This work identified aromatic amino acid hydroxylase (AAH)-like sequences from nonflowering plants. The encoded proteins were shown to be plastid targeted, to have Phe hydroxylase activity, and, unlike animal and bacterial AAHs, to use a folate as cofactor rather than a pterin. Ablating AAH in moss led to accumulation of Phe and caffeate esters.
Abstract
Tetrahydropterin-dependent aromatic amino acid hydroxylases (AAHs) are known from animals and microbes but not plants. A survey of genomes and ESTs revealed AAH-like sequences in gymnosperms, mosses, and algae. Analysis of full-length AAH cDNAs from Pinus taeda, Physcomitrella patens, and Chlamydomonas reinhardtii indicated that the encoded proteins form a distinct clade within the AAH family. These proteins were shown to have Phe hydroxylase activity by functional complementation of an Escherichia coli Tyr auxotroph and by enzyme assays. The P. taeda and P. patens AAHs were specific for Phe, required iron, showed Michaelian kinetics, and were active as monomers. Uniquely, they preferred 10-formyltetrahydrofolate to any physiological tetrahydropterin as cofactor and, consistent with preferring a folate cofactor, retained activity in complementation tests with tetrahydropterin-depleted E. coli host strains. Targeting assays in Arabidopsis thaliana mesophyll protoplasts using green fluorescent protein fusions, and import assays with purified Pisum sativum chloroplasts, indicated chloroplastic localization. Targeting assays further indicated that pterin-4a-carbinolamine dehydratase, which regenerates the AAH cofactor, is also chloroplastic. Ablating the single AAH gene in P. patens caused accumulation of Phe and caffeic acid esters. These data show that nonflowering plants have functional plastidial AAHs, establish an unprecedented electron donor role for a folate, and uncover a novel link between folate and aromatic metabolism.
INTRODUCTION
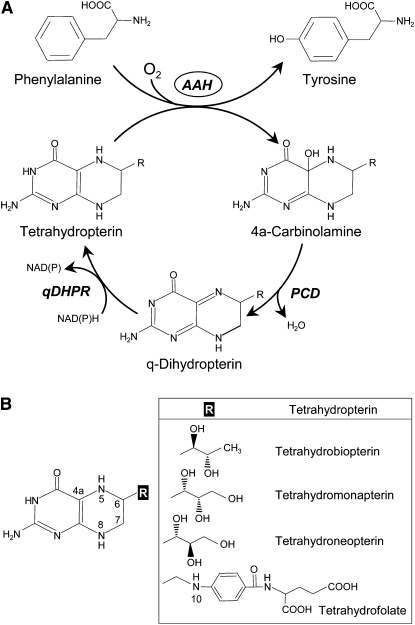
Aromatic amino acid hydroxylases (AAHs) are known from animals, protists, and some bacteria. They are iron-dependent monooxygenases that use a tetrahydropterin as electron donor and hydroxylate the ring of an aromatic amino acid (Figure 1) (Fitzpatrick, 1999). All known AAHs share sequence similarity, including two His residues and a Glu that together bind the catalytic iron atom. Animal AAHs comprise three subfamilies that prefer Phe, Tyr, or Trp, while most bacterial AAHs form a separate family that is essentially specific for Phe (Fitzpatrick, 1999; Siltberg-Liberles et al., 2008; Pey and Martinez, 2009). Tetrahydropterin-dependent enzymes that hydroxylate aromatics such as mandelate or benzoate have been reported in microorganisms but have not been cloned (e.g., Bhat and Vaidyanathan, 1976).
Figure 1.
Aromatic AAHs and Their Tetrahydropterin Cofactors.
(A) The reaction mediated by a representative AAH (Phe hydroxylase) and regeneration of the tetrahydropterin cofactor. Tyr hydroxylase converts Tyr to 3,4-dihydroxy Phe; Trp hydroxylase converts Trp to 5-hydroxy Trp.
(B) The structures of some natural tetrahydropterins and of tetrahydrofolate. Note that the differences all reside in the substituent at the 6-position. One-carbon derivatives of tetrahydrofolate have one-carbon groups attached to the N5 or N10 positions, or bridged between them.
The in vivo AAH cofactor is tetrahydrobiopterin (H4BPt) in animals and tetrahydromonapterin (H4MPt) in Pseudomonas aeruginosa (Thöny et al., 2000; Pribat et al., 2010). In vitro, however, these and other AAHs accept various natural or artificial tetrahydropterins and, in the case of Chromobacterium violaceum AAH, tetrahydrofolate (THF) (Fujisawa and Nakata, 1987; Fitzpatrick, 1999; Siltberg-Liberles et al., 2008). THF and its one-carbon derivatives can be seen as tetrahydropterins with a large 6-substituent (Figure 1). AAH reactions convert the tetrahydropterin cofactor to a 4a-carbinolamine, which is recycled by pterin-4a-carbinolamine dehydratase (PCD) and quinonoid dihydropterin reductase (Thöny et al., 2000) (Figure 1).
Tyr and Trp hydroxylases have anabolic roles in the synthesis of catecholamines and serotonin. By contrast, Phe hydroxylases can work anabolically or catabolically. The synthesis of Tyr is itself anabolic, but Tyr can be degraded via the homogentisate ring cleavage pathway to fumarate and acetoacetate and then to CO2. Phe hydroxylase-mediated funneling of Phe into the homogentisate pathway is well known in mammals and P. aeruginosa (Salter et al., 1986; Gu et al., 1998).
Although AAH activities are occasionally reported from plants (Nair and Vining, 1965; Yamamoto et al., 2001; Luthra and Singh, 2010), these studies lack adequate controls for chemical artifacts, and AAHs have generally been thought not to occur in the plant kingdom (Fitzpatrick, 1999). However, a hint that this might be wrong came from pilot comparative genomics work showing that nonflowering plant genomes encode AAH- and PCD-like proteins and that the latter have PCD activity (Naponelli et al., 2008).
The occurrence of hitherto unsuspected AAHs that metabolize aromatic compounds could be very significant. This is especially so for Phe, which gives rise to lignin, a vast array of soluble phenylpropanoids, and benzoates (Vogt, 2010) and whose breakdown has been little studied (Ellis and Towers, 1970). We accordingly made a thorough comparative analysis of plant AAH sequences, expressed and characterized selected AAH proteins, and determined the impact of ablating AAH in the moss Physcomitrella patens.
RESULTS
Bioinformatics
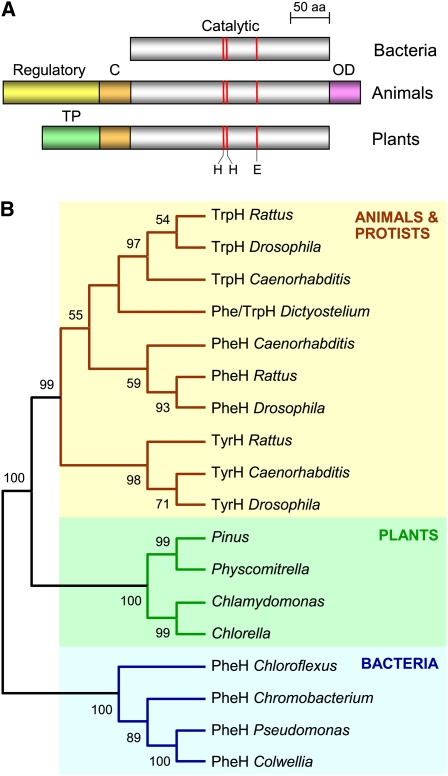
Plant genomes and ESTs were searched for AAH-like sequences using tBLASTn, with eukaryotic or bacterial AAHs as query. Significant hits were obtained for conifers, mosses, and green algae, but not angiosperms (see Supplemental Figure 1 online). Complete sequencing of EST clones from Chlamydomonas reinhardtii, P. patens, and loblolly pine (Pinus taeda), plus 5′-rapid amplification of cDNA ends, yielded full-length cDNAs encoding proteins containing the canonical catalytic region found in animal and bacterial AAHs, including the iron binding residues (Figure 2A; see Supplemental Figure 2 online). The plant proteins resemble animal AAHs in having an ~50-residue extension of the catalytic domain but differ in lacking the C-terminal domain responsible for oligomerization and the N-terminal regulatory region (Figure 2A). Instead of the latter, plant sequences have a nonconserved extension of ~60 to 120 residues that is predicted to be a plastid targeting peptide.
Figure 2.
Overall Domain Structures and Phylogenetic Relationships of Aromatic AAH Family Proteins.
(A) Comparison of the domain structures of bacterial, animal, and plant AAH proteins. Note the common catalytic domain and its conserved extension (C) in animal and plant AAHs, the unique regulatory and oligomerization (OD) domains in animal AAHs, and the putative targeting peptide (TP) in plant AAHs. Red lines mark the sites of iron-liganding His and Glu residues.
(B) Unrooted neighbor-joining tree for AAH proteins. Bootstrap values (1000 replicates) are indicated for nodes with >50% support. Only the tree topology is shown, so that branch lengths are not proportional to estimated numbers of amino acid substitutions. Enzymatic activities (based on published biochemical and/or genetic evidence): PheH, phenylalanine hydroxylase; TyrH, tyrosine hydroxylase; TrpH, tryptophan hydroxylase. Full organism names: Caenorhabditis elegans, Chlamydomonas reinhardtii, Chlorella sp NC64A, Chloroflexus aurantiacus, Chromobacterium violaceum, Colwellia psychrerythraea, Dictyostelium discoideum, Drosophila melanogaster, Physcomitrella patens, Pinus taeda, Pseudomonas aeruginosa, and Rattus rattus.
Phylogenetic analysis of the plant AAH sequences and of animal, protistan, and bacterial AAHs of known specificity placed the plant proteins with those of the other eukaryotes (Figure 2B; see Supplemental Data Set 1 online). This grouping was further supported by the presence of two introns common to plants and mammals (see Supplemental Figure 2 online). Although phylogenetic analysis confirmed that specificity for Phe, Tyr, or Trp appeared early in the animal lineage (Siltberg-Liberles et al., 2008), it did not suggest the substrate specificity of the plant proteins. However, since bacterial AAHs typically prefer Phe, and Phe hydroxylation is considered the ancestral activity in animals (Siltberg-Liberles et al., 2008) and is partially retained by Tyr and Trp hydroxylases (Tong and Kaufman, 1975; Daubner and Fitzpatrick, 1998), plant AAHs seemed most likely to have activity toward Phe.
Functional Complementation
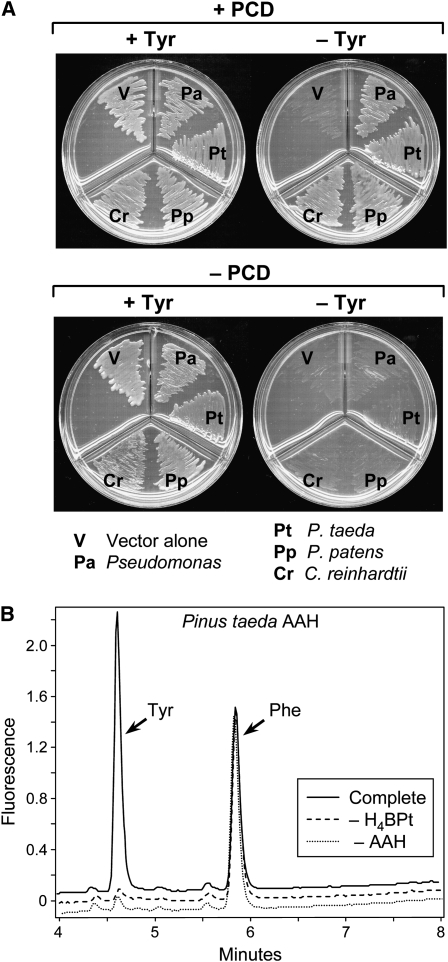
A functional complementation assay in Escherichia coli was accordingly used to test plant AAH proteins (minus their putative targeting peptides) for Phe hydroxylase activity. In this assay, an E. coli Tyr auxotroph is transformed with one expression plasmid encoding a Phe hydroxylase and another encoding PCD, both enzymes being absent from E. coli (Zhao et al., 1994; Naponelli et al., 2008). A functional Phe hydroxylase will restore Tyr prototrophy; a tetrahydropterin requirement can be demonstrated by omitting PCD, which is essential for tetrahydropterin regeneration (Figure 1A). The AAH proteins of C. reinhardtii, P. patens, and P. taeda restored Tyr prototrophy in the presence of PCD but not in its absence (Figure 3A), showing that they have tetrahydropterin-dependent Phe hydroxylase activity. This finding was confirmed by assaying Phe hydroxylase activity in extracts of E. coli cells expressing P. taeda or P. patens AAH; H4BPt-dependent conversion of Phe to Tyr was readily detected by HPLC (Figure 3B; see Supplemental Figure 3 online).
Figure 3.
Evidence that Plant AAH Proteins Have Tetrahydropterin-Dependent Phe Hydroxylase Activity.
(A) Detection of Phe hydroxylase activity by functional complementation. An E. coli Tyr auxotroph was transformed with pBluescript alone (– PCD) or containing the P. aeruginosa gene encoding PCD (+ PCD), plus the pSU18 vector alone or containing the P. aeruginosa Phe hydroxylase gene or a plant AAH cDNA (truncated to remove the putative targeting peptide). The doubly transformed cells were plated on minimal medium containing IPTG, plus or minus Tyr (+Tyr and −Tyr).
(B) Phe hydroxylase activity of recombinant P. taeda AAH. The complete reaction comprised 1 mM Phe, 0.2 mM H4BPt, 50 μM Fe(NH4)2(SO4)2, 10 mM DTT, 40 units of catalase, and extract (10 μg protein) from E. coli BL21-CodonPlus (DE3)-RIPL cells harboring pET28b containing P. taeda AAH cDNA. Controls were run without H4BPt (– H4BPt) or with extract from E. coli cells harboring pET28b alone (– AAH). Incubation was at 30°C for 15 min. Reactions were analyzed by fluorometric HPLC.
Biochemical Characterization of Pine and Moss AAHs
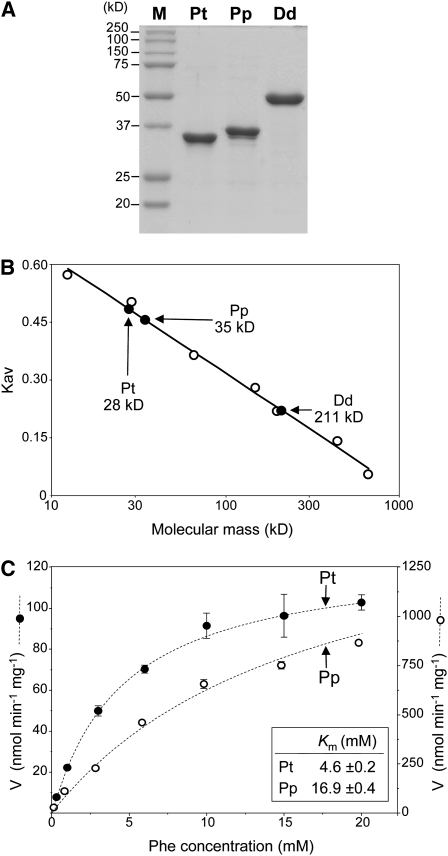
Recombinant His-tagged P. taeda and P. patens AAHs were further characterized using Dictyostelium discoideum Phe hydroxylase (Siltberg-Liberles et al., 2008) for comparison. All three proteins were purified to near-homogeneity by Ni affinity chromatography (Figure 4A). In analytical size exclusion chromatography, both plant AAHs behaved as species with a molecular mass near that of the monomer (Figure 4B), which fits with their lack of an oligomerization domain (Figure 2A). The D. discoideum control behaved as a tetramer, as expected (Siltberg-Liberles et al., 2008) (Figure 4B).
Figure 4.
Characterization of Plant AAH Proteins.
(A) Purification of recombinant P. taeda (Pt) and P. patens (Pp) AAHs and D. discoideum (Dd) Phe hydroxylase. Gel lanes contained 5 μg of Ni affinity-purified protein. Proteins were separated by denaturing gel electrophoresis and stained with Coomassie blue. M, molecular mass markers.
(B) Analytical size exclusion chromatography of P. taeda AAH, P. patens AAH, and D. discoideum Phe hydroxylase. The calculated molecular masses of the monomers are 35, 37, and 51 kD, respectively. Open circles are calibration standards (kD): cytochrome c (12.4), carbonic anhydrase (29), albumin (66), alcohol dehydrogenase (150), β-amylase (200), apoferritin (443), and thyroglobulin (669).
(C) Velocity versus Phe concentration curves for P. patens and P. taeda AAHs; the H4BPt cofactor concentration was 500 μM. Data are means and se of three replicates. Error bars smaller than the points are omitted.
Exploratory tests showed H4BPt to be a cofactor for P. taeda and P. patens AAHs, so it was used for further assays. H4BPt does not occur in plants (Kohashi et al., 1980; Díaz de la Garza et al., 2004; Hossain et al., 2004), but it is widely used to assay the activity of AAHs for which it is not the in vivo cofactor (Leiros et al., 2007; Siltberg-Liberles et al., 2008; Pey and Martinez, 2009). Tests with the natural (R) and unnatural (S) forms of H4BPt showed that only the R form was active and that the S form had no inhibitory activity (see Supplemental Figure 4 online). Various aromatic compounds were tried as substrates, using fluorometric HPLC to measure product formation (see Supplemental Table 1 online). The plant AAHs showed high activity with l-Phe; Tyr was the sole product, as in crude extracts (Figure 3B). Neither enzyme had activity with d-Phe, l-Tyr, l-Trp, cinnamate, p-coumarate, benzoate, anthranilate, mandelate, indoleacetate, caffeate, or ferulate, the detection limit being ≤1% of the activity with Phe (see Supplemental Table 1 online). Both enzymes showed Michaelian kinetics with Phe as substrate (Figure 4C); Km and Kcat values were in or close to the ranges reported for other Phe hydroxylases (0.1 to 7 mM and 0.08 to 8 s−1, respectively). Preincubation with Phe before the reaction did not change the kinetic properties of either enzyme; this contrasts with mammalian Phe hydroxylases, which are Phe activated (Døskeland et al., 1987; Shiman, 1987). The conservation of iron binding residues in the plant AAHs (see Supplemental Figure 2 online) suggested iron dependence, and this proved to be the case. Although activity was not affected by adding ferrous iron (25 μM to 1 mM), it was abolished by treating with the iron and copper chelator o-phenanthroline (50 μM), but not by treating with m-phenanthroline, which does not chelate iron, or with bathocuproine, which chelates copper but not iron (see Supplemental Figure 5 online). From bathophenanthroline complex formation, the iron contents (mean of triplicates ± sd) of the P. patens and P. taeda AAH preparations were estimated as 0.72 ± 0.01 and 0.33 ± 0.01 mol/mol protein, respectively. Such substoichiometric values are typical of purified AAHs (Fitzpatrick, 1999; Olafsdottir and Martínez, 1999).
Identity of the Physiological Cofactor
The main plant pterins are the folate synthesis intermediates dihydroneopterin, dihydromonapterin, and hydroxymethyldihydropterin (Kohashi et al., 1980; Díaz de la Garza et al., 2004; Hossain et al., 2004; Orsomando et al., 2006). These might be reduced to the tetrahydro level by a dihydropterin reductase like those in bacteria or protists (Gourley et al., 2001; Pribat et al., 2010). Conceivably, THF (Fujisawa and Nakata, 1987) or its one-carbon derivatives could serve as cofactor.
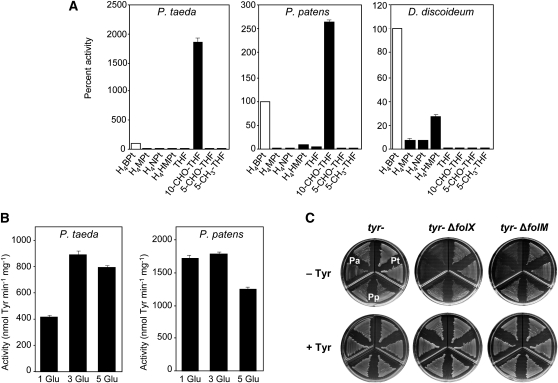
For plant AAHs, we therefore compared the cofactor activity of the following tetrahydropterins and tetrahydrofolates with that of H4BPt as a benchmark: tetrahydroneopterin, tetrahydromonapterin, hydroxymethyltetrahydropterin, THF, 10-formyltetrahydrofolate (10-CHO-THF), 5-formyltetrahydrofolate, and 5-methyltetrahydrofolate. D. discoideum Phe hydroxylase was included as a control for possible pterin contamination of folates, it being known to use H4BPt and H4MPt but not THF (Siltberg-Liberles et al., 2008). Surprisingly, of all the plant pterins and folates tested, only 10-CHO-THF was an effective cofactor for P. patens and P. taeda AAHs; the others were inactive or nearly so (Figure 5A). 10-CHO-THF was more effective than H4BPt, which, as noted above, is not a plant pterin. As expected, the D. discoideum enzyme showed high activity with H4BPt and appreciable activity with all the plant pterins, but none with 10-CHO-THF or any other folate. This result makes it unlikely that the activity of plant AAHs with 10-CHO-THF was due to a tetrahydropterin contaminant (Lloyd et al., 1971), nor was pterin contamination of 10-CHO-THF detectable chromatographically (see Methods).
Figure 5.
Cofactor Preferences of Plant AAHs in Vitro and In Vivo.
(A) Activities of recombinant AAHs with various cofactors. Each enzyme was assayed in conditions optimal for it. To maximize discrimination, the cofactor concentration used for each enzyme approximated the Km value for H4BPt. Cofactor concentrations (μM) were: P. taeda, 200; P. patens, 100; D. discoideum, 50. Data are means and se of three replicates and are expressed as percent of activities with H4BPt, which were (nmol min−1 mg−1 protein): P. taeda, 93; P. patens, 382; D. discoideum, 3115. Cofactor abbreviations: H4MPt, tetrahydromonapterin; H4NPt, tetrahydroneopterin; H4HMPt, hydroxymethyltetrahydropterin; THF, tetrahydrofolate; 10-CHO-THF, 10-formyltetrahydrofolate; 5-CHO-THF, 5-formyltetrahydrofolate; 5-CH3-THF, 5-methyltetrahydrofolate.
(B) Activities of recombinant P. taeda and P. patens AAHs with 10-CHO-THF (1 Glu) and its tri- and pentaglutamate forms (3 Glu and 5 Glu, respectively). Each enzyme was assayed in conditions optimal for it. The cofactor concentration used for each enzyme was approximately half the Km value for the natural (R) form of 10-CHO-THF (Table 1). Cofactor concentrations (μM) were as follows: P. taeda, 150; P. patens, 480. Data are means and se of three replicates. 10-CHO-THF (1, 3, or 5 Glu) was prepared from (R,S) 5-CHO-THF (1, 3, or 5 Glu) as described in Methods.
(C) Complementation of an E. coli Tyr auxotroph (tyr-) by AAH cDNAs from P. taeda (Pt) or P. patens (Pp), in the presence or absence of folX or folM genes, which are required for H4MPt synthesis. P. aeruginosa (Pa) Phe hydroxylase was included as a control. Each AAH was matched with a PCD from the same species. Cells were plated on medium plus or minus Tyr (+Tyr and −Tyr). Other conditions were as in Figure 3A. Note that deleting folX or folM caused complementation to fail only in the P. aeruginosa control.
The Km and Kcat values for 10-CHO-THF were compared with those for H4BPt for the plant AAHs (Table 1). Both Kcat values were higher for 10-CHO-THF; the Km values were the same for P. taeda but higher for 10-CHO-THF in P. patens. The Km values for both cofactors and both enzymes were in or near the range reported for Phe hydroxylases (3 to 500 μM). As 10-CHO-THF exists mainly as polyglutamates in chloroplasts (Orsomando et al., 2005; Mehrshahi et al., 2010), we compared the activity of 10-CHO-THF with that of its tri- and pentaglutamate forms. P. taeda AAH activity was twofold higher with polyglutamates, whereas P. patens AAH activity was almost the same (Figure 5B). Both enzymes can thus use the main natural forms of 10-CHO-THF.
Table 1.
Kinetic Characterization of Plant AAHs With Respect to the Cofactor
| Enzyme | H4BPt Cofactor |
10-CHO-THF Cofactor |
||
| Km (μM) | Kcat (s−1) | Km (μM) | Kcat (s−1) | |
| P. taeda | 281 ± 14 | 0.132 ± 0.002 | 293 ± 13 | 2.30 ± 0.01 |
| P. patens | 203 ± 6 | 1.16 ± 0.01 | 950 ± 34 | 6.89 ± 0.19 |
The Phe concentration was 20 mM. Values are means and se of three replicates.
To corroborate these in vitro data, we performed complementation tests as in Figure 3A, except that folX or folM was deleted from the host strain. Both folX and folM are needed to synthesize H4MPt, the tetrahydropterin cofactor in E. coli (Ikemoto et al., 2002; Pribat et al., 2010). P. aeruginosa Phe hydroxylase, which is H4MPt dependent, fails to complement Tyr auxotrophy in ΔfolX or ΔfolX strains (Pribat et al., 2010) (Figure 5C). By contrast, P. taeda and P. patens AAHs gave complementation in both these strains (Figure 5C). These results confirm that the plant enzymes cannot use H4MPt and are consistent with a 10-CHO-THF cofactor because neither folX nor folM affects folate synthesis (Pribat et al., 2010).
Subcellular Localization
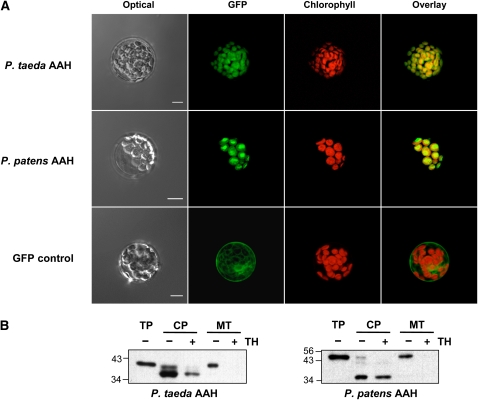
When full-length P. taeda or P. patens AAH sequences were fused to green fluorescent protein (GFP), they directed the latter exclusively to chloroplasts in transient expression experiments with Arabidopsis thaliana mesophyll protoplasts (Figure 6A). By contrast, controls using GFP alone showed no chloroplast targeting (Figure 6A). Dual import assays with mixtures of isolated pea (Pisum sativum) chloroplasts and mitochondria (Rudhe et al., 2002) substantiated these results (Figure 6B). After reactions with either full-length protein, chloroplasts contained a labeled product that was smaller than the full-length precursor and resistant to attack by thermolysin, as expected for a translocated protein. No translocated protein was found in the mitochondria.
Figure 6.
Evidence That the P. taeda and P. patens AAH Proteins Are Chloroplast Targeted.
(A) Transient expression in Arabidopsis mesophyll protoplasts of GFP fused to the C terminus of P. taeda AAH (top panels), P. patens AAH (middle panels), or GFP alone (bottom panels). GFP (green pseudocolor) and chlorophyll (red pseudocolor) fluorescence were observed by confocal microscopy. Bars = 10 μm.
(B) Protein import into isolated pea chloroplasts and mitochondria. Full-length P. taeda and P. patens AAH sequences were translated in vitro in the presence of [3H]Leu. The translation products were incubated for 15 (P. taeda) or 10 (P. patens) min in the light with chloroplasts (CP) or mitochondria (MT), which were then repurified on an 8% (v/v) Percoll gradient, without or with prior thermolysin (TH) treatment to remove adsorbed proteins. Proteins were separated by SDS-PAGE and visualized by fluorography. Samples were loaded on the basis of equal chlorophyll or mitochondrial protein content next to aliquots of the respective translation products (TP). The positions of molecular mass standards (kD) are indicated.
The chloroplastic location of AAH led us to examine that of its partner, PCD (Figure 1A). As with AAH, full-length P. taeda or P. patens PCD sequences targeted GFP to chloroplasts (see Supplemental Figure 6A online). Since plant PCD and AAH colocalize, we tested whether they form a complex as in bacteria (Song et al., 1999). P. taeda PCD was expressed in E. coli alone or together with His-tagged P. taeda AAH, and cell extracts were analyzed by Ni affinity chromatography and gel electrophoresis (see Supplemental Figure 6B online). A band in the position of PCD appeared only when PCD and AAH were expressed together. Tryptic cleavage and tandem mass spectrometric analysis of this band yielded two peptides (MKCSQANEIELAK and CSQANEIELAK) matching the N-terminal region of P. taeda PCD, confirming its identity.
Metabolic Impact of Ablating AAH in Moss
The single AAH-encoding gene in P. patens was inactivated by targeted gene replacement. Four independent lines were confirmed to have lost the native gene and to lack a copy of the AAH gene elsewhere in the genome (see Supplemental Figure 7 online). These knockout lines showed no appreciable differences from the wild type in growth rate or appearance when cultured in standard conditions (photoautotrophic growth on Knop mineral medium). However, metabolic profiling revealed characteristic metabolite level changes in the knockout lines, as summarized in the heat map of Supplemental Figure 8 online and listed in full in Supplemental Data Sets 2 and 3 online.
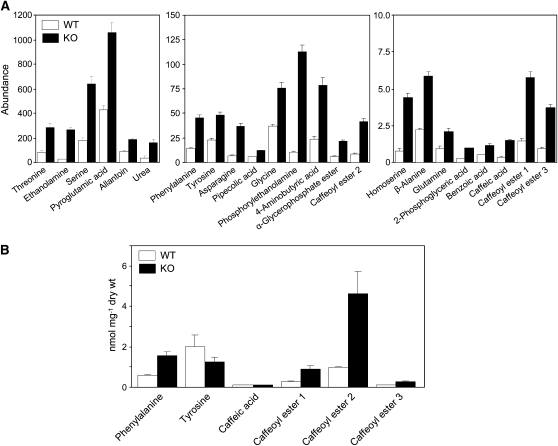
Consistent with loss of Phe hydroxylase activity, there was a significant threefold increase in the level of Phe in knockout lines. Levels of ethanolamine, Ser, and several other amino and organic acids also increased significantly (Figure 7A). Three unknown peaks (retention times 46.04, 46.62, and 47.76 min; see Supplemental Data Set 2 online) were elevated about fourfold in the knockout lines compared with the wild type. Electron ionization spectra suggested that these were esters of caffeic acid (named caffeoyl esters 1, 2, and 3 in Figure 7A). Chemical ionization gas chromatography–mass spectrometry (GC-MS) indicated a mass of 658 atomic mass units for their trimethylsilyl derivatives (see Supplemental Figures 9A and 9B online). The analysis of the molecular ion isotopic ratio, database searching, and liquid chromatography–MS analysis suggested that the unknowns are caffeoyl esters of trihydroxybutyric acids (see Supplemental Figure 9C online). This identification was confirmed by the synthesis of caffeoylthreonic acid and the good match of the GC-MS analysis of the synthesized compound with the unknown peak at 46.62 min (see Supplemental Figure 9B online). The other two unknowns are presumably different trihydroxybutyrate esters. Absolute quantification of the levels of Phe and the caffeic acid esters (Figure 7B) confirmed the relative quantification data of Figure 7A. Absolute quantification of Tyr, however, showed no significant change, as opposed to a modest accumulation in the relative quantification data set.
Figure 7.
Selected Metabolite Contents of AAH Knockout and Wild-Type P. patens.
(A) Relative abundance of amino acids, organic acids, and related metabolites that change significantly (q-value < 0.01, except for Ser q-value < 0.05; see Supplemental Methods online) and by at least twofold in the knockout lines, as estimated from GC-MS profiling of the polar fraction. Data are means and se for four independent knockout (KO) lines (a biological triplicate of each line was used) and five wild-type (WT) samples. Note the scale differences between the frames.
(B) Absolute quantification of Phe, caffeic acid, and its esters. L-Phe, caffeic acid, and chlorogenic acid were used as reference standards. Data are means and se for three independent knockout lines and three wild-type samples.
DISCUSSION
Plant AAHs Are Evolutionarily Ancient and of Eukaryote Ancestry
Our results establish that AAHs occur in plants and are of ancient origin in the plant lineage. Plant AAHs most likely share a common ancestor with those of other eukaryotes, as judged from phylogenetic analysis and from the presence of two introns in common with mammalian AAHs (Irimia and Roy, 2008). AAH genes have so far been found only in nonflowering plants, but it would be premature to infer that flowering plants as a group lack AAHs because genome sequences and deep EST databases are available only for a small and nonrepresentative subset of angiosperm taxa. Also, there exists one intriguing, classical report of the partial purification of a THF-dependent Phe hydroxylase activity from spinach (Spinacia oleracea) leaves (Nair and Vining, 1965). AAH genes are not ubiquitous in nonflowering plants, being absent from some algae and the clubmoss Selaginella (see Supplemental Figure 1 online). AAH might have a similarly sporadic distribution among flowering plants.
Plant AAHs Are Phe Hydroxylases with Unique Cofactor Specificity
Collectively, the biochemical characterization data show that plant AAHs are fairly typical monomeric Phe hydroxylases, save in one crucial respect: 10-CHO-THF is the preferred cofactor in vitro and apparently the operative one in vivo. This is not wholly exceptional inasmuch as bacterial Phe hydroxylase has weak activity with THF in vitro (Fujisawa and Nakata, 1987) and THF can replace H4BPt as cofactor for bacterial NO synthase, again in vitro (Adak et al., 2002). However, the use of folates as cofactors in vivo is unprecedented for Phe hydroxylase or any other pterin-dependent enzyme. If 10-CHO-THF is the in vivo cofactor, it follows that plant PCDs must recycle it. That this is the case is attested by the complementation data of Figure 5C, where P. taeda and P. patens AAHs were each supported by their respective PCD. A second unprecedented aspect is that neither 10-CHO-THF nor any other folate is known to serve solely as a net electron donor in a physiological reaction (Matthews, 1982).
Our tests with diverse aromatic compounds exclude all of them as alternatives to Phe but cannot rule out the possibility that other substrates exist. However, the functional complementation experiments show unequivocally that plant AAHs function as Phe hydroxylases in E. coli, making it likely that they do so in planta. That Phe is a substrate in planta is further supported by data from P. patens AAH knockouts, as discussed below.
Plant AAHs and Their PCD Partners Are Chloroplast Localized
AAHs were predicted and then proven by two methods to be chloroplast targeted. A chloroplast location is reasonable inasmuch as chloroplasts are rich in 10-CHO-THF (Orsomando et al., 2005; Mehrshahi et al., 2010) and are where the Phe substrate of AAH, and its product Tyr, are made from l-arogenate (Rippert et al., 2009). Because the 4a-carbinolamine produced by AAH must be recycled to maintain cofactor supply (Figure 1A), there is a high antecedent probability that the recycling enzyme PCD is also chloroplastic. Targeting experiments with GFP fusions indicated that this is the case for P. patens and P. taeda PCDs, and a chloroplast location was further supported by evidence that P. taeda AAH and PCD form a complex. Earlier experiments suggested a mitochondrial location for P. taeda PCD (Naponelli et al., 2008) but used protoplasts from cultured cells with fewer and less developed chloroplasts than the mesophyll protoplasts used here and so perhaps amplified a low level of mitochondrial (mis)targeting.
Moss AAH Mediates Significant Phe to Tyr Flux in Vivo
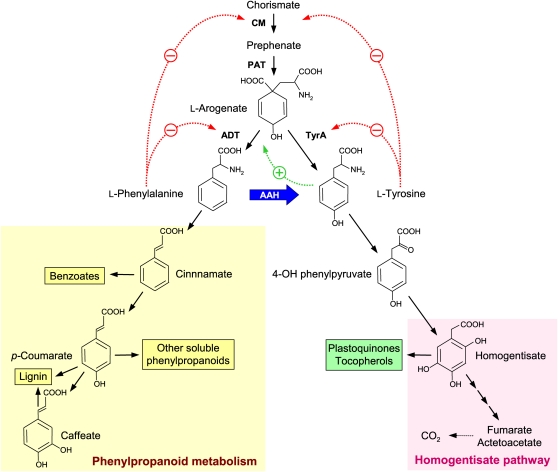
The effects of ablating P. patens AAH are consistent with its normal role being to convert Phe to Tyr. Thus, AAH knockout lines accumulated Phe and caffeic acid esters, which are Phe metabolites (Figure 8). While there was no Tyr depletion, this is not necessarily expected. First, Phe and Tyr synthesis both branch from l-arogenate and are subject to feedback inhibition and activation loops that tend to dampen the impact of perturbations (Figure 8) (Jung et al., 1986; Siehl and Conn, 1988; Rippert and Matringe, 2002; Tzin and Galili, 2010). Second, only the chloroplastic Tyr pool would be immediately affected by loss of AAH, and this pool is presumably only part of the total free Tyr pool that was measured. Other major metabolite changes in the knockout strains were consistent with loss of folate-dependent AAH activity, which would be expected to perturb chloroplast folate pools (Orsomando et al., 2005) and, hence, pools of folate-related metabolites such as Ser. Indeed, there were large accumulations of Ser, its decarboxylation product ethanolamine (Rontein et al., 2001), and of phosphorylethanolamine. The accumulation of Asp family amino acids (homoserine, Asn, and Thr) may likewise result from disturbances to folate metabolism that affect the plastidial supply of methyl groups for Met synthesis (Ravanel et al., 2004).
Figure 8.
AAH in Relation to Pathways of Aromatic Metabolism in Plants.
The broad blue arrow shows the AAH short circuit between the Phe and Tyr branches of aromatic metabolism. Dotted red and green arrows denote feedback inhibition and feedback activation, respectively. CM, chorismate mutase; PAT, prephenate aminotransferase; ADT, arogenate dehydratase; TyrA, arogenate dehydrogenase.
Plant AAH: A Short Circuit between Two Major Branches of Aromatic Metabolism
AAH in effect installs a short circuit between the Phe and Tyr branches of aromatic metabolism (Figure 8). These two branches, which are otherwise quite separate, lead to different sets of products. The Phe branch leads, via cinnamate and p-coumarate, to benzoates, to soluble phenylpropanoids and, in higher plants, to lignin; the Tyr branch leads, via homogentisate, to plastoquinones and tocopherols or to degradation of the aromatic ring (Moran, 2005; Vogt, 2010). The AAH short circuit potentially confers three unique features on aromatic metabolism. These features are of special interest in gymnosperms, in which ~30% of the carbon fixed in photosynthesis courses down the Phe branch en route to lignin, the flux down the Tyr branch being far smaller (Rippert and Matringe, 2002). First, the short circuit provides an alternative biosynthetic path to Tyr. Were only a minor fraction of the huge flux down the Phe branch in woody plants diverted to Tyr, the usual Tyr pathway from l-arogenate would become redundant (Figure 8). Second, the short circuit enables catabolism of the Phe ring, thus allowing efficient disposal of excess Phe. Third, the requirement of AAH for plastidial tetrahydrofolates and for their recycling via PCD creates a nexus at which folate metabolism interacts with, and perhaps regulates, the aromatic metabolism network. Future experiments with P. taeda will address these possibilities.
METHODS
Bioinformatics
Plant AAH genes and ESTs were sought at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), Joint Genome Institute (http://www.jgi.doe.gov/), and Sol Genomics Network (http://solgenomics.net/) databases. ESTs almost identical to animal or bacterial sequences were taken to be contaminants and excluded. Sequence alignments were made using ClustalW; phylogenetic analyses were made with MEGA 4 (Tamura et al., 2007). Targeting was predicted with TargetP (http://www.cbs.dtu.dk/services/TargetP/) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html). Enzyme data were taken from the Brenda enzyme database (http://www.brenda-enzymes.org/).
Chemicals
The following pterins and folates were obtained from Schircks Laboratories (Jona, Switzerland): (R), (S), and (R,S) tetrahydrobiopterin, (R,S) 6-hydroxymethyltetrahydropterin, (R,S) tetrahydromonapterin, (R,S) tetrahydroneopterin, (S) tetrahydrofolate, (S) 5-methyltetrahydrofolate, (S) 5-formyltetrahydrofolate, and (R,S) 5-formyltetrahydrofolate and its tri- and pentaglutamates. Merck & Cie (Schaffhausen, Switzerland) kindly donated (R) 10-formyltetrahydrofolate; fluorometric HPLC analysis plus and minus iodine oxidation (Pribat et al., 2010) revealed no detectable contamination (<0.1%) with unconjugated pterins. Near-saturated stock solutions of pterins and folates were prepared freshly in N2-sparged potassium phosphate, 100 mM, pH 7.5, containing 10 mM glutathione or 5 mM β-mercaptoethanol, excluding light; titers were determined spectrophotometrically (Pribat et al., 2010). Polyglutamates of (R,S) 10-formyltetrahydrofolate were prepared from the corresponding (R,S) 5-formyltetrahydrofolates (Baggott et al., 1995).
AAH and PCD cDNAs
Primer sequences are detailed in Supplemental Table 2 online. Full-length Chlamydomonas reinhardtii and Physcomitrella patens AAH cDNAs were amplified from ESTs GenBank numbers BG846252 and BU052179, respectively. A full-length P. taeda AAH cDNA was constructed using EST sequences, genome walking (Siebert et al., 1995), and 5′-rapid amplification of cDNA ends. The 3′ sequence (from 437 to 987 bp) of the cDNA was obtained from EST GenBank number DT628401. For isolation of genomic fragments flanking this EST, DNA was isolated from Pinus taeda (genotype 10-5 from North Carolina State University Cooperative Tree Improvement Program) using a CTAB-based method, quality-checked on a 0.8% (w/v) agarose gel, digested with DraI, EcoRV, StuI, or PvuII, and ligated to adaptors according to the GenomeWalker protocol (Clontech). Primers were designed with Netprimer (Premier BiosoftInternational) as specified in the GenomeWalker protocol. This protocol was used to amplify upstream (primers PtAAH-GSP1/PtAAH-GSP2) and downstream (primers PtAAH-GSP3/PtAAH-GSP4) regions in steps using primers matching DT628401 and adaptor primers from the GenomeWalker kit. Gel-purified amplicons were cloned in pGEM-T (Invitrogen) and sequenced. Sequences were assembled with Sequencher (Gene Codes), and open reading frames (from 286 to 436 bp and from 987 bp to the end of the cDNA) were identified using ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The missing 5′ region (from the start codon to 285 bp) was cloned by nested PCR from an unamplified cDNA library in pSL1180 (from W. Lorenz, University of Georgia). Primers PtAAH7 and PtAAH8 were located in the second exon, and primers pSL1 and pSL2 in the vector. Six independent sequences were cloned. The whole open reading frame was assembled in silico, and a full-length cDNA with added 5′ SalI and 3′ NcoI sites was synthesized by GenScript and inserted in pUC57. PCD cDNAs were as described (Naponelli et al., 2008).
Functional Complementation
P. taeda, P. patens, and C. reinhardtii cDNAs were truncated using PCR to replace their predicted targeting sequences with a start codon (see Supplemental Figure 2 online). The truncated cDNAs and the Pseudomonas aeruginosa Phe hydroxylase gene (Naponelli et al., 2008) were PCR amplified. The forward primers contained an EcoRI site, a stop codon in frame with LacZ, and a Shine-Dalgarno sequence before the start codon; reverse primers contained a SpheI site (see Supplemental Table 2 online). Amplicons were cloned between the EcoRI and SpheI sites of pSU18 and sequence verified. Electroporation was used to simultaneously transform Escherichia coli strain JP2255 (Zhao et al., 1994) with each AAH construct or pSU18, and P. aeruginosa PCD in pBluescript SK− (Naponelli et al., 2008) or pBluescript SK− alone. Complementation tests used minimal medium containing M9 salts, 0.1 mM CaCl2, 0.5 mM MgSO4, 0.4% (w/v) glucose, 17 μg/mL thiamine, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 100 μg/mL ampicillin, 25 μg/mL chloramphenicol, 100 μg/mL streptomycin, and 50 μg/mL Phe, plus or minus 50 μg/mL Tyr. Incubation was at 22°C for 2 to 4 d. For the in vivo cofactor preference complementation, E. coli strains JP2255, JP2255 ΔfolM, and JP2255 ΔfolX (Pribat et al., 2010) were transformed simultaneously with plant AAH cDNAs in pSU18 or P. aeruginosa Phe hydroxylase plasmid pJS11 as a control (Pribat et al., 2010) and the PCD from the same species (in pBluescript SK−). Complementation tests were as above except that kanamycin (50 μg/mL) replaced chloramphenicol in plate sections with P. aeruginosa Phe hydroxylase.
Production and Isolation of Recombinant AAHs
Truncated P. taeda and P. patens AAH cDNAs with flanking NcoI and XhoI sites were PCR amplified from the pSU18 constructs above (see Supplemental Table 2 online for primers), digested with NcoI and XhoI, and cloned into the matching restriction sites in pET28b (which adds a C-terminal His tag). The sequence-verified plasmids and pET21a containing D. discoideum Phe hydroxylase (Siltberg-Liberles et al., 2008) were introduced into E. coli strain BL21-CodonPlus (DE3)-RIPL (Stratagene). Cultures (300 mL) were grown at 37°C in Luria-Bertani containing 10 μM Fe(NH4)2(SO4)2 and appropriate antibiotics. When A600 reached 0.6, IPTG was added (final concentration 0.5 mM) and growth was continued for 4 h at 22°C. Subsequent steps were at 4°C. Cells were harvested by centrifugation, resuspended in 50 mM sodium phosphate, pH 8, 300 mM NaCl, 10% (v/v) glycerol and 10 mM imidazole, and sonicated. The cleared supernantant was loaded onto a 0.5-mL Ni2+-nitrilotriacetic acid agarose column (Qiagen) equilibrated with the sonication buffer. After washing with 80 mL 50 mM sodium phosphate, 300 mM NaCl, 10% (v/v) glycerol, and 50 mM imidazole, pH 8.0, proteins were eluted with 2.5 mL of the same buffer containing 250 mM imidazole, desalted twice on PD-10 columns (Pierce) equilibrated in 50 mM sodium phosphate, pH 8.0, 100 mM NaCl, and 10% (v/v) glycerol, and concentrated to ~1.5 mg/mL in Centricon Y10 units (Amicon). Purified proteins were frozen in liquid N2 and stored at −80°C. Freezing was shown not to affect activity. Protein was estimated by dye binding (Bradford, 1976) with BSA as standard. Size exclusion chromatography used a Superdex 200 HR 10/30 column eluted with 100 mM potassium phosphate, pH 7.5, containing 0.1 M KCl.
AAH Activity Assays
Assays (50 μL) contained 100 mM potassium phosphate, pH 7.5, 10% glycerol (v/v), 10 mM glutathione, 1 mM β-mercaptoethanol, 25 μM Fe(NH4)2(SO4)2, 80 units bovine liver catalase (Sigma-Aldrich), and the indicated substrate and cofactor concentrations. β-Mercaptoethanol was omitted for Phe Km determinations and substrate specificity tests. DTT (10 mM) was used in place of glutathione and β-mercaptoethanol in pilot work. Assays were at 30°C. Tests of the survival of THF (the least stable of the folates tested as cofactors) in reaction mixtures from which AAH was omitted showed no loss after 10 min, as estimated spectrophotometrically. Protein levels and reaction times were adjusted so that product formation was linearly proportional to both. Phe Km determinations used 4.9 μg (P. taeda) or 2.8 μg (P. patens) AAH in 10 min assays with 0 to 20 mM Phe and 500 μM H4BPt. Cofactor specificity tests used 1.2 to 2.1 μg (P. taeda) or 0.4 to 0.6 μg (P. patens) AAH in 1- to 2-min (10-CHO-THF) or 3- to 5-min (other cofactors) assays. Chelation tests omitted Fe(NH4)2(SO4)2 and used 1 mM Phe, 200 μM H4BPt, and 50 μM o- or m-phenanthroline, or bathocuproine. Reactions were stopped with 10 μL 5 M HCl. Ten percent of the mixture was analyzed by HPLC using a 4-μm, 250 × 4.6-mm Synergi Fusion RP80 column (Phenomenex) eluted (1.5 mL/min) with one of two gradients. Gradient 1 was 0 to 0.5 min 100% A; 0.5 to 8 min linear increase to 20% B; 8 to 12 min 100% A (A = 10 mM sodium acetate, pH 3.5; B = acetonitrile). Gradient 2 was 0 to 0.5 min 100% A; 0.5 to 20 min linear increase to 50% B; 20 to 25 min 100% A. Peaks were detected by fluorescence (see Supplemental Table 1 online for excitation and emission wavelengths) and identified using standards. Iron content was estimated colorimetrically by bathophenanthroline complex formation as described (Olafsdottir and Martínez, 1999) except that the bathophenanthroline concentration was 5 mM and cofactor was omitted.
Subcellular Localization
Primers are described in Supplemental Table 2 online. Full-length P. taeda and P. patens AAH cDNAs were cloned (between SalI and NcoI sites) in frame upstream of GFP in pTH2 (Niwa, 2003). The vector pPZP212 (GFP alone) served as control (Hajdukiewicz et al., 1994). Arabidopsis thaliana was grown in soil in a 10-h-light (~120 μmol m–2 s–1)/14-h-dark regime at 23°C. Mesophyll protoplasts isolated from 4-week-old plants were transfected via a polyethylene glycol–mediated procedure (Yoo et al., 2007) and imaged after 16 h using a Zeiss Pascal LSM5 confocal laser scanning microscope. GFP and chlorophyll fluorescence were respectively excited with a 488-nm argon and a 543-nm helium-neon laser and detected using 505- to 530-nm and 600- to 650-nm band-pass filters. Images were processed with ImageJ software (http://rsb.info.nih.gov/ij).
For dual import assays, P. taeda and P. patens AAH cDNAs were truncated as above by PCR to remove the first 56 and 60 amino acids, respectively. Forward primers included a Kozak sequence. Amplicons were cloned as EcoRI-PstI fragments into pGEM-4Z (Promega). Coupled in vitro transcription-translation was performed using a TnT kit (Promega) and [3H]leucine (108 Ci/mmol). Before use, translation products (50 μL) were diluted with 50 μL of 60 mM Leu in 2× import buffer (1× = 0.3 M sucrose, 15 mM HEPES-KOH, pH 7.4, 5 mM KH2PO4, 4 mM MgCl2, 4 mM Met, 4 mM ATP, 1 mM GTP, 0.2 mM ADP, 5 mM succinate, 4.5 mM DTT, 10 mM KCH3COO, 10 mM NaHCO3, and 0.2% [w/v] BSA). Chloroplasts and mitochondria were from 12-d-old pea shoots (Pisum sativum cv Laxton’s Progress 9 Improved). Chloroplasts were isolated as described (Cline et al., 1993) and resuspended (1 mg chlorophyll/mL) in import buffer. Mitochondria were isolated as described (Day et al., 1985) except that phosphate buffers and Cys were replaced by MOPS and glutathione, pyrophosphate was omitted from the grinding buffer, and mitochondria were washed twice after diluting 20× in resuspending medium. Mitochondrial protein was determined with the BCA assay (Pierce). Dual import assays (25 μg of mitochondrial proteins, 25 μg of chlorophyll, 86 μL of diluted translation products, final volume 200 μL in import buffer) were incubated for 15 (P. taeda) or 10 (P. patens) min at 25°C in the light. Protease treatment and repurification of the organelles were performed as described (Rudhe et al., 2002) except that the organelles were repurified using an 8% (v/v) Percoll gradient. Samples were separated by SDS-PAGE and visualized by fluorography.
To study AAH-PCD complex formation, P. taeda PCD with added NdeI and EcoRI sites was PCR amplified from a pBluescript SK− construct harboring the cDNA minus its targeting sequence (Naponelli et al., 2008) and cloned into the NdeI and EcoRI sites of pET43. For coexpression, the P. taeda PCD sequence preceded by a ribosome binding site was inserted into the XbaI site of the pET28b construct containing truncated, His-tagged P. taeda AAH (see above). E. coli protein expression and isolation were as above. Proteins were separated by SDS-PAGE and stained using Coomassie Blue or silver. The PCD gel region was excised and digested with trypsin (Sheffield et al., 2006). The digest was analyzed by liquid chromatography–MS/MS using a QStar hybrid quadrupole time-of-flight mass spectrometer (AB Sciex). MS/MS spectra were analyzed using Mascot 2.0.01 (Matrix Science), set up to search a database containing potential peptides of P. taeda PCD.
AAH Ablation in Moss
P. patens AAH was ablated by targeted gene replacement (Frank et al., 2005). Gene replacement and analyses of knockout lines were performed as described in the Supplemental Methods online.
Metabolomic Analysis
Polar and nonpolar metabolite fractions isolated from lyophilized cultures of knockout lines and wild-type controls were analyzed as described in the Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under accession numbers HQ003814 (P. taeda), HQ003815 (P. patens), and HQ003816 (C. reinhardtii). P. patens plants described in this article can be retrieved from the International Moss Stock Center (http://www.moss-stock-center.org/) using accession numbers 40390 (KO 45), 40391 (KO 46), 40392 (KO 47), and 40394 (KO 49).
Author Contributions
A.D.H., A.P., and A.N. designed the research and performed functional complementation assays and biochemical characterization. W.F., R.H., and R.R. created and validated moss knockouts. M.B. and L.W.S. performed metabolomics analyses. R.F. conducted, and K.L. and S.R. did pilot work on, targeting of GFP fusions. A.M.M. and J.M.D. participated in cloning P. taeda AAH cDNAs. A.D.H. wrote the article, and A.P. contributed to the writing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Distribution of AAH-Like Sequences in Plant Genomes and EST Collections.
Supplemental Figure 2. Multiple Sequence Alignment of AAH Proteins from Eukaryotes and Prokaryotes.
Supplemental Figure 3. Phenylalanine Hydroxylase Activity of Recombinant Physcomitrella patens AAH.
Supplemental Figure 4. Effect of H4BPt Chirality on Phenylalanine Hydroxylase Activity.
Supplemental Figure 5. Evidence That Phenylalanine Hydroxylase Activity Requires Iron.
Supplemental Figure 6. Evidence That P. taeda and P. patens PCD Proteins Are Chloroplast Targeted and That P. taeda PCD and AAH Form a Complex in Vitro.
Supplemental Figure 7. Generation and Quality Controls for Four Independent P. patens AAH Knockout Lines.
Supplemental Figure 8. Heat Map Depicting the Changes in Metabolite Levels that Characterize P. patens AAH Knockout Lines.
Supplemental Figure 9. Mass Spectral Evidence for the Identity of Caffeic Acid Esters in P. patens.
Supplemental Table 1. Separation and Detection of Potential Products of AAH Action Using Fluorometric HPLC.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. FASTA File of Aligned AAH Sequences.
Supplemental Data Set 2. Metabolomic Analysis: Polar Fraction GC-MS.
Supplemental Data Set 3. Metabolomic Analysis: Apolar Fraction GC-MS.
Supplementary Material
Acknowledgments
We thank P. Stover for advice on folate chemistry, K. Cline, C. Aldridge, and J. Waller for help with dual import experiments, C. Dervinis for statistical analysis of metabolomics data, M. Ziemak for technical support, G. Basset for pPZP212, A. Martinez for the D. discoideum Phe hydroxylase expression clone, the University of Leeds, the Chlamydomonas Center at Duke University, and the J. Craig Venter Institute for ESTs, and the Proteomics Division at the University of Florida Interdisciplinary Center for Biotechnology Research for proteomics analyses. This project was supported in part by National Research Initiative Competitive Grant 2008-35318-04589 from the USDA National Institute of Food and Agriculture and by an endowment from the C.V. Griffin, Sr. Foundation.
References
- Adak S., Bilwes A.M., Panda K., Hosfield D., Aulak K.S., McDonald J.F., Tainer J.A., Getzoff E.D., Crane B.R., Stuehr D.J. (2002). Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 99: 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggott J.E., Johanning G.L., Branham K.E., Prince C.W., Morgan S.L., Eto I., Vaughn W.H. (1995). Cofactor role for 10-formyldihydrofolic acid. Biochem. J. 308: 1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S.G., Vaidyanathan C.S. (1976). Involvement of 4-hydroxymandelic acid in the degradation of mandelic acid by Pseudomonas convexa. J. Bacteriol. 127: 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cline K., Henry R., Li C., Yuan J. (1993). Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 12: 4105–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner S.C., Fitzpatrick P.F. (1998). Mutation to phenylalanine of tyrosine 371 in tyrosine hydroxylase increases the affinity for phenylalanine. Biochemistry 37: 16440–16444 [DOI] [PubMed] [Google Scholar]
- Day D.A., Neuberger M., Douce R. (1985). Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aus. J. Plant Physiol. 12: 219–228 [Google Scholar]
- Díaz de la Garza R., Quinlivan E.P., Klaus S.M., Basset G.J., Gregory J.F., III, Hanson A.D. (2004). Folate biofortification in tomatoes by engineering the pteridine branch of folate synthesis. Proc. Natl. Acad. Sci. USA 101: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Døskeland A.P., Døskeland S.O., Flatmark T. (1987). Phenylalanine 4-monooxygenase from bovine liver. Methods Enzymol. 142: 35–44 [DOI] [PubMed] [Google Scholar]
- Ellis B.E., Towers G.H.N. (1970). Degradation of aromatic compounds by sterile plant tissues. Phytochemistry 9: 1457–1461 [Google Scholar]
- Fitzpatrick P.F. (1999). Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 68: 355–381 [DOI] [PubMed] [Google Scholar]
- Frank W., Decker E.L., Reski R. (2005). Molecular tools to study Physcomitrella patens. Plant Biol (Stuttg) 7: 220–227 [DOI] [PubMed] [Google Scholar]
- Fujisawa H., Nakata H. (1987). Phenylalanine 4-monooxygenase from Chromobacterium violaceum. Methods Enzymol. 142: 44–49 [DOI] [PubMed] [Google Scholar]
- Gourley D.G., Schüttelkopf A.W., Leonard G.A., Luba J., Hardy L.W., Beverley S.M., Hunter W.N. (2001). Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nat. Struct. Biol. 8: 521–525 [DOI] [PubMed] [Google Scholar]
- Gu W., Song J., Bonner C.A., Xie G., Jensen R.A. (1998). PhhC is an essential aminotransferase for aromatic amino acid catabolism in Pseudomonas aeruginosa. Microbiology 144: 3127–3134 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hossain T., Rosenberg I., Selhub J., Kishore G., Beachy R., Schubert K. (2004). Enhancement of folates in plants through metabolic engineering. Proc. Natl. Acad. Sci. USA 101: 5158–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto K., Sugimoto T., Murata S., Tazawa M., Nomura T., Ichinose H., Nagatsu T. (2002). (6R)-5,6,7,8-tetrahydro-L-monapterin from Escherichia coli, a novel natural unconjugated tetrahydropterin. Biol. Chem. 383: 325–330 [DOI] [PubMed] [Google Scholar]
- Irimia M., Roy S.W. (2008). Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res. 36: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E., Zamir L.O., Jensen R.A. (1986). Chloroplasts of higher plants synthesize L-phenylalanine via L-arogenate. Proc. Natl. Acad. Sci. USA 83: 7231–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohashi M., Tomita K., Iwai K. (1980). Analysis of unconjugated pterins in food resources and human urine. Agric. Biol. Chem. 44: 2089–2094 [Google Scholar]
- Leiros H.K., Pey A.L., Innselset M., Moe E., Leiros I., Steen I.H., Martinez A. (2007). Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J. Biol. Chem. 282: 21973–21986 [DOI] [PubMed] [Google Scholar]
- Lloyd T., Mori T., Kaufman S. (1971). 6-Methyltetrahydropterin. Isolation and identification as the highly active hydroxylase cofactor from tetrahydrofolate. Biochemistry 10: 2330–2336 [DOI] [PubMed] [Google Scholar]
- Luthra P.M., Singh S. (2010). Identification and optimization of tyrosine hydroxylase activity in Mucuna pruriens DC. var. utilis. Planta 231: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Matthews R.G. (1982). Are the redox properties of tetrahydrofolate cofactors utilized in folate-dependent reactions? Fed. Proc. 41: 2600–2604 [PubMed] [Google Scholar]
- Mehrshahi P., et al. (2010). Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J. 64: 267–279 [DOI] [PubMed] [Google Scholar]
- Moran G.R. (2005). 4-Hydroxyphenylpyruvate dioxygenase. Arch. Biochem. Biophys. 433: 117–128 [DOI] [PubMed] [Google Scholar]
- Nair P.M., Vining L.C. (1965). Phenylalanine hydroxylase from spinach leaves. Phytochemistry 4: 401–411 [Google Scholar]
- Naponelli V., Noiriel A., Ziemak M.J., Beverley S.M., Lye L.F., Plume A.M., Botella J.R., Loizeau K., Ravanel S., Rébeillé F., de Crécy-Lagard V., Hanson A.D. (2008). Phylogenomic and functional analysis of pterin-4a-carbinolamine dehydratase family (COG2154) proteins in plants and microorganisms. Plant Physiol. 146: 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y. (2003). A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol. 20: 1–11 [Google Scholar]
- Olafsdottir S., Martínez A. (1999). The accessibility of iron at the active site of recombinant human phenylalanine hydroxylase to water as studied by 1H NMR paramagnetic relaxation. Effect of L-Phe and comparison with the rat enzyme. J. Biol. Chem. 274: 6280–6284 [DOI] [PubMed] [Google Scholar]
- Orsomando G., Bozzo G.G., de la Garza R.D., Basset G.J., Quinlivan E.P., Naponelli V., Rébeillé F., Ravanel S., Gregory J.F., III, Hanson A.D. (2006). Evidence for folate-salvage reactions in plants. Plant J. 46: 426–435 [DOI] [PubMed] [Google Scholar]
- Orsomando G., de la Garza R.D., Green B.J., Peng M., Rea P.A., Ryan T.J., Gregory J.F., III, Hanson A.D. (2005). Plant γ-glutamyl hydrolases and folate polyglutamates: characterization, compartmentation, and co-occurrence in vacuoles. J. Biol. Chem. 280: 28877–28884 [DOI] [PubMed] [Google Scholar]
- Pey A.L., Martinez A. (2009). Iron binding effects on the kinetic stability and unfolding energetics of a thermophilic phenylalanine hydroxylase from Chloroflexus aurantiacus. J. Biol. Inorg. Chem. 14: 521–531 [DOI] [PubMed] [Google Scholar]
- Pribat A., Blaby I.K., Lara-Núñez A., Gregory J.F., III, de Crécy-Lagard V., Hanson A.D. (2010). FolX and FolM are essential for tetrahydromonapterin synthesis in Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 192: 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S., Block M.A., Rippert P., Jabrin S., Curien G., Rébeillé F., Douce R. (2004). Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279: 22548–22557 [DOI] [PubMed] [Google Scholar]
- Rippert P., Matringe M. (2002). Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana. Eur. J. Biochem. 269: 4753–4761 [DOI] [PubMed] [Google Scholar]
- Rippert P., Puyaubert J., Grisollet D., Derrier L., Matringe M. (2009). Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol. 149: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W.I., Voelker D.R., Basset G., Hanson A.D. (2001). Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 276: 35523–35529 [DOI] [PubMed] [Google Scholar]
- Rudhe C., Chew O., Whelan J., Glaser E. (2002). A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 30: 213–220 [DOI] [PubMed] [Google Scholar]
- Salter M., Knowles R.G., Pogson C.I. (1986). Quantification of the importance of individual steps in the control of aromatic amino acid metabolism. Biochem. J. 234: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield J., Taylor N., Fauquet C., Chen S. (2006). The cassava (Manihot esculenta Crantz) root proteome: Protein identification and differential expression. Proteomics 6: 1588–1598 [DOI] [PubMed] [Google Scholar]
- Shiman R. (1987). Purification and assay of rat liver phenylalanine 4-monooxygenase. Methods Enzymol. 142: 17–27 [DOI] [PubMed] [Google Scholar]
- Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehl D.L., Conn E.E. (1988). Kinetic and regulatory properties of arogenate dehydratase in seedlings of Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 260: 822–829 [DOI] [PubMed] [Google Scholar]
- Siltberg-Liberles J., Steen I.H., Svebak R.M., Martinez A. (2008). The phylogeny of the aromatic amino acid hydroxylases revisited by characterizing phenylalanine hydroxylase from Dictyostelium discoideum. Gene 427: 86–92 [DOI] [PubMed] [Google Scholar]
- Song J., Xia T., Jensen R.A. (1999). PhhB, a Pseudomonas aeruginosa homolog of mammalian pterin 4a-carbinolamine dehydratase/DCoH, does not regulate expression of phenylalanine hydroxylase at the transcriptional level. J. Bacteriol. 181: 2789–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thöny B., Auerbach G., Blau N. (2000). Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347: 1–16 [PMC free article] [PubMed] [Google Scholar]
- Tong J.H., Kaufman S. (1975). Tryptophan hydroxylase. Purification and some properties of the enzyme from rabbit hindbrain. J. Biol. Chem. 250: 4152–4158 [PubMed] [Google Scholar]
- Tzin V., Galili G. (2010). The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Yamamoto K.i., Kobayashi N., Yoshitama K., Teramoto S., Komamine A. (2001). Isolation and purification of tyrosine hydroxylase from callus cultures of Portulaca grandiflora. Plant Cell Physiol. 42: 969–975 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao G., Xia T., Song J., Jensen R.A. (1994). Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc. Natl. Acad. Sci. USA 91: 1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.