., Horos R., Von Lindern M., Enright A.J., O’Carroll D. 2010. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 207:1351–1358 10.1084/jem.20100458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T.D., Chohan M., Barber D.L. 2005. Turning cells red: signal transduction mediated by erythropoietin. Trends Cell Biol. 15:146–155 10.1016/j.tcb.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K.E., Guyot B., Heck A.J., Vyas P., de Boer E., Grosveld F., Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366 10.1038/sj.emboj.7600702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Kim J., Rooke H.M., Orkin S.H. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell. 27:562–572 10.1016/j.molcel.2007.06.039 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. 2007. Genome regulation by polycomb and trithorax proteins. Cell. 128:735–745 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Snow J.W., Kim J., Currie C.R., Xu J., Orkin S.H. 2010. Sumoylation regulates interaction of FOG1 with C-terminal-binding protein (CTBP). J. Biol. Chem. 285:28064–28075 10.1074/jbc.M109.096909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E., Andrieu-Soler C., de Boer E., Bryne J.C., Thongjuea S., Stadhouders R., Palstra R.J., Stevens M., Kockx C., van Ijcken W., et al. 2010. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 24:277–289 10.1101/gad.551810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack M.R., Whitington T., Yuen W.S., Wainwright E.N., Keys J.R., Gardiner B.B., Nourbakhsh E., Cloonan N., Grimmond S.M., Bailey T.L., Perkins A.C. 2010. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20:1052–1063 10.1101/gr.106575.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C.R., Gregory G.D., Hardison R.C., Blobel G.A. 2009. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 113:2191–2201 10.1182/blood-2008-07-169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grütz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L., deBoer E., Grosveld F. 1988. The human beta-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 2:1089–1100 10.1101/gad.2.9.1089 [DOI] [PubMed] [Google Scholar]
- Weiss M.J., Yu C., Orkin S.H. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17:1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.K., Miranda-Saavedra D., Kinston S., Bonadies N., Foster S.D., Calero-Nieto F., Dawson M.A., Donaldson I.J., Dumon S., Frampton J., et al. 2009. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 113:5456–5465 10.1182/blood-2009-01-200048 [DOI] [PubMed] [Google Scholar]
- Yu M., Riva L., Xie H., Schindler Y., Moran T.B., Cheng Y., Yu D., Hardison R., Weiss M.J., Orkin S.H., et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 36:682–695 10.1016/j.molcel.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., dos Santos C.O., Zhao G., Jiang J., Amigo J.D., Khandros E., Dore L.C., Yao Y., D’Souza J., Zhang Z., et al. 2010a. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 24:1620–1633 10.1101/gad.1942110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Ji W., Zhang H., Renda M.J., He Y., Lin S., Cheng E.C., Chen H., Krause D.S., Min W. 2010b. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J. Exp. Med. 207:1183–1195 10.1084/jem.20092215 [DOI] [PMC free article] [PubMed] [Google Scholar]