Abstract
Mutations in superoxide dismutase 1 (SOD1) are associated with familial cases of amyotrophic lateral sclerosis (fALS). Studies in transgenic mice have suggested that wild-type (WT) SOD1 can modulate the toxicity of mutant SOD1. In the present study, we demonstrate that the effects of WT SOD1 on the age at which transgenic mice expressing mutant human SOD1 (hSOD1) develop paralysis are influenced by the nature of the ALS mutation and the expression levels of WT hSOD1. We show that regardless of whether WT SOD1 changes the course of disease, both WT and mutant hSOD1 accumulate as detergent-insoluble aggregates in symptomatic mice expressing both proteins. However, using a panel of fluorescently tagged variants of SOD1 in a cell model of mutant SOD1 aggregation, we demonstrate that the interactions between mutant and WT SOD1 in aggregate formation are not simply a co-assembly of mutant and WT proteins. Overall, these data demonstrate that the product of the normal SOD1 allele in fALS has potential to influence the toxicity of mutant SOD1 and that complex interactions with the mutant protein may influence the formation of aggregates and inclusion bodies generated by mutant SOD1.
INTRODUCTION
Familial amyotrophic lateral sclerosis (fALS) is a severe motor neuron disease caused, in ∼20% of cases, by mutations in superoxide dismutase 1 (SOD1) (1). To date, over 150 mutations in the coding sequence of SOD1 have been identified in ALS patients; most mutations are point substitutions but a few produce truncated proteins due to early termination (for most recent list, see http://alsod.iop.kcl.uk, last date accessed 09/30/2010). ALS-associated mutations in SOD1, both missense and premature termination, induce misfolding and aggregation of the protein (2,3). In studies of G93A transgenic mice that model SOD1-fALS, sedimentable detergent-insoluble aggregates of mutant SOD1 first begin to accumulate at approximately the time that symptoms become first noticeable (at ∼80 days of age) (4,5). Similar data have been observed in multiple lines of ALS mice, with the levels of insoluble SOD1 in the spinal cord dramatically accumulating over the last few weeks of life (6). Both theoretical models (7) and cell culture models (3) have produced data to suggest that mutations imparting a high propensity to aggregate are disproportionately associated with disease of shorter duration. Thus, we have proposed that large aggregates may modulate the rate of disease progression in humans with forms of mutant SOD1 that are soluble being responsible for initiating disease (3,6).
The molecular features of the mutant SOD1 responsible for initiating disease have yet to be defined. Recent studies have focused our attention on the possible role of WT SOD1 as a modulator of disease initiation. In transgenic mice, co-expression of high levels of WT human SOD1 (hSOD1) with mutant hSOD1 (A4V, G85R, G93A, T116X or L126Z) accelerates the course of disease by decreasing the age to onset and paralysis with no obvious change in the rate of progression from onset to paralysis (8–11). In mice that express mutants in which the mutant protein is easily distinguished from WT SOD1, such as the G85R, T116X and L126Z mutants, it has been possible to determine that earlier disease onset is accompanied by the formation of detergent-insoluble aggregates that appear to contain both WT hSOD1 and mutant hSOD1 (8,9,11). However, in cultured cell models, WT hSOD1 does not promote the aggregation of mutant SOD1, but rather seems to slow the process (12,13). In vitro, small amounts of immature mutant SOD1 can seed the aggregation of WT SOD1 (14). Collectively, these studies indicate a potential for WT hSOD1 to play a role in the aggregation of mutant SOD1 and to modulate the course of disease.
Although the foregoing evidence is compelling, initial studies to investigate the role of WT hSOD1 as a modulator of disease produced conflicting outcomes. Bruijn et al. (15) reported that co-expressing WT and G85R hSOD1 had no effect on disease onset or survival. In the present study, we have directly compared the WT hSOD1 mouse line used in the Bruijn study [designated line 76 (16)] with the line used in the studies by Deng et al. (described in 17) to determine the degree to which the source of WT hSOD1 expression affects the age at which co-expressed mutant SOD1 causes paralysis. We have also used cell models to examine interactions between WT and mutant hSOD1 in aggregate formation. Our findings demonstrate that the effect of WT hSOD1 on age to paralysis in mutant SOD1 transgenic mice is influenced by the nature of the mutation and the expression level of WT SOD1. We also demonstrate that the assembly of large aggregates that include WT protein, in mice co-expressing WT and mutant SOD1, is secondary to the processes that are responsible for accelerating the age to paralysis.
RESULTS
Studies involving matings of mice that over-express WT hSOD1 to mice that over-express mutant hSOD1 have reported differing outcomes depending on the strain of WT hSOD1 mice used. A study by Bruijn et al. (15) reported that WT hSOD1 expressed from a line of mice developed by Wong et al. [line 76; (16)] had no effect on the course of disease caused by G85R hSOD1. In contrast, multiple studies have reported that WT hSOD1 expressed from a line of mice developed by Gurney et al. (17) accelerates the course of disease caused by mutant hSOD1 (8–10), including the G85R variant of hSOD1 (11). To determine whether the different lines of WT hSOD1 mice used in these studies account for the different outcomes, we crossed three different strains of WT hSOD1 mice to mice expressing either G37R or L126Z hSOD1 proteins. The three WT hSOD1 strains of mice used were: (i) B6SJL-Tg(SOD1)2Gur/J hybrid line (SJLWT) (17), (ii) B6/L76WT SOD1 Wong congenic line (L76WT) (16) and (iii) a variant of the B6SJL-Tg(SOD1)2Gur/J Gurney hybrid line (17), here termed congenic (Cg) WT, that was backcrossed to C57BL/6J to create a strain possessing an equivalent background to that of the B6/L76WT SOD1 congenic line.
Expression of WT SOD1 in the Gurney and L76 lines of transgenic mice
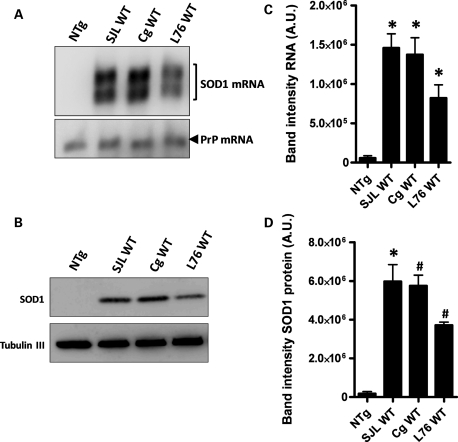
We compared side by side the levels of hSOD1 mRNA and protein in the different WT hSOD1 mouse strains used in this study, using northern and immunoblot analyses (Fig. 1). In young mice (∼3–4 months of age), the levels of hSOD1 mRNA in spinal cords from the Gurney strains of WT SOD1 mice, both the SJLWT and the CgWT lines, were found to be ∼30% higher than those of L76WT hSOD1 mice (Fig. 1A and B). Correspondingly, the levels of hSOD1 protein in spinal cords of the Gurney strains were also 30% higher than those of L76WT mice (Fig. 1C and D). In all strains of mice, most of the WT SOD1 in the spinal cord appeared to be properly post-translationally modified by an intramolecular disulfide bond (Supplementary Material, Fig. S1).
Figure 1.
Expression of WT hSOD1 in Gurney and L76 lines of mice. (A) Northern blot of hSOD1 mRNA in spinal cords of WT mice used in this study. A probe for mRNA of the endogenous mouse prion protein (PrP) gene was used as a loading control. (B) Quantification of the mRNA levels normalized to the PrP control. (C) Immunoblot analysis of the same spinal cords with a hSOD1 antibody. Antibody recognizing β-tubulin III was used as a loading protein control. (D) Quantification of the human WT protein levels normalized to loading control. Significant differences were found in RNA and protein levels between L76WT and each of the Gurney strains of mice (P ≤ 0.05) but not between the SJL and Cg WT Gurney strains. (B) and (D) were assessed by unpaired Student's t-tests: *P ≤ 0.05, #P ≤ 0.005. Bars represent mean ± SEM of three different spinal cords for each mouse line.
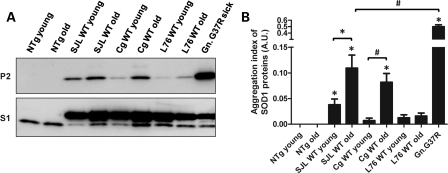
In studies of L76WT mice and various lines of mutant SOD1 mice, we have viewed WT hSOD1 as being relatively resistant to aggregation (2,6,18,19). Jonsson et al. (5) reported that spinal cords of older Gurney SJLWT mice accumulate detergent-insoluble forms of WT hSOD1. However, the concentrations of detergent used in Jonsson's study were lower than our protocol, and insoluble forms of WT hSOD1 were detected in young and older mice (5). Our methodology, which uses higher concentrations of non-ionic detergent and possibly higher detergent to protein ratios (6), routinely solubilizes nearly all mutant hSOD1 in spinal cords of asymptomatic mice expressing mutant hSOD1 (6) and all WT hSOD1 in spinal cords of adult L76WT mice (2,6). Applying our methodology, we detected small amounts of detergent-insoluble WT hSOD1 in spinal cords of older Gurney WT SOD1 mice but not in L76WT mice (Fig. 2). Specifically, in the Gurney line of WT mice, in both SJL and Cg WT strains, significant accumulations of aggregated WT SOD1 protein were found as early as 8 months (SJLWT), and increased to higher levels at 11 (SJLWT) and 17 (CgWT) months of age (Fig. 2A and B). However, in L76WT SOD1 mice, the accumulation of detergent-insoluble protein was negligible at 17 months of age (Fig. 2A and B). In an analysis of total protein levels in all three strains of mice, we noted that the total levels of WT SOD1 were higher in spinal cords of the older mice (Supplementary Material, Fig. S2). Thus, even though we did not detect detergent-insoluble aggregates in spinal cords of the L76 mice, there was evidence of an accumulation of the protein. Importantly, the relative aggregation index of WT hSOD1 protein in all of the WT SOD1 lines of mice was not as high as mutant G37R hSOD1 in symptomatic G37R Line 29 hSOD1 mice [genomic hSOD1 construct (16)] (Gn.G37R, Fig. 2A and B), which develop disease at ∼7 months of age and which accumulate relatively low levels of aggregated hSOD1 at disease endstage (6). Thus, the Gurney strains of mice exhibit a spontaneous aggregation of WT hSOD1, but the levels appear to be below the threshold to induce ALS phenotypes or such assemblies of the WT protein do induce the full spectrum of disease phenotypes.
Figure 2.
Low levels of WT hSOD1 aggregates in spinal cords of older Gurney mice. Spinal cords from Gurney strains (SJL and Cg) of mice, but not from L76 mice, contain detergent-insoluble aggregates of WT SOD1. (A) Immunoblots of S1 and P2 fractions of mice expressing WT hSOD1 at varied ages. An antibody that recognizes mouse and hSOD1 was used. NTg, non-transgenic mice. Gn.G37R denotes symptomatic aged mice (∼7 months) that harbor human genomic fragments of the SOD1 gene (line 29)—mice that were originally described by Wong et al. (16). (B) Quantification of the aggregation index. Unpaired Student's t-tests were performed to establish significant differences as indicated: *P ≤ 0.05, #P ≤ 0.005. Bars represent mean ± SEM.
Co-expression of WT SOD1 decreases the age at which mice expressing the G37R fALS variant develop paralysis
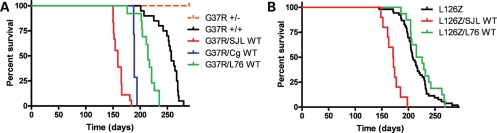
The three different strains of WT mice were bred to two different lines of mutant SOD1 mice. One mutant line expresses the G37R variant of hSOD1 from a vector that utilizes the mouse prion promoter (PrP.SOD1-G37R line 110, designated hereafter simply as G37R), which drives transgene expression primarily in muscle and neural tissue (20). The expression levels of the transgene in heterozygous G37R mice are known to be too low to produce an ALS phenotype in mice (20). However, when G37R expression levels are increased by breeding the mice to homozygosity, the resulting mice manifest typical ALS symptoms and pathology (20). Thus, whereas heterozygous G37R mice do not develop ALS over the course of their 2 year lifespan, homozygous G37R mice develop paralysis at ∼8 months of age (Fig. 3A). The other mutant SOD1 mouse line that we used expresses the L126Z SOD1 transgene, which is controlled by the normal hSOD1 gene promoter and consists of modified genomic SOD1 (19). The age at which these mice develop paralysis is also 8 months (Fig. 3B). We obtained a total of five different types of double-transgenic mice expressing WT and mutant hSOD1 proteins: G37R/SJLWT, G37R/CgWT, G37R/L76WT, L126Z/SJLWT and L126Z/L76WT. In the case of crosses of G37R mice to the three strains of WT hSOD1 mice, co-expression of both the WT and G37R protein induced paralysis earlier than what occurs in homozygous G37R mice (paralyzed at 8 months: 252.6 ± 4.75 days, N = 20). In G37R/SJLWT mice, we observed paralysis by 5 months of age (160.1 ± 2.40 days, N = 18); the G37R/CgWT SOD1 became paralyzed at ∼6 months of age (190.1 ± 1.01 days, N = 7); whereas in G37R/L76WT mice, we observed paralysis at ∼7 months of age (213.8 ± 4.35 days, N = 13) (Fig. 3A). In this experiment, littermate mice that were heterozygous for the G37R SOD1 transgene were aged only to 12 months (N > 25), remaining free of disease. In prior experiments, heterozygous G37R mice have been shown to reach ages of >20 months without developing disease (20).
Figure 3.
Paralysis develops earlier in G37R or L126Z mice that co-express WT hSOD1. (A and B) Plots of age to paralysis of mice from G37R (A) and L126Z (B) crosses to Gurney and L76 mice. Endpoints were defined as the point at which hind limb paralysis impedes access to food and water. Log-rank tests demonstrated statistical differences in lifespan between homozygous G37R mice and G37R/SJLWT (P ≤ 0.0001), between G37R/CgWT (P ≤ 0.0005) and G37R/L76WT (P ≤ 0.05) and between L126Z mice and L126Z/SJLWT (P ≤ 0.005), but not with L126Z/L76WT (P = 0.8987). The number of mice of each genotype is provided in Results.
Similar to the outcome described above, in double-transgenic mice harboring WT and L126Z hSOD1 proteins, we observed earlier onsets of paralysis in L126Z/WT mice in which the WT hSOD1 transgene was derived from the Gurney WT strain of mice. L126Z/SJLWT mice developed disease between 5 and 6 months of age (170.1 ± 4.74 days, N = 10; Fig. 3B), which was ∼1–2 months earlier than heterozygous mice expressing only L126Z SOD1 (214.1 ± 3.74 days, N = 53). However, L126Z/L76WT double-transgenic mice did not develop paralysis until 8 months of age (224.8 ± 6.41 days, N = 16; Fig. 3B), which was the same age at which heterozygous mice transgenic for only L126Z SOD1 developed paralysis. Thus, in this experiment, it would appear that a higher level of WT hSOD1 expression was required to alter age to paralysis in the L126Z mice.
Accumulation of WT SOD1 in detergent-insoluble aggregates in symptomatic mice co-expressing mutant SOD1
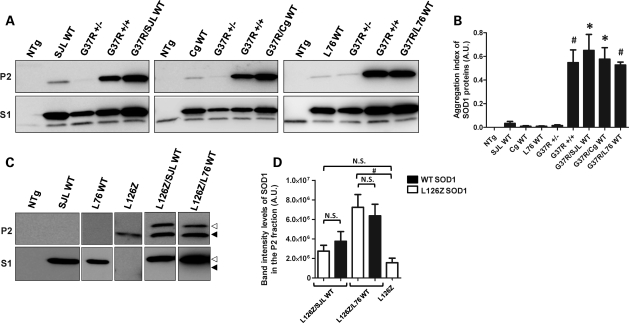
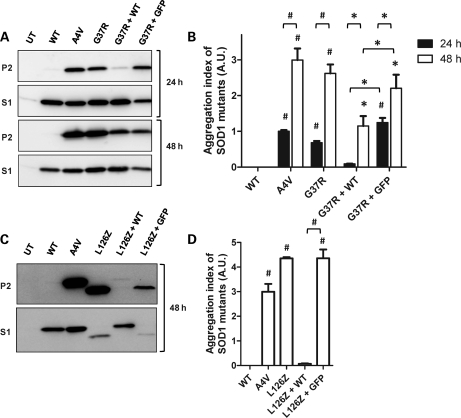
Previous studies have established that spinal cords of paralyzed mice co-expressing WT and mutant hSOD1 accumulate detergent-insoluble forms of both mutant and WT proteins (8). Similar to this previous report, we detected detergent-insoluble forms of SOD1 in spinal cords of endstage G37R/WT mice (Fig. 4A, P2). The levels of SOD1 in the detergent-insoluble (P2) and soluble (S1) fractions were quantified and graphed as the ratio of SOD1 in P2 versus S1; we refer to this value as aggregation index (see Materials and Methods). In all symptomatic G37R/WT mice, the accumulation of detergent-insoluble SOD1 species was significantly higher than asymptomatic heterozygous G37R mice, which accumulate little or no detergent-insoluble aggregates (Fig. 4B). The aggregation index for total SOD1 in spinal cords of symptomatic G37R/WT mice was similar to that of symptomatic homozygous G37R mice (Fig. 4B). We also determined that most of the SOD1 protein in spinal cords of the G37R/WT mice possessed the normal intramolecular disulfide bond, indicating that the excess burden of WT SOD1 did not greatly alter the ability of the system to properly process both WT and mutant SOD1 (Supplementary Material, Fig. S3). Thus, symptomatic mice co-expressing G37R and WT SOD1 accumulated detergent-insoluble forms of SOD1.
Figure 4.
Symptomatic mice accumulate significant amounts of detergent-insoluble SOD1 aggregates at endstage. (A and C) Immunoblots of detergent-insoluble (P2) and detergent-soluble (S1) fractions of spinal cord from transgenic mice. SOD1 protein was detected with an antibody that recognizes mouse and hSOD1 (A), or an antibody specific for hSOD1 (C). Note that the truncation mutant migrates more rapidly (black arrowhead) than WT hSOD1 (open arrowheads). (B and D) Quantification of aggregation index of total SOD1 in G37R/WT crosses (B), and quantification of WT and L126Z protein levels in P2 fraction of L126Z/WT mice (D). *P ≤ 0.05, #P ≤ 0.005. Bars represent mean ± SEM.
Similarly, symptomatic mice co-expressing L126Z and WT SOD1 accumulated detergent-insoluble forms of SOD1. In these animals, the WT and L126Z proteins can be distinguished on immunoblots due to the smaller size of the truncated mutant protein. In all symptomatic L126Z/WT mice, we detected detergent-insoluble forms of WT and mutant SOD1 proteins (Fig. 4C). Note, as described previously (19), we were unable to detect detergent-soluble forms of the L126Z protein (Fig. 4C; Supplementary Material, Fig. S1). In spinal cords of both the L126Z/SJLWT and L126Z/L76WT, the amount of insoluble WT hSOD1 was approximately equivalent to insoluble L126Z protein (Fig. 4D). In comparison with symptomatic heterozygous mice that express the L126Z alone, spinal cords of symptomatic mice co-expressing WT and L126Z SOD1 accumulated higher total levels of insoluble SOD1 (Fig. 4D, add values for L126Z and WT in the insoluble fractions). In L126Z/L76WT mice, the levels of insoluble L126Z protein were significantly higher than the levels of that in heterozygous mice expressing L126Z alone. This finding indicates that the higher levels of total SOD1 in spinal cords of these animals stimulate the generation of these aggregates. However, and importantly, insoluble WT hSOD1 was detected in spinal cords of symptomatic L126Z/L76WT mice (Fig. 4C) even though these mice show an age to paralysis that is identical to that of mice expressing the L126Z mutant alone. This finding indicates that induced aggregation of WT hSOD1 is not sufficient to decrease the age to paralysis.
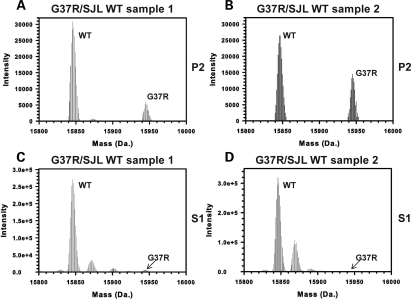
In our immunoblot analyses of detergent-insoluble SOD1 in the G37R/WT mice, it was not possible to determine whether WT acquired insolubility by standard immunoblot of SDS–PAGE. In order to determine whether detergent-insoluble forms of WT hSOD1 accumulate in G37R mice, we analyzed the detergent-insoluble fractions of spinal cords from symptomatic G37R/SJLWT mice by hybrid linear ion-trap Fourier-transformation ion cyclotron resonance mass spectrometry (FTMS) analyses. In two different pooled samples of spinal cords from G37R/SJLWT mice we detected both WT and G37R SOD1 in the detergent-insoluble fraction (Fig. 5A and B). As expected, WT and G37R SOD1 proteins were detected in the soluble S1 fractions (Fig. 5C and D). However, in the soluble fraction, the level of the mutant protein detected by mass spectroscopy was much lower than expected. Based on the quantification of the immunoblots shown in Figure 4A (Supplementary Material, Fig. S4), we would have expected the signal intensity for G37R to be 50% of that of the signal for WT SOD1. However, the signal for G37R SOD1 in the soluble fractions was <10% of the WT protein signal. Because the detection of the G37R mutant in soluble fractions was less than expected, we suspect that the detection of the G37R protein by mass spectroscopy is less efficient than the detection of WT protein. Therefore, the relative amount of the G37R protein in the insoluble fraction could be much greater than indicated by the mass spectroscopic analysis of the insoluble fractions. From these data, we can conclude that WT SOD1 is present in the insoluble fractions from spinal cords but we cannot be sure of the relative amount of each protein.
Figure 5.
WT hSOD1 is present in detergent-insoluble fractions of spinal cords of G37R/SJLWT mice. (A–D) FTMS analyses of P2 (A and B) and S1 (C and D) fractions from two different sets of spinal cord samples. For each sample, three spinal cords were combined (from different symptomatic G37R/SJLWT mice) and extracted in detergent as described in Materials and Methods. Mass peaks indicative of WT and mutant SOD1 were found in both the S1 and P2 fractions.
Formation of detergent-insoluble aggregates of WT and mutant SOD1 in cell culture models
The foregoing findings, along with prior work by Deng et al. (8), demonstrate that high-level expression of WT SOD1 in some manner heightens the toxicity of mutant SOD1 to decrease the age at which paralysis develops. In cell culture models of aggregation, we have noted that co-expression of WT and mutant SOD1 at 1:1 ratios inhibits the aggregation of mutant SOD1 (12). Here, we evaluated the ability of WT hSOD1 to modulate the aggregation of G37R and L126Z hSOD1 mutant proteins in cell culture. Co-expression of WT and G37R hSOD1 proteins (24 h incubations) did not produce detectable levels of aggregated hSOD1 proteins (Fig. 6A and B, 24 h). At 48 h after transfection, similar to what we have observed previously with other ALS mutants (A4V, G85R, G93A) (12), we observed significant accumulation of aggregated SOD1 proteins in cells co-transfected with WT + G37R hSOD1 expression plasmids (Fig. 6A). However, the total amount of aggregated SOD1 in the insoluble fraction was lower in the cells expressing WT and G37R compared with cells co-expressing G37R SOD1 and green fluorescent protein (GFP; GFP serves as a control for non-specific effects of co-transfection) (Fig. 6A and B, 48 h). In co-transfections of WT hSOD1 with the L126Z hSOD1 truncation mutant, we evaluated aggregation index only at the 48 h transfection interval because the L126Z mutant was inherently slower to produce aggregates in the cell model. Cells expressing only L126Z hSOD1 accumulated aggregated protein and small quantities of soluble protein (Fig. 6C and D). However, in cells co-transfected with L126Z + WT hSOD1, detergent-insoluble forms of either protein were virtually undetectable (Fig. 6C and D). A difficult aspect of this experiment is that in contrast to WT protein, the L126Z protein was not readily detected in the soluble fraction of the co-transfected cell lysates as one might expect if aggregation was inhibited (Fig. 6C). This result was repeatedly observed in multiple transfections (Supplementary Material, Fig. S5). The L126Z mutant is relatively rapidly degraded and fails to accumulate to appreciable levels except as insoluble aggregates. It appears that when the aggregation of the L126Z protein is inhibited by WT SOD1, then the mutant is rapidly degraded. In the co-transfection control (L126Z hSOD1 + GFP), the aggregation of the L126Z protein was reduced due to a reduced expression of total mutant protein, but the levels of aggregated L126Z protein remained significantly higher than those of cells co-expressing L126Z and WT hSOD1 (Fig. 6C and D). These findings indicate that, in our cell model, the presence of WT SOD1 produces a pronounced slowing of the formation of the large sedimentable aggregates for both the G37R and L126Z mutants.
Figure 6.
WT hSOD1 slows the aggregation of both G37R and L126Z hSOD1 in cultured cells. (A and C) Aggregate formation determined by immunoblot of transiently transfected cells for 24 (A, upper panels) or 48 h (A, lower panels; and C). In co-transfections of WT and mutant hSOD1 or mutant hSOD1 and GFP (the latter as a control for co-transfection), we used equimolar amounts of each plasmid, with the total amount of transfected protein remaining the same for all reactions (4 µg). (B and D) Relative aggregation index of SOD1 proteins was calculated as P2/S1 ratios, and unpaired Student's t-test were performed as statistical analyses. Symbols over the bars indicate differences with WT hSOD1-transfected cells, or as indicated in the figure: *P ≤ 0.05, #P ≤ 0.0005. Bars represent mean ± SEM of a minimum of three independent transfection experiments.
Complex interactions between WT and mutant SOD1 in aggregate assembly
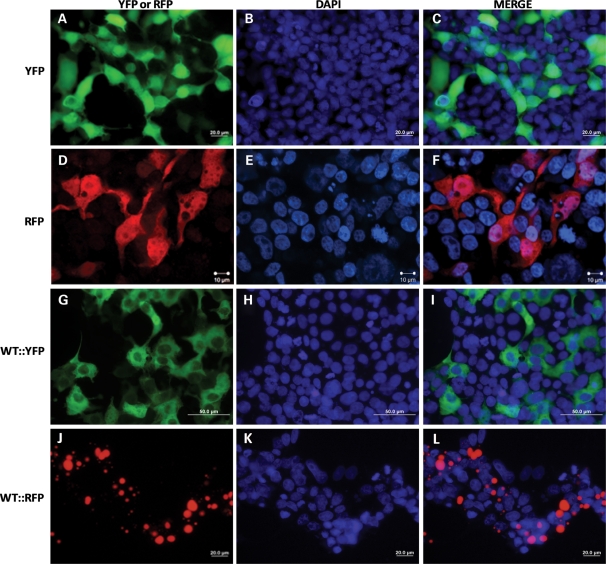
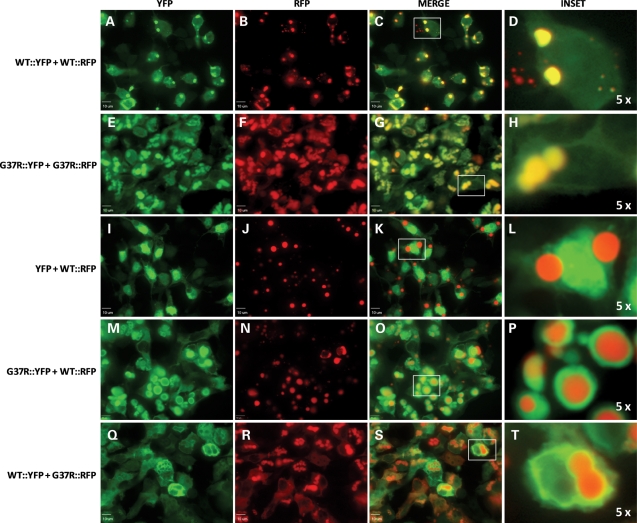
To visualize interactions between WT and mutant hSOD1 in the formation of inclusions, we created a series of hSOD1 proteins fused to the YFP or RFP variants of GFP. Normally, YFP and RFP are diffusely distributed throughout the cell cytoplasm (Fig. 7A–F). A fusion of WT hSOD1 with YFP (WT::YFP) produced a protein that was diffusely distributed throughout the cell similar to YFP alone (Fig. 7G–I). In contrast, WT hSOD1 fused to RFP (WT::RFP) rapidly formed large inclusion structures (Fig. 7J–L). In cells expressing WT::RFP, nearly all of the protein was concentrated in an inclusion, making it difficult to visualize the boundaries of the cell. Co-expression of WT::YFP and WT::RFP induced the formation of inclusion structures that contained both proteins, with the two fluorophores completely overlapping in localization (Fig. 8A–D). Similarly, co-expression of G37R proteins fused to YFP (G37R::YFP) and RFP (G37R::RFP) fluorescent tags leads to the formation of inclusions in which both fluorophores precisely co-localized (Fig. 8E–H). The co-localization of SOD1::YFP with SOD1::RFP (WT or G37R SOD1) appeared to be mediated by the SOD1 portion of the protein because co-expression of YFP alone with WT::RFP did not produce inclusions containing both fluorophores (Fig. 8I–L). Co-expression of G37R::YFP with WT::RFP produced structures in which the G37R::YFP protein appeared to be depositing on the surface of an inclusion formed by the rapidly aggregating WT::RFP protein (Fig. 8M–P). Reversal of the fluorophores in which WT::YFP was co-expressed with G37R::RFP produced the same pattern, but in this case the central inclusion was formed by the G37R::RFP, with WT::YFP depositing on the surface (Fig. 8Q–T).
Figure 7.
Morphology of WT hSOD1 tagged with YFP (WT::YFP) and RFP (WT::RFP). WT::YFP produces a diffusely localized protein, whereas WT::RFP produces inclusions. (A–I) HEK293FT transfected for 24 h with YFP alone (A–C), WT::YFP (D–F) or WT::RFP (G–I) were fixed and visualized as described in Materials and Methods. All images were captured using a 40x objective using epifluorescence on an Olympus BX60 microscope with a color camera; bar 20 (A–C and G–I) or 50 (D–F) µm.
Figure 8.
Visualization of interactions between WT and G37R hSOD1. (A–T) HEK293FT were co-transfected with the indicated SOD1::YFP and SOD1::RFP constructs. Although expression of the same SOD1 protein with different tags (YFP and RFP tags) produced inclusions in which both fluorophores co-localize (WT::YFP + WT::RFP in A–D, G37R::YFP + G37R::RFP in E–H), YFP did not co-localize in inclusions containing WT::RFP (I–L). Co-expression of WT- and G37R-tagged proteins (G37R::YFP + WT::RFP in M–P, and WT::YFP + G37R::RFP in Q–T) produced inclusion structures consisting of a core of the RFP-tagged protein surrounded by the YFP-tagged protein. All images were captured with a 60x water immersion objective and a spinning disk confocal microscope; bars 10 µm.
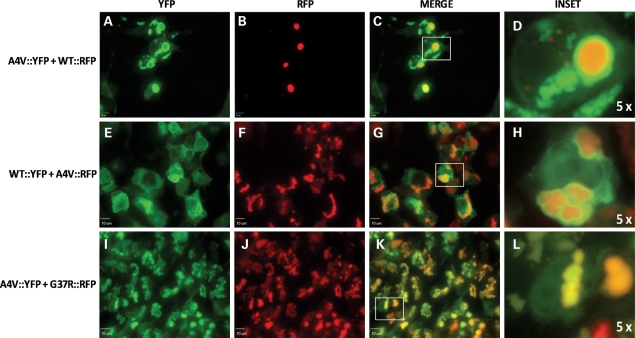
These patterns of interaction between WT and mutant SOD1 fusion proteins were also seen when A4V::YFP was co-expressed with WT::RFP (Fig. 9). We observed some instances in which an inclusion formed by the WT::RFP fusion was surrounded by the A4V::YFP fusion protein (Fig. 9A–D). More rarely, we observed cells in which it appeared that inclusions formed by A4V::YFP fusion protein were adjacent to inclusions formed by WT::RFP fusion protein (not shown). In the converse experiment, at 48 h post-transfection, all cells that exhibited inclusions formed by A4V::RFP fusion proteins were surrounded by WT::YFP fusion proteins that appeared to be deposited on the surface of the A4V::RFP inclusions (Fig. 9E–H). In contrast, co-transfections of two mutant SOD1::YFP fusion proteins that were differentially tagged (A4V::YFP with G37R::RFP) produced inclusions in which the two fusion proteins appeared to be homogenously co-localized (Fig. 9I–L). Collectively, these data suggest that WT and mutant SOD1 adopt conformations that do not favor co-assembly into higher order structures. In contrast, the conformations adopted by A4V and G37R SOD1 are similar enough to facilitate a more intimate co-mingling of the two proteins within the inclusion structure.
Figure 9.
WT and A4V SOD1 fusion proteins do not form homogenous inclusions in which both fluorophores co-localize. HEK293FT cells were co-transfected with the indicated SOD1 constructs for 24 (A–D) or 48 (E–L) h. Co-expression of A4V::YFP and G37R::RFP produces homogenous inclusions containing both proteins (I–L). Images were captured with a 60x water immersion objective in a spinning disc confocal microscope; bars 10 µm.
DISCUSSION
In the present study, we demonstrate that co-expression of WT hSOD1 with fALS mutant hSOD1 accelerates the course of disease in matings of three different strains of mice expressing WT hSOD1 with two different lines of mutant mice. The effect of WT hSOD1 mice on disease in mice expressing mutant hSOD1 was influenced by the nature of the mutation and the level of the mutant and WT hSOD1 proteins expressed. Symptomatic mice co-expressing WT and mutant SOD1 accumulate detergent-insoluble aggregates of both proteins regardless of whether the age to paralysis is altered. In cell models, co-expression of WT hSOD1 with G37R or L126Z hSOD1 diminished the capacity of the mutant protein to form detergent-insoluble aggregates. Using variants of both WT and mutant hSOD1 fused to fluorescent tags, we visualized inclusions generated in cells co-expressing WT and mutant SOD1, finding evidence that WT and mutant SOD1 do not readily interact to form homogenous aggregates. From these observations, we cannot easily conclude that the earlier development of paralysis observed in mice co-expressing WT and mutant SOD1 is a consequence of more rapid formation of aggregated SOD1 proteins.
Acceleration of disease by co-expression of WT hSOD1
In matings of G37R and WT mice, we observed surprisingly that double-transgenic progeny of G37R mice crossed with the Gurney and L76WT strains of mice developed disease earlier than mice homozygous for the G37R transgene, with the age to paralysis appearing to be related to the levels of WT hSOD1 expression. Within the data on the crosses of these strains of mice, we can observe that the background strain of the animals moderates the effect of the WT hSOD1 on age to paralysis. G37R/SJLWT mice show a slightly earlier development of paralysis than G37R/CgWT mice. G37R/L76WT mice (congenic on the C57BL6/J strain) show an age to paralysis that is somewhat slower than that shown by G37R/CgWT mice. In this latter case, we assume that the slower course of G37R/L76WT mice is due to slightly lower expression of the WT hSOD1 transgene (Fig. 1). Importantly, all of the double-transgenic animals in these studies were compared with littermates born of the same breeders and thus the littermates harboring only the G37R transgene would have similar mixtures of the background strains as their double-transgenic littermates. Thus, the dramatically accelerated course of disease in G37R/WT mice of all three strains must be due to an ability of the WT protein to either acquire a toxic property or augment the toxicity of the G37R variant.
Given the magnitude of the effect of WT hSOD1 co-expression on the age to paralysis of G37R mice, it appears that WT proteins possess the capacity to: (i) double the toxicity of a given unit of G37R hSOD1, (ii) double the steady-state level of the G37R protein by some means of stabilization or (iii) acquire a level of toxicity essentially equivalent to mutant SOD1. We know from previous work that WT and G37R hSOD1 are able to form a dimeric enzyme, and that at equilibrium we could expect that at least 50% of the G37R protein would be heterodimerized with WT protein (21). One possibility is that such heterodimers protect the mutant protein from degradation and simply increase the total dose of mutant SOD1 (13). However, in previous studies, we have demonstrated that the co-expression of mutant SOD1 with a rapid rate of turnover with WT protein does not result in a more rapid turnover of WT protein or a slower turnover of the mutant protein (21). For the G37R mutant, it seems unlikely that the WT protein would be able to stabilize mutant SOD1 protein to a sufficient degree to increase the levels of the mutant protein by more than 2-fold as would be required to explain why co-expression of the WT protein in G37R mice causes paralysis earlier than observed in the homozygous G37R mice. However, because of the difficulty in distinguishing the G37R mutant from WT protein in spinal cord extracts, we cannot be certain that some type of stabilization has not occurred. In the crosses of L126Z mice with WT SOD1 mice, the detection of both proteins is much easier. In spinal cords of double-transgenic mice, the aggregation of the mutant and WT proteins confounds determination of whether the presence of the WT protein changes the steady-state level of the mutant protein. However, in the forebrain of symptomatic L126Z mice, detergent-insoluble forms of the mutant protein are not detected (see Supplementary Material, Fig. S3 of reference 22). Immunoblots of total protein in the forebrain do not produce evidence that the steady-state levels of L126Z protein increase when WT protein is co-expressed (Supplementary Material, Fig. S6). Thus, although we cannot completely rule out the possibility that the presence of WT SOD1 in some manner stabilizes the mutant protein to increase steady-state levels in spinal cord, we think that the weight of evidence favors another explanation. Thus, we propose that WT SOD1 acquires toxic properties through some type of interaction with mutant SOD1. Recent demonstrations that various forms of WT SOD1 possess toxicity lend credibility to such a hypothesis (23,24).
In our study involving mice expressing L126Z/WT hSOD1, we observed that the L126Z/SJLWT showed earlier paralysis than mice expressing L126Z alone, as Deng et al. (8) reported in a similar cross, whereas L126Z/L76WT mice develop disease at the same time as L126Z mice, similar to what Bruijn et al. (15) reported in crosses of L76WT mice to G85R hSOD1 mice. In this cross, we are comparing SJLWT mice with L76WT mice, which adds a complication in regard to potential influences of background strain. Unfortunately, the breeding of the CgWT Gurney strain to L126Z mice failed to produce an adequate cohort of double-transgenic animals. However, as noted above, the comparisons in age to paralysis between L126Z/SJLWT mice included littermates that were transgenic only for the L126Z transgene and which would possess the same mixture of background strains. Hence, we are confident that the earlier development of paralysis in L126Z/SJLWT mice is due to an effect of the co-expressed WT hSOD1. Because expression of L126Z hSOD1 is above the threshold to induce disease, we are looking only for an augmentation of toxicity. Similarly, G85R mice used in the study by Bruijn et al. (15) develop disease spontaneously and thus assessed augmentation of toxicity by WT hSOD1. We conclude that the simplest explanation for the accelerated disease in mice generated from matings of Gurney WT and L126Z mice is that higher levels of WT SOD1 expression in Gurney WT mice simply increase the strength and frequency of interactions between WT and mutant proteins and thus increase the opportunities to produce toxic structures. We propose that this explanation could also hold to explain how crosses of Gurney WT mice with a line of G85R mice resulted in double-transgenic progeny that developed paralysis earlier than mice expressing the G85R mutant alone (11), whereas crosses of G85R mice with L76WT mice produced no effect (15). We note that the age to paralysis of G85R mice described by Wang et al. (11) and that of G85R mice described by Bruijn et al. (15) were approximately the same, ∼12 months. Thus, we conclude that the discrepancy between the report by Wang et al. (11) and Bruijn et al. (15) is due to the differing levels of expression by the lines of WT SOD1 mice used in the respective studies.
In reviewing the literature that describes mice co-expressing WT and mutant hSOD1, it is clear that the magnitude of the effect of WT protein on the toxicity of mutant SOD1 is related to the level of mutant SOD1 expression. There are three examples of crosses between WT mice and mutant mice in which the level of the mutant protein is below the threshold to develop disease. These include a line of mice that express the A4V mutant (8), mice that express G93A mutant via a Thy1 promoter (10) and mice that express an experimentally truncated SOD1 ending at residue 116 (T116X) (9). In each of these cases, mice heterozygous for the mutant transgene do not develop disease, but mice doubly transgenic for these mutant SOD1 transgenes and the WT SOD1 (derived from the Gurney WT line) develop paralysis by 12–18 months (8–10). Mice that express L126Z SOD1 at levels to induce paralysis by 12 months of age develop paralysis by 7 months of age when doubly transgenic with the Gurney WT SOD1 mice (8). Our L126Z mice express mutant protein at a level that induces paralysis by 7 months of age with the co-expression of WT SOD1 (via mating to the Gurney WT) reducing the age to paralysis by 1–2 months. In contrast, the effect of mating Gurney WT SOD1 mice to Gurney G93A mice, which develop paralysis at 130 days, was a change in age to paralysis of only 10 days earlier in mice transgenic for both mutant and WT SOD1 (8). Thus, the magnitude of the effect WT SOD1 has on the toxicity of mutant SOD1 is inversely proportional to the level of mutant SOD1 that is expressed.
Another important point is that among the examples of large effects of WT SOD1 on age to paralysis, T116X and L126Z mutants would not be expected to be able to form normal heterodimers with WT SOD1 due to loss of portions of the dimer interface. Similarly, the A4V mutant has been reported to be less able to form normal dimeric enzyme (25,26). If normal dimeric interactions between WT and mutant SOD1 were by some manner responsible for stabilizing the mutant protein to increase steady-state levels, then one might expect that these poorly dimerizing mutants would be less affected by co-expression of WT SOD1. Clearly, however, the toxicity of these mutants is dramatically augmented by the presence of WT protein. These observations also suggest that mutant proteins are probably interacting with WT SOD1 via some mechanism other than the normal dimer interface to produce the augmented toxicity.
Earlier disease in mutant/WT mice is associated with aggregated SOD1
In all our double-transgenic mutant/WT hSOD1 mice, hind limb weakness and paralysis were accompanied by the accumulation, in the spinal cord, of detergent-insoluble species of WT hSOD1. Thus, the earlier appearance of paralytic disease led to a coincident earlier appearance of detergent-insoluble SOD1 aggregates. In tissues from L126Z/WT double-transgenic mice, it is possible to detect the WT protein in detergent-insoluble fractions by immunoblot due to the smaller size of the truncation mutant, relative to WT hSOD1. In the tissues from G37R/WT double-transgenic mice, we utilized mass spectroscopy approaches to identify WT and mutant proteins in insoluble fractions. Thus, in both cases, WT SOD1 acquired insolubility in symptomatic mice.
In spinal cords of symptomatic L126Z/WT mice, we easily detected detergent-insoluble forms of WT SOD1 by immunoblotting. Importantly, we demonstrate that symptomatic mice derived from mating L126Z SOD1 mice with L76WT SOD1 mice accumulate detergent-insoluble forms of WT hSOD1 despite the fact that there was no change in the age to paralysis. Thus, in this example, the aggregation of the WT protein was purely incidental to the disease. This finding disconnects the co-aggregation of WT and mutant SOD1 to the processes responsible for decreasing the age to paralysis since L126Z/L76WT mice develop paralysis at an age similar to heterozygous mice expressing only the L126Z variant.
Our findings in cell models of aggregation are consistent with the idea that the capture of WT hSOD1 in insoluble aggregates could be secondary to the assembly of mutant SOD1 into insoluble structures. Studies involving fluorescently tagged versions of WT and G37R SOD1 produce images that delineate interactions between WT and mutant proteins in aggregate assembly. WT::YFP fusion proteins do not readily form inclusions, whereas G37R::RFP fusion proteins readily generate inclusions. When inclusions are rapidly formed by G37R::RFP or A4V::RFP, the co-expressed WT::YFP deposits around the outer edge of the core of the inclusion. In contrast, inclusions formed in cells co-expressing WT::YFP and WT::RFP, or G37R::YFP and G37R::RFP, appear to contain interlaced assemblies of both proteins. The co-expression of A4V::YFP with G37R::RFP also produced perfectly interlaced inclusions. Thus, the exclusion of WT::YFP to the periphery of inclusions formed by mutant SOD1::RFP fusion proteins indicates that WT and mutant SOD1 do not interact effectively as aggregates assemble.
In a recent study of mice that co-express WT SOD1 with G85R hSOD1, immunoblots of detergent-insoluble protein in the spinal cord demonstrated immunoreactive bands that showed electrophoretic migrations that were suggestive of aberrant disulfide cross-links between WT and G85R mutant SOD1 (11). The banding pattern was also interpreted to indicate abnormal disulfide-linked homodimers of G85R and homodimers of WT hSOD1 were present. The relative amount of WT and G85R that were cross-linked in aberrant dimers was not quantified but it appeared to be a relatively minor percentage. Still, this finding would suggest that some mutant and WT proteins are in close proximity within aggregates formed in spinal cords of these mice. In our cell model, we show images in which WT SOD1 appears to be deposited on the surface of mutant SOD1 inclusions and vice versa. We suggest that if the structures we observe in the cell model occur between WT and mutant SOD1 in vivo, then there would be ample opportunity for WT and mutant SOD1 to be in close enough contact to permit aberrant disulfide cross-links. It is also possible that the much slower kinetics of aggregation that occur in vivo provide greater opportunity for WT protein to be incorporated into growing aggregates of the mutant protein. We suggest that our findings in the cell model indicate that such interactions are probably not favored but we cannot exclude the possibility that they occur.
Mechanisms of toxicity
The foregoing discussion paints a view in which the aggregation of mutant SOD1 is proposed to be secondary to toxicity. However, we do not believe that this view is entirely correct. The outcomes of our study of the G37R mice are potentially very informative regarding aggregation and toxicity. Mice homozygous for the G37R transgene develop disease, whereas mice with 50% less expression, due to heterozygosity for the transgene, do not (20). Increasing total SOD1 expression in G37R mice by co-expressing WT hSOD1 at levels equivalent to or slightly above the G37R mutant protein also produces disease. In this later case, mice reach endstage paralysis sooner than mice homozygous for G37R. Importantly, the level of expression of mutant SOD1 or mutant/WT SOD1 that is required to induce disease is identical to the level required to induce the formation of large detergent-insoluble aggregates. Because we know from a variety of studies of protein aggregation that oligomeric assemblies are precursors to larger aggregates (for review, see 27,28), we assume that the large detergent-insoluble aggregates detected in symptomatic mice are derived from some type of oligomeric precursor. Our findings in the G37R mice suggest that the threshold of mutant or mutant/WT SOD1 that is required to induce toxicity is linked to the threshold required to induce aggregation and presumably oligomerization. The simplest scenario is that a population of oligomeric structures is generated early in the course of disease, with some of these structures possessing toxicity (Fig. 10).
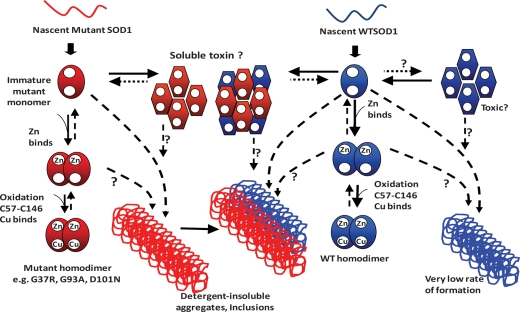
Figure 10.
A model for the maturation and aggregation of WT and fALS mutant SOD1. The steps in the normal maturation of mutant and WT SOD1 are not completely understood. It is likely that the binding of zinc (Zn) is one of the first steps in maturation. With the binding of Zn, the protein may be able to form a weakly interacting dimer. Oxidation of the normal C57 to C146 disulfide bond may or may not occur prior to the binding of copper (Cu); however once Cu is bound and the disulfide is formed, a very stable dimeric enzyme is created. Some of the fALS mutants appear to be able to properly mature, whereas others, such as the L126Z truncation mutant, cannot. We suggest that toxic oligomeric forms of mutant SOD1 may be generated early in the course of disease, with WT protein possibly being able to co-oligomerize with the mutant protein to generate toxic species responsible, decreasing the age to paralysis by initiating disease earlier. Late in disease, WT SOD1 may be captured in larger aggregates of mutant SOD1. WT SOD1, when over-expressed in transgenic mice, can produce large detergent-insoluble aggregates. Whether WT SOD1 can also produce toxic oligomeric assemblies is unknown.
At present, we lack evidence for the existence of toxic oligomeric structures and thus the model we present is speculative. It is possible that WT SOD1 has some indirect effect on the biology of the system to change the way that mutant SOD1 is processed, leading to a heightened generation of forms of the mutant protein that are toxic. However, our cell culture studies indicate that WT protein can modulate the aggregation of mutant SOD1 and it is thus tempting to suggest that one mechanism by which WT SOD1 accelerates the course of disease is by oligomerizing with mutant SOD1 to produce toxic species that in some manner mediate early events in disease course. We are reasonably sure that early in the course of disease, when the first pathologic features first appear, the vast majority of the mutant protein is soluble and yet to have assembled into large aggregates (6) and that the formation of the large aggregates, which occurs in the interval between first visible symptoms and paralysis (4–6), may potentially mediate events involved in disease progression (Supplementary Material, Fig. S7) (3,7). Thus, we have proposed that more than one molecular form of mutant SOD1 mediates toxic events in disease pathogenesis, with large detergent-insoluble aggregates playing roles late in disease and detergent-soluble forms of the protein involved in events that initiate disease (3,6). If, as indicated in these prior studies, soluble forms of mutant SOD1 are involved in events that initiate disease, then it appears that co-expression of WT SOD1 enhances the generation of these toxic soluble species.
What do these studies tell us about the human disease? The collection of studies involving transgenic mice that co-express mutant and WT hSOD1 indicate that WT protein has the potential to either influence the toxicity of the mutant protein or to acquire toxic properties. In either case, the age to paralysis is noticeably decreased in mice when WT hSOD1 is co-expressed with mutant hSOD1. However, in the mouse models, it is clear that the mutant protein need not partner with WT hSOD1 for disease to occur because all examples of transgenic mice that model SOD1-linked ALS were initially made by expressing the mutant human gene alone. In humans, the extent to which WT SOD1 influences the onset or progression of disease is unclear. The vast majority of patients with SOD1-linked fALS are heterozygous for the disease-causing mutation. One notable exception is disease caused by the mutation of Asp 90 to Ala (D90A) (29). In most families, this mutation only causes disease when individuals are homozygous for the mutation. However, there are a few rare families in which the D90A mutation acts as a dominant mutation to cause disease in a heterozygous state (30). Thus, if the WT protein plays some role in modulating the toxicity of D90A SOD1, we have examples in which the WT protein would seem to be negating toxicity in pedigrees in which disease is present only when homozygous for the mutation and examples in which WT protein could promote the toxicity of the mutant protein in pedigrees in which disease is present in patients heterozygous for the D90A mutation. There are a few rare examples of patients who are homozygous for a mutation that is otherwise dominantly inherited. In one case (N86S mutation), the homozygous patient developed a rapidly progressing disease at a very young age (31). However, in another case (L126S mutation), the homozygous patient developed disease at an age typical for ALS, but the rate of progression was extremely rapid (32). Collectively, these studies indicate that, similar to the mouse studies, the presence of WT hSOD1 is not obligatory for the mutant protein to exert toxicity. The unresolved questions are whether WT protein may be involved in the generation of toxic forms of the mutant protein in humans and whether the involvement WT protein varies with different mutations.
Conclusions
Our findings indicate that the effect of WT SOD1 on the toxicity of mutant SOD1 is influenced by both the nature of the fALS mutation with which it is co-expressed and the level of WT SOD1 expression. Although WT SOD1 is present in insoluble aggregates found in symptomatic double-transgenic mice, we provide the first evidence that dissociates WT SOD1 co-aggregation from the earlier development of paralysis. From cell culture models of aggregation, we find that WT SOD1 modulates the aggregation of the mutant protein to slow the formation of large detergent-insoluble structures. Our study of SOD1::YFP fusion proteins indicates that WT and mutant SOD1 do not intimately co-assemble in cellular aggregates. Together, these findings implicate soluble assemblies of WT and mutant SOD1 as possible mediators of toxic processes involved in initiating motor neuron disease. The extent to which such assemblies are involved in human disease, where expression levels of both mutant and WT SOD1 protein are much lower than the mouse models, is likely to be dependent on the nature of the fALS mutation.
MATERIALS AND METHODS
Transgenic mice
All the strains of transgenic mice used in this study have been previously described. The PrP.SOD1-G37R SOD1 line 110 mice were generated by a vector that utilized the mouse prion protein promoter (MoPrP.Xho) containing a hSOD1 cDNA with the G37R mutation (20). These mice were maintained in a hybrid background of C57BL/6J and C3H/HeJ. All other strains of mice used in this study were generated with genomic fragments of the hSOD1 gene. The L126Z SOD1 line 45 used a modified genomic fragment in which exons 3–5 of the human gene were fused (19) and a stop codon was inserted at codon 127. These mice were maintained in a hybrid background of C57BL/6J and C3H/HeJ. Mice expressing WT hSOD1 that we used included a line developed by Gurney et al. (17), maintained in both hybrid backgrounds of C57BL/6J × SJL and in a congenic background of C57BL/6J (these strains are designated SJLWT and CgWT, respectively), and WT line 76 mice (designated L76WT) (16) maintained in a congenic background of C57BL/6J mice. The congenic Tg(SOD1)Gur2/J mice were graciously provided by Dr Gregory Cox (Jackson Laboratories, Bar Harbor, ME, USA). All studies involving mice were approved by the Institutional Animal Care and Use Committee at the University of Florida.
For the identification of genotype, DNA from mouse tail biopsy was extracted by digesting tissue (0.6 cm length) in 600 μl of TNES buffer (50 mm Tris base, pH 7.5; 400 mm NaCl; 100 mm EDTA, pH 8.0; 0.5% SDS) with 18 μl of Proteinase K at 20 mg/ml. After being kept overnight in a 55°C water bath, 167 μl of supersaturated 6 m NaCl was added to each sample, mixed and then centrifuged at 16 100 g for 5 min. A total of 650 μl of supernatant was removed and placed into a new microfuge tube, then centrifuged again before 600 μl of supernatant was moved to a new tube and mixed with an equal volume of cold 100% ethanol. DNA was then pelleted by centrifuging at 14 000 r.p.m. for 5 min and washed with 1 ml of cold 70% ethanol before centrifuging again for 2 min. The final DNA pellet was resuspended in 200 μl of TE buffer (10 mm Tris base, pH 7.5; 1 mm EDTA, pH 8.0).
The DNA extracted from mouse tail was analyzed by PCR using the following primers. For the L126Z and WT hSOD1 lines of mice, we used two primers: Hu-S primer (5′-TCA AGC GAT TCT CCT GCC T); H/M-AS primer (5′-CAC ATT GCC CAR GTC TCC A; R = A/G). For the G37R line 110 hSOD1 mice, we used three primers: PrP-S primer (5′-GGG ACT ATG TGG ACT GAT GTC GG); PrP-AS primer (5′-CCA AGC CTA GAC CAC GAG AAT GC); HuSOD1-S (5′-GTC GAC AAG CAT GGC CAC GAA GGC CGT GTG C). PCR reactions were performed using Taq DNA polymerase and PCR reagents from New England Biolabs (Ipswich, MA, USA) following the manufacturer's protocol. The settings for the thermocycler were as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s; 60°C for 1 min; 72°C for 5 min; 72°C for 10 min. The PCR products were the following: the L126Z hSOD1 transgene produced a fragment of ∼500 bp, the WT hSOD1 transgene produced a fragment of ∼1200 bp and G37R hSOD1 transgene produced a fragment of ∼500 bp. Endogenous mouse prn-p genes produce fragments of ∼750 bp and served as a positive control.
SOD1 cDNA expression plasmids
All of the non-tagged WT SOD1 and ALS-associated mutant hSOD1 cDNAs were expressed from plasmids based on the mammalian pEF-BOS expression vector, and have been previously described (19,33). GFP was expressed in pcDNA3.1 as described previously (12).
SOD1 fusion protein cDNA variants were created from an expression vector (pPD30.38) that contains WT hSOD1 fused to eYFP (yellow fluorescent protein), which was kindly provided by Dr Rick Morimoto (Northwestern University). This SOD1::eYFP construct contains a 27 bp linker (translated sequence—LQLKLQASA) between SOD1 and YFP that was modified to include an SalI restriction site (translated sequence—LQSTLQASA). This SOD1::YFP DNA fusion construct was then cloned into our mammalian pEF-BOS expression vector. From this initial SOD1::YFP expression plasmid, we generated vectors for A4V::YFP and G37R::YFP. Additionally, we also created a control plasmid in pEF-BOS for eYFP (termed YFP). Similar to SOD1::YFP fusion proteins, we created SOD1 fusion proteins with a red fluorescent protein tag [RFP, Turbo RFP cDNA obtained from the pTRIPZ empty vector available at Open Biosystems (Huntsville, AL, USA)] by replacing the YFP tag with the RFP tag. In this way, we created WT::RFP, A4V::RFP and G37R::RFP constructs along with a vector to express RFP alone. All cell culture studies of SOD1 aggregation used HEK293FT cells (Invitrogen, Carlsbad, CA, USA).
Northern blotting
Fresh spinal cords were harvested from 4-month-old mice, with one-half of the cord immediately used for RNA extraction and the other half frozen for analyses of protein levels. Extraction of spinal cord mRNA was performed using TRIzol (Invitrogen) as described by the manufacturer. Five micrograms of total RNA was electrophoresed in formaldehyde–agarose gels and then transferred onto a nylon membrane by capillary action using 10x SSC buffer (1.5 m NaCl, 150 mm sodium citrate, in distilled water treated with diethyl pyrocarbonate). The nylon membrane was then cross-linked, and pre-hybridized for 1 h at 65°C with hybridization buffer (1% BSA, 1 mm EDTA at pH 8.0, 0.5 m NaHPO4 at pH 7.2, 7% SDS) followed by hybridization at 65°C with the 32P-labeled cDNA probes (at ∼1.4 × 106 c.p.m./ml) for 12 h. As probes, we generated a fragment of hSOD1 cDNA and a fragment that recognizes endogenous mouse PrP mRNA, the latter serving as a loading control. Probes were labeled using the DNA-Ready to Go-label beads 32P-dCTP from GE Healthcare (Pittsburgh, PA, USA) and purified with Illustra ProbeQuant G-50 Micro Columns from GE Healthcare as described by the manufacturer. After hybridization, membranes were washed three times for 30 min with wash buffer 1 (0.1% BSA, 1 mm EDTA at pH 8.0, 40 mm NaHPO4 at pH 7.2, 5% SDS) at 65°C. Two more washes were then performed with wash buffer 2 (1 mm EDTA at pH 8.0, 40 mm NaHPO4 at pH 7.2, 1% SDS). Membranes were then exposed to film and developed after 24 h or imaged by phosphorimaging on a Molecular Dynamics Instrument (Bio-Rad Laboratories, Hercules, CA, USA) . Northern blot analyses were performed on a total of three spinal cords from each of WT mouse strains. Band intensities from the northern blots quantified by phosphorimaging were calculated using Quantity One 4.6.5 software (Bio-Rad Laboratories). In the quantification of SOD1 mRNA levels, we normalized the values of endogenous prion protein mRNA, which served as a loading control. The statistical significance of differences in mRNA levels was determined by unpaired Student's t-tests.
Determination of total SOD1 protein levels in the spinal cord
Spinal cords were dissolved by sonication in five volumes (by weight) of 1x TEN buffer (10 mm Tris, pH 7.5; 1 mm EDTA, pH 8.0; 100 mm NaCl) with protease inhibitor cocktail at 1:100. A brief 800g centrifugation for 10 min was performed to discard cell debris. Protein concentrations in the supernatant fraction, which represents total extracted protein, were measured by BCA assay (Pierce Biotechnology, Rockford, IL, USA). Five microgram of protein was electrophoresed in 18% SDS–PAGE gels, transferred to membranes and immunoblotted with an antibody (1:2500, overnight) that recognizes mouse and hSOD1 (34) or an antibody (1:2500 overnight) that recognizes only hSOD1 (33) as indicated in the figure legends. Blots were also probed (1:5000, overnight) with an antibody that recognizes β-tubulin-III for loading control. A secondary goat anti-rabbit IgG (1:5000, 1 h) was used to detect primary antibodies. Band intensities from the immunoblots were calculated using the Fujifilm imaging system (FUJIFILM Life Science, Valhalla, NY, USA). In the quantification of SOD1 protein levels, we normalized the values to the tubulin-loading control.
Detergent extraction of SOD1 aggregates
This assay generates two protein fractions termed S1 (detergent-soluble) and P2 (detergent-insoluble), the latter containing aggregated forms of mutant SOD1, as described previously (3,35). Detergent extraction of cultured cells was performed as described in the foregoing reports; however, for spinal cord samples, slight modifications of the protocol were followed to obtain higher levels of extracted protein. A total of 300 μl of spinal cord crude supernatant was extracted by adding an equal volume of buffer to produce a final concentration of 10 mm Tris, pH 7.5; 1 mm EDTA, pH 8.0; 100 mm NaCl; 0.5% NP-40, 1:100 v/v protease inhibitor cocktail. The mixture was sonicated three times for 10 s each before centrifugation at >100 000g for 5 min in a Beckman AirFuge (Brea, CA, USA) to produce supernatant 1 (S1) and pellet (P1) fractions. The P1 fractions were then washed with the same extraction buffer (sonicated twice, 15 s each) and centrifuged at >100 000g for 5 min. The supernatant was discarded and the remaining pellet represented the detergent-insoluble (P2) fraction, which was resuspended in a buffer containing SDS and deoxycholate (10 mm Tris, pH 7.5; 1 mm EDTA, pH 8.0; 100 mm NaCl; 0.5% NP-40, 0.25% SDS, 0.5% deoxycholate, 1:100 v/v protease inhibitor cocktail). The total volumes corresponding to S1 and P2 fractions were of 600 and 100 μl, respectively. Protein concentrations of S1 and P2 fractions were determined by BCA assay, as described by the manufacturer (Pierce Biotechnology). For SDS–PAGE and immunoblot analyses, 5 μg of protein from S1 and 20 μg of protein from P2 were used. In some of the SDS–PAGE analyses, reducing agents were omitted from the loading and running buffers in order to separate and visualize reduced and oxidized forms of SOD1 following procedures described previously (6,36).
Analyses of S1 and P2 fractions by FTMS
FTMS analyses of S1 and P2 of spinal cords of two different G37R/SJLWT samples were performed as described previously (12). Here, each spinal cord sample was extracted in detergent to obtain S1 and P2 fractions from three combined spinal cords (1.2 ml of S1 and 600 μl of P2). The entire P2 fraction was analyzed by HPLC followed by FTMS to identify WT and mutant hSOD1 mass signatures.
Immunocytochemistry
Transfection of HEK293FT for immunocytochemistry was performed on glass coverslips that were previously coated with 0.5 mg/ml poly-l-lysine in 1x phosphate-buffered saline (PBS) solution. Lipofectamine 2000 was used to transfect 2 μg of expression vector per well following the manufacturer's protocol (Invitrogen). Transfected cells were then fixed with 4% paraformaldehyde in 1x PBS solution for 15 min. Additional staining was performed to visualize the nuclei, using DAPI solution (4′,6-diamidino-2-phenylindole, dihydrochloride, stock 14.3 mm from Invitrogen) at 1:2000 for a minimum of 10 min. Fluorescence was visualized by epifluorescence on an Olympus BX60 microscope with a color camera and by confocal microscopy on an Olympus IX81-DSU spinning disk confocal microscope indicated in the figure legends.
Quantitation of immunoblots
Quantification of the SOD1 protein in detergent-insoluble and detergent-soluble fractions was performed by measuring the band intensity of SOD1 in each lane using a Fuji Imaging system (FUJIFILM Life Science, Stamford, CT USA). For immunoblots of S1 fractions, we load 5 μg of total protein, and for the P2 fractions, we load 20 μg of total protein. Protein concentrations of S1 and P2 fractions were determined by BCA assay, as described by the manufacturer (Pierce Biotechnology). The untransfected cells served as a control. Aggregation index was calculated as the ratio of the band intensity in the detergent-insoluble fraction (P2) to that of the detergent-soluble fraction (S1). Note that the amount of protein loaded on the gel for P2 fractions is 4-fold more than the S1 fraction. The mean and standard error of the mean (SEM) were calculated for the aggregation index of each sample in each experiment. In each cell culture experiment, we included a positive control, which consisted of transfection of expression vectors for A4V SOD1. To normalize variation in data between experiments, relative values of aggregation index are normalized to P2/S1 value of A4V at 24 h transfection (set to 1) (3).
Statistical analyses
All statistical analyses were performed using GraphPad PRISM 5.01 Software (La Jolla, CA, USA). The specific statistical method used in analyzing the data is noted in the figure legends.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by a grant from the National Institutes of Neurologic Disease and Stroke (P01 NS049134—Program Project award to Drs Joan S. Valentine, P. John Hart, D.R.B. and J.P.W.). Funding to pay the Open Access Charge was provided by the University of Florida.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Celeste Karch for thoughtful discussions and help in the preparation of this manuscript. We also thank Ms Hilda Brown for her help in generating mutant SOD1 cDNA vectors. We thank Dr Richard Morimoto for providing plasmids with cDNA constructs for SOD1::YFP fusion proteins. We are grateful to Professor Joseph A. Loo for providing access to the LTQ-FT ultra-high resolution mass spectrometer. We thank Dr Gregory Cox for providing the congenic Tg(SOD1)Gur2/J strain of mice before general release by the Jackson Laboratories. We thank Drs P. John Hart and Joan Valentine for thoughtful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.-X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. doi:10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. doi:10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 3.Prudencio M., Hart P.J., Borchelt D.R., Andersen P.M. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum. Mol. Genet. 2009;18:3217–3226. doi: 10.1093/hmg/ddp260. doi:10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso M., Massignan T., Samengo G., Cheroni C., De Biasi S., Salmona M., Bendotti C., Bonetto V. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J. Biol. Chem. 2006;281:33325–33335. doi: 10.1074/jbc.M603489200. doi:10.1074/jbc.M603489200. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Lindberg M., Oliveberg M., Marklund S.L. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. doi:10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 6.Karch C.M., Prudencio M., Winkler D.D., Hart P.J., Borchelt D.R. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl Acad. Sci. USA. 2009;106:7774–7779. doi: 10.1073/pnas.0902505106. doi:10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Johnson J.L., Agar N.Y., Agar J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng H.X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G.H., Khan M.S., Hung W.Y., Bigio E.H., et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. doi:10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng H.X., Jiang H., Fu R., Zhai H., Shi Y., Liu E., Hirano M., Dal Canto M.C., Siddique T. Molecular dissection of ALS-associated toxicity of SOD1 in transgenic mice using an exon-fusion approach. Hum. Mol. Genet. 2008;17:2310–2319. doi: 10.1093/hmg/ddn131. doi:10.1093/hmg/ddn131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaarsma D., Teuling E., Haasdijk E.D., De Zeeuw C.I., Hoogenraad C.C. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J. Neurosci. 2008;28:2075–2088. doi: 10.1523/JNEUROSCI.5258-07.2008. doi:10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Deng H.X., Grisotti G., Zhai H., Siddique T., Roos R.P. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum. Mol. Genet. 2009;18:1642–1651. doi: 10.1093/hmg/ddp085. doi:10.1093/hmg/ddp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prudencio M., Durazo A., Whitelegge J.P., Borchelt D.R. Modulation of mutant superoxide dismutase 1 aggregation by co-expression of wild-type enzyme. J. Neurochem. 2009;108:1009–1018. doi: 10.1111/j.1471-4159.2008.05839.x. doi:10.1111/j.1471-4159.2008.05839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witan H., Gorlovoy P., Kaya A.M., Koziollek-Drechsler I., Neumann H., Behl C., Clement A.M. Wild-type Cu/Zn superoxide dismutase (SOD1) does not facilitate, but impedes the formation of protein aggregates of amyotrophic lateral sclerosis causing mutant SOD1. Neurobiol. Dis. 2009;36:331–342. doi: 10.1016/j.nbd.2009.07.024. doi:10.1016/j.nbd.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Chattopadhyay M., Durazo A., Sohn S.H., Strong C.D., Gralla E.B., Whitelegge J.P., Valentine J.S. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl Acad. Sci. USA. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. doi:10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruijn L.I., Houseweart M.K., Kato S., Anderson K.L., Anderson S.D., Ohama E., Reaume A.G., Scott R.W., Cleveland D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. doi:10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 16.Wong P.C., Pardo C.A., Borchelt D.R., Lee M.K., Copeland N.G., Jenkins N.A., Sisodia S.S., Cleveland D.W., Price D.L. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. doi:10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 17.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.-X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. doi:10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Xu G., Borchelt D.R. High molecular weight complexes of mutant superoxide dismutase 1: age- dependent and tissue-specific accumulation. Neurobiol. Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. doi:10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Xu G., Li H., Gonzales V., Fromholt D., Karch C., Copeland N.G., Jenkins N.A., Borchelt D.R. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: {alpha}B-crystallin modulates aggregation. Hum. Mol. Genet. 2005;14:2335–2347. doi: 10.1093/hmg/ddi236. doi:10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Xu G., Slunt H.H., Gonzales V., Coonfield M., Fromholt D., Copeland N.G., Jenkins N.A., Borchelt D.R. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol. Dis. 2005;20:943–952. doi: 10.1016/j.nbd.2005.06.005. doi:10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Borchelt D.R., Guarnieri M., Wong P.C., Lee M.K., Slunt H.S., Xu Z.-S., Sisodia S.S., Price D.L., Cleveland D.W. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J. Biol. Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. doi:10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 22.Karch C.M., Borchelt D.R. An examination of alpha B-crystallin as a modifier of SOD1 aggregate pathology and toxicity in models of familial amyotrophic lateral sclerosis. J. Neurochem. 2010;113:1092–1100. doi: 10.1111/j.1471-4159.2010.06572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ezzi S.A., Urushitani M., Julien J.P. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J. Neurochem. 2007;102:170–178. doi: 10.1111/j.1471-4159.2007.04531.x. doi:10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 24.Witan H., Kern A., Koziollek-Drechsler I., Wade R., Behl C., Clement A.M. Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum. Mol. Genet. 2008;17:1373–1385. doi: 10.1093/hmg/ddn025. doi:10.1093/hmg/ddn025. [DOI] [PubMed] [Google Scholar]
- 25.Ray S.S., Nowak R.J., Strokovich K., Brown R.H., Jr, Walz T., Lansbury P.T., Jr. An intersubunit disulfide bond prevents in vitro aggregation of a superoxide dismutase-1 mutant linked to familial amytrophic lateral sclerosis. Biochemistry. 2004;43:4899–4905. doi: 10.1021/bi030246r. doi:10.1021/bi030246r. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Caruano-Yzermans A., Rodriguez A., Scheurmann J.P., Slunt H.H., Cao X., Gitlin J., Hart P.J., Borchelt D.R. Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J. Biol. Chem. 2007;282:345–352. doi: 10.1074/jbc.M604503200. doi:10.1074/jbc.M604503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobson C.M. Principles of protein folding, misfolding and aggregation. Semin. Cell Dev. Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. doi:10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Frieden C. Protein aggregation processes: in search of the mechanism. Protein Sci. 2007;16:2334–2344. doi: 10.1110/ps.073164107. doi:10.1110/ps.073164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen P.M., Nilsson P., Ala-Hurula V., Keränen M.-L., Tarvainen I., Haltia T., Nilsson L., Binzer M., Forsgren L., Marklund S.L. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat. Genet. 1995;10:61–66. doi: 10.1038/ng0595-61. doi:10.1038/ng0595-61. [DOI] [PubMed] [Google Scholar]
- 30.Robberecht W., Aguirre T., Van Den Bosch L., Tilkin P., Cassiman J.J., Matthijs G. D90A heterozygosity in the SOD1 gene is associated with familial and apparently sporadic amyotrophic lateral sclerosis. Neurology. 1996;47:1336–1339. doi: 10.1212/wnl.47.5.1336. [DOI] [PubMed] [Google Scholar]
- 31.Hayward C., Brock D.J., Minns R.A., Swingler R.J. Homozygosity for Asn86Ser mutation in the CuZn-superoxide dismutase gene produces a severe clinical phenotype in a juvenile onset case of familial amyotrophic lateral sclerosis. J. Med. Genet. 1998;35:174. doi: 10.1136/jmg.35.2.174. doi:10.1136/jmg.35.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato M., Aoki M., Ohta M., Nagai M., Ishizaki F., Nakamura S., Itoyama Y. Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci. Lett. 2001;312:165–168. doi: 10.1016/s0304-3940(01)02212-1. doi:10.1016/S0304-3940(01)02212-1. [DOI] [PubMed] [Google Scholar]
- 33.Borchelt D.R., Lee M.K., Slunt H.H., Guarnieri M., Xu Z.-S., Wong P.C., Brown R.H., Jr, Price D.L., Sisodia S.S., Cleveland D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl Acad. Sci. USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. doi:10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardo C.A., Xu Z., Borchelt D.R., Price D.L., Sisodia S.S., Cleveland D.W. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl Acad. Sci. USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. doi:10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karch C.M., Borchelt D.R. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. doi:10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zetterstrom P., Stewart H.G., Bergemalm D., Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Oliveberg M., Marklund S.L. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl Acad. Sci. USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. doi:10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.