Abstract
Spinal muscular atrophy (SMA) is a neurological disorder characterized by motor neuron degeneration and progressive muscle paralysis. The disease is caused by a reduction in survival of motor neuron (SMN) protein resulting from homozygous deletion of the SMN1 gene. SMN protein is also encoded by SMN2. However, splicing of SMN2 exon 7 is defective, and consequently, the majority of the transcripts produce a truncated, unstable protein. SMN protein itself has a role in splicing. The protein is required for the biogenesis of spliceosomal snRNPs, which are essential components of the splicing reaction. We now show that SMN protein abundance affects the splicing of SMN2 exon 7, revealing a feedback loop inSMN expression. The reduced SMN protein concentration observed in SMA samples and in cells depleted of SMN correlates with a decrease in cellular snRNA levels and a decrease in SMN2 exon 7 splicing. Furthermore, altering the relative abundance or activity of individual snRNPs has distinct effects on exon 7 splicing, demonstrating that core spliceosomal snRNPs influence SMN2 alternative splicing. Our results identify a feedback loop in SMN expression by which low SMN protein levels exacerbate SMN exon 7 skipping, leading to a further reduction in SMN protein. These results imply that a modest increase in SMN protein abundance may cause a disproportionately large increase in SMN expression, a finding that is important for assessing the therapeutic potential of SMA treatments and understanding disease pathogenesis.
INTRODUCTION
Proximal spinal muscular atrophy (SMA) is an autosomal recessive disorder characterized by progressive muscle weakness and paralysis, resulting from the specific degeneration of lower motor neurons in the spinal cord. SMA affects approximately one in 6000 live births and is a leading genetic cause of infant mortality (1,2).
SMA is caused by homozygous mutation or deletion of the survival of motor neuron 1 (SMN1) gene that codes for SMN protein (3). Two genes, SMN1 and SMN2, code for SMN protein in humans, and the copy number of SMN2 is a determinant of disease severity (4,5). SMN1 and SMN2 are nearly identical, and both are ubiquitously expressed. However, SMN2 produces less SMN protein than SMN1 due to a C-to-T change in exon 7 of SMN2 (6,7) that compromises exon 7 recognition by the splicing machinery (Fig. 1A). As a result of this nucleotide difference, the majority of SMN2 mRNA transcripts lack exon 7 and code for truncated SMN protein that is unstable and rapidly degraded (8). Thus, SMN2 cannot fully compensate for the loss of SMN1 in SMA because the amount of full-length mRNA and functional SMN protein produced from SMN2 is considerably lower than that from SMN1. For unknown reasons, the reduced abundance of SMN protein results in the specific degeneration of motor neurons (9).
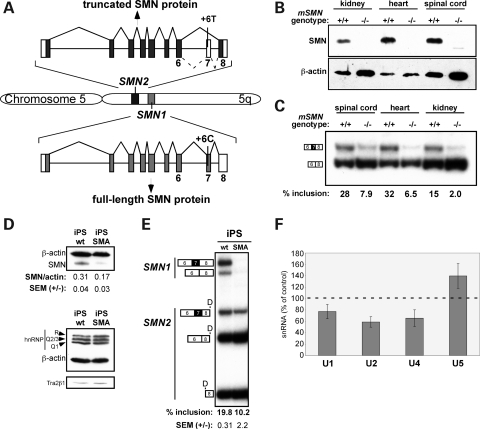
Figure 1.
Low SMN protein abundance correlates with low SMN2 exon 7 splicing in SMA. (A) SMN1 and SMN2 gene structures. Boxes indicate exons and horizontal lines are introns. Dominant splicing pattern is shown with solid diagonal lines, and minor alternative splicing is indicated with hatched lines. Shaded boxes indicate the coding region of the most abundant mRNA. (B) Immunoblot analysis of SMN protein in lysates from tissues of normal (Smn+/+) or SMA (Smn−/−) hSMN2 transgenic mice. Blotting of β-actin protein was used as a loading control. (C) Semi-quantitative radiolabeled RT–PCR analysis of human SMN2 mRNA from tissues of human SMN2 transgenic mice that are either homozygous (+/+) or (−/−) for murine Smn. Products were separated on a 6% native polyacrylamide gel. Transcripts that include or skip exon 7 are indicated, and results are quantitated and shown as % exon 7 inclusion [included/(included + skipped)×100]. (D) Immunoblot analysis of SMN, hnRNP Q/R, Tra2β1 (SFRS10) and β-actin (loading control) in cell lysates from human-induced pluripotent stem (iPS) cells derived from an SMA patient or an unaffected family member (wt) (42). The SMN/β-actin ratio is shown beneath the blot. (E) RT–PCR analysis of RNA from iPS cells. Reaction products were incubated with DdeI (D) to digest SMN2 RNA and identities of each transcript are indicated. The percent of SMN2 transcripts that include exon 7 is shown beneath the gel and reflect the average of three RT–PCR reactions from independent RNA isolations. (F) Real-time PCR analysis of snRNAs from iPS-WT and iPS-SMA cells. The percent change in snRNA quantity in SMA samples compared with WT samples was normalized to Rpl39 and graphed as the percentage of WT. Results are the average of three independent tests of the WT-iPS and SMA-iPS samples, which were repeated in triplicate for each reaction. Error bars represent the standard error of the means (SEM).
SMN protein is essential for the biogenesis of spliceosomal snRNPs U1, U2, U4, U4atac, U5, U11 and U12 (10–12). SMN, in a complex with other proteins (SMN complex), assembles the Sm proteins B/B', D1, D2, D3, E, F and G, onto the snRNA in the first steps of snRNP biogenesis. The decrease in SMN protein resulting from the loss of SMN1 protein alters the repertoire of snRNAs in the cell and leads to deficits of fully assembled snRNPs (13–15). In fact, a decrease in SMN protein has been reported to have an effect on a number of splicing events (13–15), although it is unclear whether splicing changes are a cause or consequence of SMA disease pathology (16). The effect that SMN protein abundance has on splicing could explain the motor neuron degeneration in SMA if splicing of a transcript that encodes a protein with critical motor neuron-specific function is altered by the decrease in SMN protein (17). Motor neurons may also have a higher demand for spliceosomal snRNPs, and thus changes in the abundance of snRNPs may result in more dramatic changes in splicing in motor neurons than in other cell types.
One question that has not yet been addressed is whether splicing of SMN2 exon 7 itself is sensitive to changes in snRNP levels. Splicing of this exon is under the control of a number of splicing factors. The C-to-T change in SMN2 that results in an increase in exon 7 skipping compared with SMN1 disrupts an exonic splicing enhancer (ESE) motif recognized by the SR protein SF2/ASF (18). The loss of this ESE weakens exon 7 recognition, making its splicing more sensitive to control by a number of splicing factors. For example, in the absence of this ESE, inhibitory interactions between splicing silencer elements and hnRNP A1 predominate and result in exon skipping (19,20). Additional proteins and sequence elements have also been identified that can influence SMN2 exon 7 splicing (21–32). For example, the Tra2 family of SR-like proteins (33) and the splicing factor hnRNP Q/R (25) influence exon 7 inclusion. RNA secondary structure is another determinant of exon 7 splicing (34,35).
One way that cis-acting sequences influence exon 7 splicing is by binding splicing factors that help to recruit spliceosomal snRNPs to exon 7 (22). The importance of efficient recruitment of snRNPs to the exon suggests that splicing of the exon may be sensitive to alterations in the snRNP abundance caused by a decrease in SMN protein levels in the cell. In addition, the 5' splice site of exon 7 is suboptimal, suggesting that U1 snRNP recruitment to the exon may be a limiting factor in the efficiency of exon 7 splicing (36,37). The importance of snRNP recruitment to exon 7 splice sites suggests that a change in the snRNP abundance, as is seen when SMN protein is reduced in SMA, may influence exon 7 splicing.
A correlation between SMN2 exon 7 splicing and SMN protein abundance is apparent in transgenic mice (25,38) as well as in human cells (8,39,40). However, this correlation has not been previously attributed to a feedback mechanism, and the direct effect of reduced SMN protein levels on exon 7 splicing has not been investigated. We now demonstrate that a decrease in SMN protein results in a decrease in SMN2 exon 7 splicing. We also show that splicing of SMN2 exon 7 is sensitive to changes in the relative abundance of spliceosomal snRNPs. Our results indicate that SMN expression is controlled, in part, by a feedback mechanism that allows homeostatic control of exon 7 splicing as a means to potentially regulate snRNP biogenesis and other functions of SMN. Feedback regulation of SMN expression has implications for understanding cellular defects in SMA. This mechanism also suggests that a small increase in SMN2 exon 7 splicing would result in a disproportionately high increase in SMN protein levels, a concept that will be important when assessing the potential therapeutic effect of compounds and other small molecules in SMA.
RESULTS
Low SMN protein levels correlate with decreased exon 7 splicing in SMA
Based on the premise that a decrease in SMN protein levels in cells results in changes in the relative snRNP abundance, which can affect alternative splicing in general (13–15), we analyzed RNA from an SMA patient and a mouse model of SMA to determine whether splicing of SMN2 exon 7 is affected by a decrease in the SMN protein abundance in vivo. We first compared SMN protein abundance in mice transgenic for human SMN2 either with both copies of the mouse Smn gene (hSMN2; Smn+/+) or with a homozygous deletion of the mouse Smn gene (hSMN2; Smn −/−) at post-natal day 1. Transgenic mice with homozygous deletion of the mouse SMN gene are referred to as SMA mice as they exhibit an SMA phenotype (41). SMN protein was not detectable in the SMA mouse tissues analyzed, indicating that very little SMN protein is produced from the hSMN2 gene (Fig. 1B). A protein with lower mobility than full-length SMN was detected in the spinal cord of SMA mice (Fig. 1B). This product may be truncated SMN protein arising from SMN2 mRNA lacking exon 7. The analysis of exon 7 splicing in the human SMN2 gene RNA transcript revealed that <10% of the hSMN2 transcripts include exon 7 (Fig. 1C). It has been shown previously that the protein product encoded by the SMN mRNA transcripts lacking exon 7 is unstable and undetectable by immunoblot in mice (8), which is consistent with the lack of SMN protein detected in the SMA tissues (Fig. 1B). In contrast, hSMN2 transgenic mice that have an intact mouse SMN gene (hSMN2, mSMN+/+) produce SMN protein from the endogenous gene and exhibit a 4–8-fold increase in SMN2 transcripts that include exon 7 compared with the mSMN −/− samples (Fig. 1C). These results demonstrate a correlation between SMN2 exon 7 splicing and SMN protein levels.
To further test whether low SMN protein abundance correlates with a decrease in SMN2 exon 7 splicing, we analyzed SMN protein and exon 7 splicing in induced pluripotent stem (iPS) cells generated from an SMA patient (iPS-SMA) or from an unaffected family member (iPS-WT) (42). SMN1 and SMN2 spliced RNA products were analyzed by reverse transcription followed by polymerase chain reaction (PCR) amplification. SMN1 and SMN2 products can be distinguished from one another by digestion with the restriction enzyme DdeI, which has a unique site in exon 8 of SMN2 that is not present in SMN1. Following digestion, SMN2 spliced products are smaller than SMN1 as visualized by gel electrophoresis. We found that in the iPS-SMA cells, SMN protein levels are reduced by 45% compared with WT (Fig. 1D), reflecting the loss of the SMN1 gene in these cells. The low SMN protein levels in the iPS-SMA cells corresponded with a 48% reduction in the percent of SMN2 transcripts that include exon 7 splicing in iPS-SMA cells compared with WT cells (Fig. 1E). This result further demonstrates the correlation between SMN protein abundance and exon 7 splicing, suggesting a possible auto-regulatory feedback mechanism.
Feedback regulation of exon 7 splicing could reflect either a direct or an indirect effect of SMN protein on SMN2 exon 7 splicing. We also analyzed by immunoblot whether depletion of SMN protein in cells causes a change in known regulators of SMN2 exon 7 splicing. We found that neither hnRNP Q/R nor Tra2β1 abundance changed between the WT and SMA iPS cells, suggesting that the reduction in SMN protein levels does not cause exon 7 skipping indirectly due to the reduction of these factors, which have been previously shown to affect exon 7 splicing (25,33) (Fig. 1D).
Because a major function of SMN protein involves assembly of Sm proteins on snRNAs (12), which stabilizes the snRNAs (14), we next analyzed snRNA levels in the iPS cells to determine whether reduced SMN protein abundance in iPS-SMA cells correlates with a reduction in snRNAs. A reduction in snRNAs could account for alterations in SMN2 exon 7 splicing that is observed when SMN protein abundance is low. Exon 7 is spliced via the major spliceosomal pathway involving U1, U2, U4, U5 and U6 snRNPs. We focused on U1, U2, U4 and U5, because SMN assembles Sm proteins onto these, whereas the U6 snRNA assembly involves a different pathway (10). We measured snRNA levels by reverse transcription followed by real-time PCR. We observed a 23, 41and 34% reduction in U1, U2and U4 snRNA abundance, respectively, in iPS-SMA relative to iPS-WT when normalized to a control transcript (Fig. 1F), whereas U5 snRNA levels were slightly higher in iPS-SMA compared with iPS-WT cells. Our results suggest that the decrease in exon 7 splicing triggered by low SMN protein levels may be a result of lowered snRNA abundance or a change in the relative abundance of the snRNAs (13,14). Overall, our results reveal that iPS cells derived from SMA patients have a deficit in the abundance of some U snRNA species as well as lower SMN2 exon 7 splicing.
Cellular depletion of SMN leads to an increase in SMN2 exon 7 skipping
To test directly whether SMN protein abundance affects SMN2 exon 7 splicing, we assayed SMN2 splicing in HEK-293T cells depleted of SMN using an siRNA that targets both SMN1 and SMN2. RNAi against SMN resulted in a 64% reduction in SMN protein (Fig. 2A). To verify that any effect of the RNAi-mediated knockdown of SMN is a direct result of lowered SMN protein levels, we introduced an RNAi rescue plasmid expressing full-length SMN2 cDNA with silent point mutations that render its mRNA insensitive to RNAi. Expression of the rescue SMN plasmid replenishes SMN protein in the siRNA-treated cells in a dose-dependent manner (Fig. 2A).
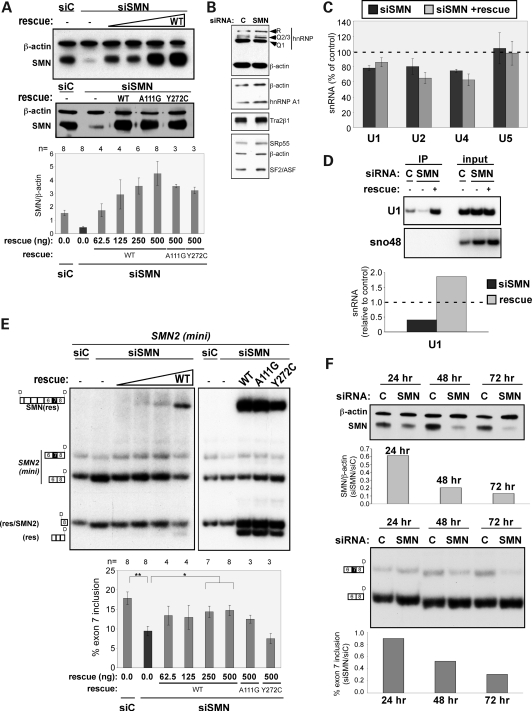
Figure 2.
SMN protein reduction causes an increase in SMN2 exon 7 skipping. (A) Immunoblot analysis of SMN protein from HEK-293T cells transfected with a minigene expressing SMN2 and either a scrambled control (siC) siRNA or an siRNA targeted to SMN1/2 (siSMN). Rescue refers to samples from cells transfected with different quantities of a plasmid expressing SMN2 cDNA with silent point mutations in the siRNA target sequence. Blots were probed with SMN or β-actin (loading control)-specific primary antibodies followed by a fluorescent and HRP-conjugated secondary antibody. Bands were quantitated by phosphorimage analysis. Graph shows results as the ratio of SMN to β-actin. Error bars represent SEM. The number of independent experimental measurements (n) is shown above the graph. For all data points, P < 0.0005 relative to siSMN as determined by the Student's t-test. (B) Immunoblots probed with antibodies against hnRNP R/Q, hnRNP A1, SFRS10 (Tra2β1), SRp55, SF2/ASF and β-actin. (C) Quantitation of snRNAs by real-time PCR. The snRNA levels of the SMN RNAi cells are normalized to Rpl39 and plotted as percentage of control (siC). Error bars represent SEM, n = 4 independent experiments for the siSMN/siC comparison and n = 3 for siSMN + rescue/siC. (D) PCR analysis of 3' end-linkered U1 snRNAs immunoprecipitated from cell lysates using an anti-Sm antibody. U1 snRNA levels are normalized to snRNA in the input lysate. SnoRNA48 was used as a control. (E) RT–PCR analysis of SMN2 minigene transcript splicing in cells treated as in (A). Reaction products were digested with DdeI as in Figure 1. Results are represented graphically as percent exon 7 inclusion. Error bars represent SEM; n is shown above the graph. *P < 0.05; **P < 0.005. (F) Immunoblot (top) and RT–PCR (bottom) analysis of SMN protein expression and exon 7 splicing as a function of time following siRNA-mediated depletion of SMN protein from cells. Quantitation was performed as in (A) and (C) above.
We also analyzed by immunoblot analysis the effect of SMN depletion on the expression of a number of known regulators of exon 7 splicing, including hnRNP Q/R and A1, Tra2β1, SF2/ASF and SRp55 (Fig. 2B) (25,43,44). There was no change in the abundance of these proteins in SMN-depleted cells compared with control cells.
Analysis of snRNA levels in SMN-depleted cells confirmed that the snRNA levels are affected by a decrease in SMN protein. Real-time PCR indicated a reduction in U1, U2 and U4, but not in U5 snRNA levels in cells with reduced SMN protein (Fig. 2C). Expression of SMN protein in siSMN-treated cells partially restored U1 snRNA, but not U2 or U4 snRNA levels (Fig. 2C). Overall, there was a change in the snRNA abundance in cells with reduced SMN protein, with some snRNAs more affected than others. These changes result in an overall alteration in the relative abundance of individual snRNPs, which could influence the dynamics of splice site recognition.
The relatively subtle effect that restoration of SMN protein expression had on snRNA levels led us to test the degree to which the abundance of mature snRNPs, which require SMN for assembly, was affected by SMN protein depletion and rescue in this system. We focused on U1 snRNP for the purpose of comparing the two snRNA assays. Mature snRNPs were immunoprecipitated from lysates of treated cells using an Sm protein-specific antibody. The immunoprecipitated snRNAs were subsequently linkered at the 3' end and reverse-transcribed using a linker-specific primer. PCR was carried out with an snRNA-specific primer and a linker-specific primer. The amount of U1 snRNA in the starting cell lysate was measured as an input control. SnoRNA48 was also measured to normalize input RNA concentration and to demonstrate the specificity of the immunoprecipitation for Sm-bound snRNAs. In this assay, U1 snRNP abundance was reduced in cells depleted of SMN and restored in cells expressing the SMN rescue protein (Fig. 2D). These results suggest that although total snRNA abundance is altered by the amount of cellular SMN protein, the effect on mature snRNPs is more dramatic and that U1 snRNP abundance is restored by expression of the SMN rescue protein in SMN-depleted cells.
We next analyzed the effect of SMN depletion on exon 7 splicing. Depletion of endogenous SMN1 and 2 from the cells resulted in a reduction in mRNA and protein levels, precluding analysis of endogenous SMN2 exon 7 splicing in these experiments because it is eliminated in the siSMN-treated cells. Instead, a minigene plasmid expressing the 3' comprised exons 6, 7 and 8 and the intervening introns and recapitulates SMN2 exon 7 alternative splicing in a manner similar to the endogenous transcript (20). Reverse transcription (RT)–PCR analysis of SMN2 exon 7 splicing from the minigene showed a nearly 48% reduction in SMN2 mRNA containing exon 7 in the cells depleted of SMN (Fig. 2E). This effect was specific to the reduction in SMN protein levels as evidenced by the restoration of exon 7 splicing when the SMN2 rescue plasmid (WT) is expressed to restore SMN protein levels (Fig. 2E).
We also rescued SMN knockdown cells with an SMN gene that is insensitive to RNAi and codes for the SMN mutant proteins A111G, which has a moderately decreased affinity for Sm proteins, and Y272C, which has more severe defects in the Sm core assembly (Fig. 2A) (2,45–47). The SMN/A111G protein partially rescued the splicing defect, whereas the SMN/Y272C protein was completely inactive (Fig. 2E). These results are consistent with the documented activity of these mutant proteins and support the idea that SMN2 exon 7 splicing is affected by SMN protein abundance and activity related to snRNP assembly and abundance. Overall, our results strongly support a role for SMN protein in SMN exon 7 splicing by altering the abundance of a subset of snRNPs.
To test the dynamics of the feedback loop, we performed a time-course experiment in which cells were treated with siSMN or the control siRNA, and exon 7 splicing of SMN2 minigene transcripts was measured at different time points following SMN depletion. A correlation between SMN protein levels and SMN2 exon 7 splicing was observed with maximum exon 7 skipping occurring when SMN protein levels are lowest at 72 h post-transfection (Fig. 2F). These results demonstrate that SMN protein abundance correlates with the degree of exon 7 splicing.
snRNP abundance influences exon 7 splicing
The differences in snRNA levels in SMA compared with WT iPS cells and in SMN knockdown cells compared with control cells suggest that the abundance of some snRNA species may be more sensitive to a decrease in SMN protein abundance than others. This differential change in snRNA species could explain the effect of SMN protein abundance on SMN2 exon 7 splicing if splicing is regulated by changes in the relative abundance of snRNPs. To directly test the idea that changes in individual snRNPs influence exon 7 splicing, we used a cell-free in vitro splicing assay. In this assay, in vitro transcribed SMN1 or SMN2 pre-mRNAs comprised exons 6 and 7 and a portion of exon 8, and the intervening introns (18) were spliced in the HeLa cell nuclear extract. U1, U2, U4 and U5 snRNAs were targeted by oligonucleotide-directed RNase H cleavage, a method commonly enlisted to digest snRNAs in order to test their requirement in splicing (48–52). Oligonucleotides were tested at different concentrations that were optimized for each snRNA in order to assess a dose-dependent effect of snRNA depletion on splicing.
RNase H cleavage of each species of snRNA resulted in strikingly different effects on exon 7 splicing. At the lowest concentration of U1 snRNA-targeting oligonucleotides, there was a greater than 50% reduction in spliced products that include exon 7, whereas transcripts lacking exon 7 did not change dramatically relative to reactions with untreated extracts (Fig. 3A). This change in alternative splicing dynamics is reflected in the dramatic decrease in the percent exon 7 inclusion (Fig. 3A). In contrast, targeting U2 snRNA resulted in a nearly 50% reduction in exon 7 skipping and an increase in exon 7 inclusion at the lowest concentration of oligonucleotide (Fig. 3B). This results in an overall increase in exon 7 inclusion, suggesting that when U2 snRNA is limiting, exon 7 splicing out-competes exon 7 skipping (Fig. 3B). A similar effect was observed when U4- and U5-targeted oligonucleotides were tested (Fig. 3C and D).
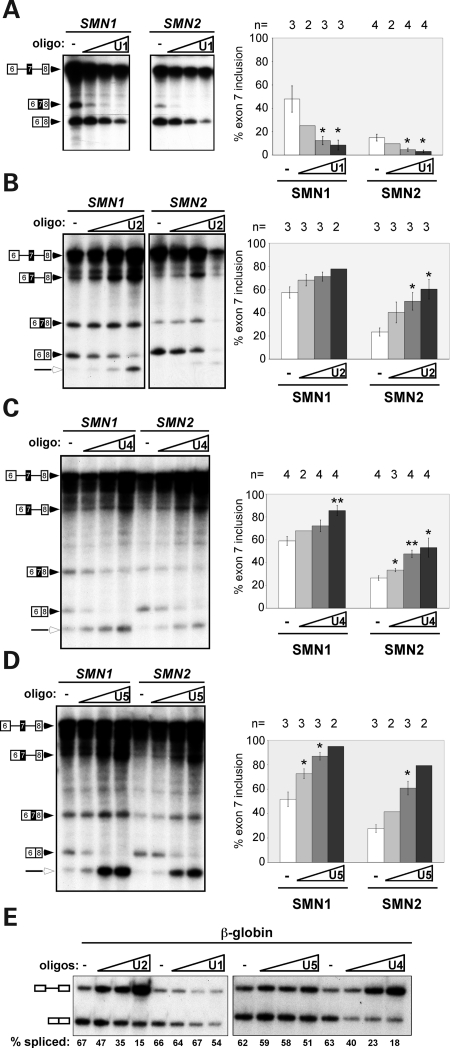
Figure 3.
snRNAs differentially affect exon 7 splicing in vitro. In vitro splicing analysis of SMN1 and SMN2 transcripts in HeLa cell nuclear extracts. Extracts were untreated (−) or treated with an increasing concentration of antisense oligonucleotides (oligo) targeting (A) U1 (0.3125, 0.625, 1.25 μm), (B) U2 (1.25, 2.5, 5 μm), (C) U4 (0.156, 0.3125, 0.625 μm) and (D) U5 (1.25, 2.5, 5 μm) snRNAs for RNase H-mediated cleavage. Unspliced pre-mRNA and spliced products are indicated. Open arrowheads represent debranched intron lariats. Quantitation of in vitro splicing, shown as the percent exon 7 inclusion [included/(included + pre-mRNA)×100], is graphed to the right in each panel. Error bars represent ± SEM with n indicated above the bars, *P < 0.05, **P < 0.005 relative to untreated (−). Bars with no error represent the average of two independent experiments. (E) In vitro splicing of β-globin transcripts. Splicing reactions were carried out as in (A).
Splicing of both SMN1 and SMN2 exon 7 was similarly responsive to the RNase H depletion of individual snRNAs, suggesting that the effect that low snRNA levels have on splicing is not dependent on the C-to-T difference in exon 7 between SMN1 and SMN2. To demonstrate that the RNase H-depleted extracts were effectively targeting snRNAs, we tested splicing of a constitutively spliced β-globin transcript. The RNase H depletion caused a dose-dependent inhibition of β-globin splicing (Fig. 3E), demonstrating a progressive block to constitutive splicing after degradation of snRNAs.
Depletion of snRNP proteins reduces SMN2 exon 7 splicing
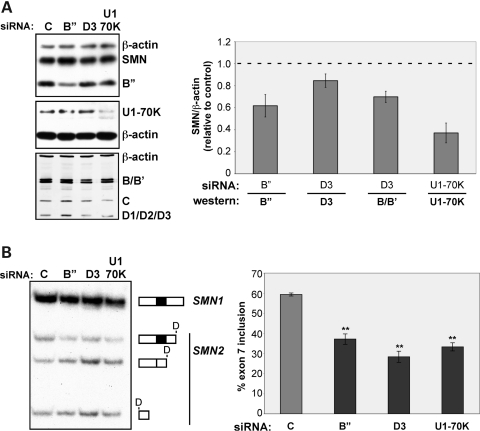
To further test whether altering snRNP levels can affect exon 7 splicing, we depleted snRNPs from cells using siRNAs directed against several protein components of the mature snRNP. RNAi-mediated knockdown of either the U2 snRNP-associated protein, B" (SNRPB2) by 51%, or the U1 snRNP-associated protein, U170K (SNRNP70) by 63%, led to a 37% and 44% decrease in SMN2 exon 7 splicing, respectively (Fig. 4A and B). Similarly, a 15% knockdown of the Sm protein D3 (SNRPD3), a direct target of SMN in the assembly onto U1, U2, U4 and U5 snRNAs (53), resulted in a 52% reduction in exon 7 splicing. Cellular depletion of D3 also resulted in a 30% reduction in another Sm protein, B/B' (SNRPB). This effect likely reflects the instability of Sm proteins when not complexed together (54). Together, these results demonstrate that SMN2 exon 7 splicing is sensitive to changes in the abundance of snRNP components.
Figure 4.
Reduction of snRNP proteins causes SMN2 exon 7 skipping. (A) Immunoblot analysis of HEK-293T cells transfected with either a scrambled control (C) siRNA or an siRNA targeted to U2-B", Sm-D3 or U1-70K. Blots were probed with primary antibodies to indicated proteins. Fluorescent and HRP-conjugated secondary antibodies were used for detection, bands were quantitated by phosphorimage analysis and results are graphed as the ratio of indicated protein to β-actin signal normalized to control samples. (B) RT–PCR analysis of SMN1 and SMN2 endogenous transcripts in cells treated as in (A). Reaction products were digested with DdeI. Results are represented graphically as the percent exon 7 inclusion. For both (A) and (B), error bars represent the SEM, n = 3, **P < 0.005.
Sequestering U1 snRNP in cells reduces exon 7 splicing
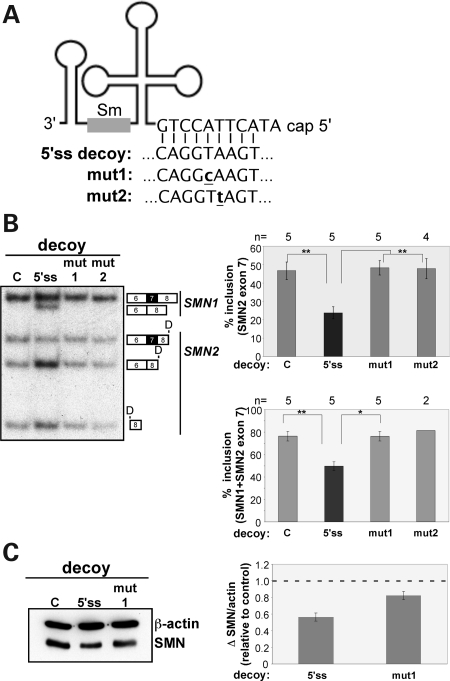
Our results indicate that U1 snRNA abundance goes down in cells with low SMN protein levels and that a decrease in U1 snRNPs dramatically reduces exon 7 splicing. We further explored the role of U1 snRNP abundance in exon 7 splicing through the use of RNA decoys. The expression of RNA decoys with sequence similarity to a 5' splice site has been shown to sequester U1 snRNP and to alter the splicing of exon 7 from SMN2 minigene-expressed pre-mRNAs (37) (Fig. 5A). This system offers a means to independently test the effect of lowered U1 snRNP levels on the splicing of endogenous SMN2 exon 7 to determine whether the modulation of U1 snRNP activity alters splicing in a manner similar to that seen with the SMN or snRNP component knockdown experiments.
Figure 5.
Overexpression of U1 snRNP binding sites causes SMN2 exon 7 skipping. (A) Diagram of U1 snRNA with 5' sequence base-pairing to the target sequence of the U1 5' splice site (5'ss) decoy or U1 mutant decoys (mut1, mut2) with one nucleotide change in the U1-recognition sequence (bold and underlined). (B) RT–PCR analysis of endogenous SMN1 and SMN2 transcripts from HEK-293T cells transfected with plasmids expressing 5'ss decoys (5'ss) or mutated decoys. Reactions were digested with DdeI and separated on a 6% native polyacrylamide gel. Bands were quantitated and graphed as the percent exon 7 inclusion [included/(included + skipped)×100] (top); ±SEM, or the change in total full-length SMN (SMN1 + SMN2 transcripts including exon 7) (bottom); ±SEM, *P < 0.05, **P < 0.01. (C) Immunoblot analysis of SMN protein levels in cells transfected with decoy plasmids. Quantitation is shown to the right with the ratio of SMN to β-actin signal normalized to mock transfected (C) samples; ±SEM, n = 3.
Expression of the 5'ss decoy in HEK-293T cells resulted in a 50% reduction in endogenous SMN2 exon 7 splicing (Fig. 5B). SMN1 exon 7 splicing was also reduced. Expression of decoys with point mutations in the 5' splice site sequence did not affect exon 7 splicing (Fig. 5B). The 5' splice site decoys caused a nearly 50% overall reduction in full-length exon 7, including transcripts from both SMN1 and SMN2. This reduction in full-length transcripts leads to a 42% reduction in SMN protein levels (Fig. 5C). These results demonstrate the specific sensitivity of exon 7 splicing to alterations in the U1 snRNP abundance and also show that a reduction in full-length SMN1/2 transcripts leads to a reduction in SMN protein. Our findings further link a modulation of snRNP abundance to the control of SMN2 exon 7 splicing and SMN protein expression.
DISCUSSION
A major goal in SMA research has been to identify approaches to improve expression of SMN protein from SMN2 for the treatment of the disease. Increasing SMN2 exon 7 splicing has been studied intensely as a means to elevate full-length SMN protein levels in SMA. One key question is how much of an increase in SMN2 exon 7 splicing is required for therapeutic value. Because SMN protein itself functions in the pre-mRNA splicing pathway, it is important to understand how the protein may influence splicing of its own pre-mRNA. We now demonstrate that the abundance of SMN protein determines, in part, the outcome of SMN2 alternative splicing. The discovery of a feedback loop in SMN protein expression has important implications for SMA by suggesting that treatment strategies that lead to modest increases in SMN protein may have significant therapeutic value for the disease.
The SMN feedback loop likely involves the key function of SMN in the snRNP assembly. We (Figs 1 and 2) and others (13,14) have found that some spliceosomal snRNA species decrease, following a reduction in the SMN protein abundance. This change in snRNAs consequently lowers snRNP abundance and correlates with changes in alternative splicing of a number of gene transcripts, presumably due to a change in the relative concentration of snRNPs (14). It is not clear how changes in the relative abundance of snRNPs lead to changes in alternative splicing. Using an in vitro splicing assay, we tested directly the possibility that subtle changes in the relative abundance of the snRNPs can have effects on alternative splicing of SMN2 exon 7 (Fig. 3). We found that altering the abundance of individual snRNPs in vitro had dramatically differing effects on exon 7 splicing, demonstrating that shifts in the balance of snRNPs can regulate alternative splicing (Fig. 3). Our results suggest that a change in the relative amount of individual snRNPs alters SMN2 exon 7 splicing, which, in turn, affects SMN protein levels which feeds back to further disrupt snRNP abundance.
SMN2 exon 7 splicing is particularly sensitive to U1 snRNP levels. We present data showing that U1 snRNA is reduced when SMN protein abundance is low (Figs 1 and 2) and that exon 7 splicing decreases following a decrease in U1 snRNP activity or abundance (Figs 3–5). U1 snRNP recognizes the 5' splice site through direct base-pairing interactions between the pre-mRNA and the snRNA (55). U1 snRNP binding is an early determinant of splice site selection (56,57). Thus, the 5' splice site sequence, and likely also the abundance of U1 snRNP, plays an important role in the initial recognition of the site. 5' splice sites with weak base-pairing potential to U1 snRNA are not recognized as efficiently as those with high base-pairing potential (58). The efficiency of 5' splice site selection is important when 5' splice sites are in competition with each other in alternatively spliced exons. For example, in the case of SMN2, the 5' splice site of exon 7 is an important determinant of exon inclusion (36,37). Exon skipping will occur if exon 6 splicing to exon 8 occurs before exon 7 can splice to exon 8. Improvement of the base-pairing potential of its 5' splice site to U1 improves exon 7 splicing, likely due, in part, to its improved ability to compete with the 5' splice site of exon 6 for splicing to exon 8. We observed that a decrease in U1 snRNP abundance in vitro results in a decrease in exon 7 inclusion relative to exon skipping (Fig. 3). This may be due to the weak exon 7 5' splice site, which is further weakened under conditions of low U1 snRNP. This site may also not be strong enough to promote exon definition across exon 7 for enhancement of the 3' splice site of exon 7, which has also been demonstrated to be a relatively weak splice site (22,59). Limiting U1 snRNP may further weaken exon 7 definition, thereby lowering the occurrence of splicing from the 5' splice site of exon 6 to the 3' splice site of exon 7. In this case, splicing from exon 6 to the 3' splice site of exon 8 may be selected over the exon 7 3' splice site, resulting in exon 7 skipping.
Interestingly, targeted reduction of individual snRNAs had differential effects on exon 7 splicing. Unlike U1 snRNA depletion, exon 7 inclusion was much more resistant to the depletion of U2, U4 and U5 snRNA species in vitro compared with skipping (Fig. 3). These results suggest that exon 7 skipping and inclusion are differentially affected by alterations in the relative abundance of individual snRNPs. This phenomenon, in which the relative abundance of individual snRNPs dictates exon 7 splicing, could result in differences in the relative amount of SMN2 exon 7 inclusion in different cell types and at different stages of development. In the case of SMA, in which SMN1 is lost, this feedback regulation of SMN2 splicing could be crucial for cell survival. It is possible that SMN2 exon 7 splicing in motor neurons is especially sensitive to alteration in snRNPs, further depleting SMN protein and contributing to motor neuron degeneration in SMA. Indeed, snRNP levels and the abundance and activity of SMN protein fluctuate in the spinal cord throughout early development (60).
Modulation of individual snRNAs did not always have the expected outcome on SMN2 exon 7 splicing. The relative resistance of exon 7 inclusion to U2 snRNA depletion in vitro (Fig. 3), for example, was surprising. U2 snRNP binds to the 3'ss and is a determinant of 3'ss selection. U2 snRNP binding to the 3' splice site of exon 7 is impaired in SMN2 compared with SMN1 (22,59). Based on these results, it might be predicted that the depletion of U2 snRNP would impair SMN2 exon 7 3' splice site recognition and lead to an increase in skipping. Our results, however, are consistent with our previous finding that depletion of U2AF65 or PUF60, which recruit U2 snRNP to the 3' splice site, results in an increase in exon 7 inclusion (28). Modulation of 3' splice site recognition may weaken the use of the distal exon 8 3' splice site and thereby improve the competitiveness of the exon 7 3' splice site for splicing. Contrary to this interpretation, depletion of the U2 snRNP protein SNRP B" resulted in an increase in exon 7 skipping (Fig. 4). This difference may be due to secondary effects of U2 snRNP depletion in cells or may indicate a functional difference in U2 snRNP when the B" protein is limiting.
The effect of snRNP abundance on splicing outcome is more complex in the context of alterations of multiple snRNP species, as we observed in the iPS cells (Fig. 1) and SMN knockdown experiments (Fig. 2). Our in vitro results (Fig. 3) indicate that a decrease in U2, U4 or U5 snRNP abundance causes an increase in exon 7 inclusion. However, in cells, a decrease in SMN protein always correlates with a decrease in exon 7 inclusion (Figs 1 and 2). It is possible that early recognition of the 5'ss by U1 snRNP is the determining step in splice site selection, and thus U1 snRNP abundance has a dominant role in exon 7 splicing.
The SMN feedback loop is likely an important regulator of SMN expression in SMA where SMN1 is mutated or deleted and thus the equilibrium of the feedback loop is disrupted and splicing of SMN2 exon 7 is reduced. The ability of a feedback loop to potentially impact endogenous SMN1 and SMN2 splicing and SMN protein production is demonstrated using the in vitro splicing assay and in experiments with the U1 snRNA decoy where both SMN1 and SMN2 exon 7 splicing were affected by alterations in the snRNP abundance (Figs 3 and 5). However, in cells, splicing of endogenous or minigene SMN1 exon 7 was not affected by changes in the SMN protein abundance. This insensitivity may be due to the high efficiency of SMN1 exon 7 splicing in combination with the relatively modest reduction in SMN protein and snRNAs. Our results suggest that the feedback loop could play a regulatory role when SMN1 is present in a non-diseased state in situations.
Regulation of exon 7 splicing could reflect an indirect effect of SMN protein on SMN2 exon 7 splicing. Reduction in SMN protein levels has been reported to result in changes in alternative splicing of a large number of transcripts (14). Alternative splicing changes are likely to have an impact on the abundance or activity of the resulting proteins. A number of protein factors have been described that alter the splicing of SMN2 exon 7. The reduction in SMN protein may cause a change in the splicing of one of these regulators and thus indirectly alter SMN2 exon 7 splicing. Microarray experiments examining global changes in splicing in SMA mice have not revealed changes in alternative splicing of any known regulators of splicing (14,16). To address this possibility more directly, we tested whether depletion of SMN protein in cells causes a change in known regulators of SMN2 exon 7 splicing and did not observe quantitative changes in the abundance of a number of these regulatory proteins including SF2/ASF, hnRNPA1, hnRNP Q/R or Tra2β1 (Figs 1D and 2B) (19,20,25,33). However, we cannot rule out the possibility that the effect of lowering the SMN protein levels on exon 7 inclusion results in part from alterations in the abundance or activity of other effectors of exon 7 splicing. Nonetheless, our demonstration that alterations in snRNP levels can alter exon 7 splicing in vitro suggests that the decrease in exon 7 splicing upon reduction of SMN protein levels is due, at least in part, to change in snRNP levels in the cell. We also provide evidence that changes in the relative abundance of core spliceosomal snRNPs can regulate alternative splicing.
Our results are a first demonstration of feedback regulation whereby an alteration in SMN protein levels controls expression of the protein itself by affecting alternative splicing of its pre-mRNA transcripts. These finding lay the groundwork for future studies to understand the degree to which an initially small increase in exon 7 splicing can result in a disproportionately larger increase in SMN protein levels. From a more broad perspective, our results indicate that the relative abundance of individual snRNPs can regulate alternative splicing.
MATERIALS AND METHODS
Plasmids and constructs
The pCI-SMN2cDNA rescue plasmid was constructed by reverse transcription of HEK-293T total cellular RNA with oligodT primers using Superscript III cDNA synthesis kit (Invitrogen, Carlsbad, CA). SMN2 full-length cDNA was amplified by PCR using the primers SMNex1XhoI and SMNex8NotI. The PCR product and the plasmid pCI (Promega, Madison, WI) were digested with XhoI and NotI and ligated together using T4 DNA ligase (New England Biolabs, Ipswich, MA). The resulting plasmid was used as a template for mutagenesis using QuikChange Lightning site-directed mutagenesis kit (Stratagene, Santa Clara, CA) and the primers SMNmisF and SMNmisR, according to the manufacturer's instructions.
Oligonucleotides
Primer and RNAi sequences are provided in Supplementary Material, Table S1.
Cell-free in vitro splicing
Plasmids pCI-SMN1 and pCI-SMN2 (61) were used as templates in PCR reactions using the forward primer, T7SMNex6, that is specific to the 5' end of SMN exon 6 with a T7 promoter sequence at the 3' end and a reverse primer, SMNex8-75R+5', specific to SMN exon 8 (20). PCR products were used as templates for in vitro transcription with T7 RNA polymerase (Promega) to make SMN1 and SMN2 E678 transcripts. β-globin transcripts were made using the template, pSP64-HβΔ6 linearized with BamHI and transcribed with SP6 RNA polymerase (48).
Transcription reactions were carried out in the presence of 32P-UTP and 7Me-GpppG cap analog (NEB) to make pre-mRNAs for in vitro splicing analysis. RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE). RNase H degradation of snRNAs in HeLa nuclear extract dialyzed in buffer D [20 mm HEPES-KOH, pH 8; 100 mm KCl; 0.2 mm EDTA; 20% (v/v) glycerol] was achieved by incubating extract with antisense oligonucleotides targeting the snRNA for 15 min at 30°C (48). Treated extracts were used immediately in a 10 μl splicing reaction in which 10 fmol of RNA was incubated with 3 μl of nuclear extract, 1.3% (w/v) polyvinyl alcohol (PVA), 0.5 mm ATP, 20 mm creatine phosphate, 1.6 mm MgCl2 and 30% (v/v) buffer D at 30°C for 3 h (62).
Cell culture, transfections and RNAi
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For RNAi experiments, 1.5 × 105 cells were plated into a six-well dish 24 h prior to treatment. DsiRNAs were synthesized by Integrated DNA Technologies, Inc. (IDT, Coralville, IA). The sequences of the siRNA duplexes are listed in Supplementary Material, Table S1. The siC sequence was the Dicector DS scrambled negative control duplex (IDT). For SMN knockdown experiments, cells were transfected with 1 μg of SMN2 or SMN1 minigene reporter (20) and 20 nm siRNA duplex using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. For rescue of SMN knockdown, the pCI-SMN2 cDNA rescue plasmid was also transfected. After 24 h, cells were split 1:2. Cells were transfected with an additional 20 nm of siRNA 48 h after the initial treatment and grown for an additional 48 h at which time total RNA and protein were collected. For the time-course experiment, cells were treated similarly except that cells were treated with 20 nm siRNA initially and then every 48 h throughout the time-course. The SMN2 minigene plasmid was transfected at time 0 h for the 24 and 48 h time points and at 48 h for the 72 h time point. For RNAi-mediated depletion of U170K, snRNP B" and SmB, cells were treated with 20 nm siRNA and total RNA and protein were isolated after 72 h. All RNA was collected from cells using Trizol (Invitrogen). Protein samples were prepared by lysing cells in Laemmli buffer and by heating at 99°C for 10 min.
U1 decoy plasmids were transfected into HEK-293T cells using Optifect (Invitrogen), according to the manufacturer's protocol. Total RNA was collected using Trizol (Invitrogen) 48 h post-transfection.
iPS cells were cultured and collected as neurospheres, as described previously (42).
Immunoblot analysis
Protein samples were separated by sodium dodecyl sulfate (SDS)–PAGE and transferred to Immobilon-FL membrane (Millipore, Billerica, MA). Blots were probed with mouse monoclonal antibodies specific for mouse and human SMN (BD Biosciences), β-actin (Sigma, St Louis, MO), hnRNP-Q (Sigma), Sm proteins (mAb Y12), U1-70K (anti-RNP), U2-B" snRNP protein (mAb 4G3) (63), SF2/ASF (mAb96), hnRNP A1 (mAb A1/55) (64) and SRp55 (gifts from Adrian Krainer) or rabbit polyclonal antibody against SFRS10/Tra2β (Sigma), followed by Alexafluor 594-conjugated anti-mouse or anti-rabbit secondary antibody (Invitrogen) or horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit secondary antibody. Detection and quantitation of the signal was performed using a Typhoon 9400 Variable Mode Imager (GE Healthcare, Waukesha, WI) and ImageQuant T software for fluorescently detected blots or with Lumi-Light Western Blotting Substrate (Roche Diagnostics, Indianapolis, IN) for HRP-labeled blots.
RNA analysis
RNA was collected using Trizol reagent (Invitrogen). Reverse transcription was performed using Superscript III cDNA synthesis kit (Invitrogen) with oligo dT primers except for snRNA quantitation experiments in which RNA was reverse-transcribed using random primers. PCR with GoTaq polymerase (Promega) was carried out for 25 amplification cycles in reactions containing (α-32P)-dCTP. Exon 7 splicing of hSMN2 RNA from transgenic mouse tissues was analyzed using human-specific primers E4-33to55-F and E8-15to36-R (30). Endogenous SMN1 and SMN2 RNA transcripts from human cells were amplified using primers SMNex6Xho and SMNex8NotI. SMN2 transcripts from pCI-SMN2cDNA rescue and pCI-SMN1 and SMN2 minigenes were amplified using pCI-FwdB and pCI-Rev. Following PCR of endogenous SMN1 and SMN2, reactions were treated with DdeI for 1h, which digests at a unique site within SMN2 exon 8 that is not present in SMN1. Products were separated on 6% native polyacrylamide gels. Quantitation is based on phosphorimage analysis (Typhoon 9400; GE Healthcare).
Real-Time PCR experiments for snRNA quantitation were carried out on an Applied Biosystems (ABI) 7500 Real-Time PCR System using Power SYBR Green (Applied Biosystems, Foster City, CA). Each snRNA was measured in triplicate. Absolute quantification was performed using the ABI 7500 detection software with correction for amplification efficiency based on an exponential model of PCR (65,66).
Immunoprecipitation
Cells (∼1 × 106) were harvested in 1 ml of IP-500 buffer [500 mm NaCl, 10 mm Tris-Cl, pH 7.4, 0.1% Triton X-100, 50 mm NaF, 0.2 mm sodium vanadate, protease inhibitor cocktail (Sigma)] and sonicated. Lysates were centrifuged, and supernatant was collected and used in the assay. Protein G Dynabeads were incubated with anti-Sm antibody (Y12, Abcam, Cambridge, MA) in IP-100 buffer (100 mm NaCl, 10 mm Tris–Cl pH 7.4, 0.1% Triton X-100, 50 mm NaF, 0.2 mm sodium vanadate) for 4 h at 4°C. Beads (5 μl) were subsequently washed with IP-100 and added to 400 μl of cell lysate and rotated for 4 h at 4°C. Proteinase K was then added to the lysates and incubated at 37°C for 20 min followed by phenol extraction and ethanol precipitation of RNA. For input control, cell lysate without beads was treated in a similar manner.
RNA linker ligation
For linker-ligated RT–PCR, the 3' end of RNA was ligated to linker 1 (IDT, Coralville, IA, USA), according to manufacturer's instructions for miRCat (IDT), except for the substitution of Ligation Enhancer with PVA, and truncated RNA ligase 2 (NEB) to promote polar ligation. Modban primer was used for reverse transcription and PCR.
Mice
Transgenic mice containing human SMN2 and the mouse Smn knockout are as described previously (41). Genotyping and SMN2 copy number determination were performed as described previously. Mouse genotypes are hSMN2+/+; mSmn+/+or hSMN2+/+ and mSmn−/−. Mice were maintained in accordance with the Cold Spring Harbor Laboratory Animal Care and Use regulations. Mouse tissues were snap-frozen in liquid nitrogen and stored at −70°C. RNA was extracted from livers using Trizol reagent (Invitrogen) according to the manufacturer's protocol. To prepare protein lysates, 100 mg of liver tissue was sonicated in 900 ml of RIPA buffer [1× phosphate-buffered saline, 0.25% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 1 mm EDTA and complete mini protease inhibitor cocktail (Roche)].
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Families of SMA and the National Institutes of Health (NS069759 to M.L.H.). Funding to pay the Open Access publication charges for this article was provided by NINDS/NIH.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Adrian Krainer for the mouse tissue samples and antibodies, Xavier Roca for the U1 decoy constructs and Clive Svendsen for the iPS cells used in this study. We are grateful to David Horowitz, Judy Potashkin, Mallory Havens and Shan-Qing Gu for comments on this manuscript and Anthony Hinrich for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Frugier T., Nicole S., Cifuentes-Diaz C., Melki J. The molecular bases of spinal muscular atrophy. Curr. Opin. Genet. Dev. 2002;12:294–298. doi: 10.1016/s0959-437x(02)00301-5. doi:10.1016/S0959-437X(02)00301-5. [DOI] [PubMed] [Google Scholar]
- 2.Burghes A.H., Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. doi:10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. doi:10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 5.Elsheikh B., Prior T., Zhang X., Miller R., Kolb S.J., Moore D., Bradley W., Barohn R., Bryan W., Gelinas D., et al. An analysis of disease severity based on SMN2 copy number in adults with spinal muscular atrophy. Muscle Nerve. 2009;40:652–656. doi: 10.1002/mus.21350. doi:10.1002/mus.21350. [DOI] [PubMed] [Google Scholar]
- 6.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. doi:10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. doi:10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 8.Vitte J., Fassier C., Tiziano F.D., Dalard C., Soave S., Roblot N., Brahe C., Saugier-Veber P., Bonnefont J.P., Melki J. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. doi:10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. doi:10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Neuenkirchen N., Chari A., Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. doi:10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Paushkin S., Gubitz A.K., Massenet S., Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. doi:10.1016/S0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 12.Chari A., Paknia E., Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 2009;21:387–393. doi: 10.1016/j.ceb.2009.02.004. doi:10.1016/j.ceb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Gabanella F., Butchbach M.E., Saieva L., Carissimi C., Burghes A.H., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. doi:10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. doi:10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campion Y., Neel H., Gostan T., Soret J., Bordonne R. Specific splicing defects in S. pombe carrying a degron allele of the survival of motor neuron gene. EMBO J. 2010;29:1817–1829. doi: 10.1038/emboj.2010.70. doi:10.1038/emboj.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. doi:10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talbot K., Davies K.E. Is good housekeeping the key to motor neuron survival? Cell. 2008;133:572–574. doi: 10.1016/j.cell.2008.05.002. doi:10.1016/j.cell.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. doi:10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 19.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. doi:10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 20.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. doi:10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrotti S., Bielli P., Paronetto M.P., Ciccosanti F., Fimia G.M., Stamm S., Manley J.L., Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. doi:10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martins de Araujo M., Bonnal S., Hastings M.L., Krainer A.R., Valcarcel J. Differential 3′ splice site recognition of SMN1 and SMN2 transcripts by U2AF and U2 snRNP. RNA. 2009;15:515–523. doi: 10.1261/rna.1273209. doi:10.1261/rna.1273209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Chen X.H., Xiao P.J., Li L., Lin W.M., Huang J., Xu P. Expression pattern and splicing function of mouse ZNF265. Neurochem. Res. 2008;33:483–489. doi: 10.1007/s11064-007-9461-3. doi:10.1007/s11064-007-9461-3. [DOI] [PubMed] [Google Scholar]
- 24.Novoyatleva T., Heinrich B., Tang Y., Benderska N., Butchbach M.E., Lorson C.L., Lorson M.A., Ben-Dov C., Fehlbaum P., Bracco L., et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. doi:10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 25.Chen H.H., Chang J.G., Lu R.M., Peng T.Y., Tarn W.Y. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol. Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. doi:10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. doi:10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann Y., Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum. Mol. Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. doi:10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 28.Hastings M.L., Allemand E., Duelli D.M., Myers M.P., Krainer A.R. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS ONE. 2007;2:e538. doi: 10.1371/journal.pone.0000538. doi:10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gladman J.T., Chandler D.S. Intron 7 conserved sequence elements regulate the splicing of the SMN genes. Hum. Genet. 2009;126:833–841. doi: 10.1007/s00439-009-0733-7. doi:10.1007/s00439-009-0733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. doi:10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyaso H., Okumura M., Kondo S., Higashide S., Miyajima H., Imaizumi K. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J. Biol. Chem. 2003;278:15825–15831. doi: 10.1074/jbc.M209271200. doi:10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. doi:10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh N.N., Singh R.N., Androphy E.J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. doi:10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R.N. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- 36.Singh N.N., Androphy E.J., Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. doi:10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roca X., Krainer A.R. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. doi:10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. doi:10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 39.Gavrilov D.K., Shi X., Das K., Gilliam T.C., Wang C.H. Differential SMN2 expression associated with SMA severity. Nat. Genet. 1998;20:230–231. doi: 10.1038/3030. doi:10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 40.Helmken C., Hofmann Y., Schoenen F., Oprea G., Raschke H., Rudnik-Schoneborn S., Zerres K., Wirth B. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum. Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. doi:10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- 41.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. doi:10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 42.Ebert A.D., Yu J., Rose F.F., Jr, Mattis V.B., Lorson C.L., Thomson J.A., Svendsen C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. doi:10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mourelatos Z., Abel L., Yong J., Kataoka N., Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. doi:10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossoll W., Kroning A.K., Ohndorf U.M., Steegborn C., Jablonka S., Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. doi:10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 45.Shpargel K.B., Matera A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl Acad. Sci. USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. doi:10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. doi:10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Workman E., Saieva L., Carrel T.L., Crawford T.O., Liu D., Lutz C., Beattie C.E., Pellizzoni L., Burghes A.H. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. doi:10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hastings M.L., Krainer A.R. Functions of SR proteins in the U12-dependent AT–AC pre-mRNA splicing pathway. RNA. 2001;7:471–482. doi: 10.1017/s1355838201002552. doi:10.1017/S1355838201002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Black D.L., Steitz J.A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986;46:697–704. doi: 10.1016/0092-8674(86)90345-4. doi:10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- 50.Berget S.M., Robberson B.L. U1, U2, and U4/U6 small nuclear ribonucleoproteins are required for in vitro splicing but not polyadenylation. Cell. 1986;46:691–696. doi: 10.1016/0092-8674(86)90344-2. doi:10.1016/0092-8674(86)90344-2. [DOI] [PubMed] [Google Scholar]
- 51.Kramer A., Keller W., Appel B., Luhrmann R. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. doi: 10.1016/0092-8674(84)90551-8. doi:10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- 52.Wyatt J.R., Sontheimer E.J., Steitz J.A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. doi:10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 53.Friesen W.J., Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J. Biol. Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. doi:10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 54.Chari A., Golas M.M., Klingenhager M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. doi:10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. doi:10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Horowitz D.S., Krainer A.R. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. doi:10.1016/0168-9525(94)90233-X. [DOI] [PubMed] [Google Scholar]
- 57.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. doi:10.1016/S0959-437X(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 58.Roca X., Sachidanandam R., Krainer A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. doi:10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim S.R., Hertel K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. doi:10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 60.Gabanella F., Carissimi C., Usiello A., Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. doi:10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 61.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. doi:10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hastings M.L., Berniac J., Liu Y.H., Abato P., Jodelka F.M., Barthel L., Kumar S., Dudley C., Nelson M., Larson K., et al. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci. Transl. Med. 2009;1:5ra12. doi: 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Habets W.J., Hoet M.H., De Jong B.A., Van der Kemp A., Van Venrooij W.J. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- 64.Allemand E., Hastings M.L., Murray M.V., Myers M.P., Krainer A.R. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 2007;14:630–638. doi: 10.1038/nsmb1257. doi:10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- 65.Liu W., Saint D.A. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem. 2002;302:52–59. doi: 10.1006/abio.2001.5530. doi:10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 66.Cikos S., Bukovska A., Koppel J. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. doi:10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.