Abstract
MicroRNA expression profiling and quantitative reverse transcription-PCR analysis of the superior temporal gyrus and the dorsolateral prefrontal cortex revealed a significant schizophrenia-associated increase in global microRNA expression. This change was associated with an elevation of primary microRNA processing and corresponded with an increase in the microprocessor component DGCR8. The biological implications for this extensive increase in gene silencing are profound, and were exemplified by members of the miR-15 family and other related microRNA, which were significantly upregulated in both brain regions. This functionally convergent influence is overrepresented in pathways involved in synaptic plasticity and includes many genes and pathways associated with schizophrenia, some of which were substantiated in vitro by reporter gene assay. Given the magnitude of microRNA changes and their wide sphere of influence, this phenomenon could represent an important dimension in the pathogenesis of schizophrenia.
Keywords: schizophrenia, microRNA, miRNA, DGCR8, dicer
Introduction
Schizophrenia is a debilitating neuropsychiatric disorder characterized by a diverse range of symptoms and neurocognitive impairments. Although its exact pathogenesis remains obscure, there is a broad consensus that schizophrenia is of neurodevelopmental origin, arising through the complex interplay of numerous genetic and environmental factors.1 Some insight into the molecular interactions within this matrix has been provided by high-throughput gene expression analyses of post-mortem brain tissues.2, 3, 4, 5, 6 These investigations have consistently shown that numerous genes are affected in schizophrenia. Although some of these changes reflect alterations in known candidate genes and their downstream influences, most are inexplicable and their origins may lie well beyond the reach of these well-known mechanisms. Despite the perplexing array of findings, there are patterns in schizophrenia-associated gene expression that are indicative of systematic regulatory dysfunction. In contexts where these coincide with functional pathways, for example, in neurotransmitter systems and neural development, they support plausible hypotheses that correspond with our limited understanding of schizophrenia pathophysiology. Efforts to understand the underlying mechanisms driving these changes in gene expression have focused predominantly on genetic and epigenetic influences on transcription, mediated by alterations in signal-transduction pathways, their transcription factors, or gene promoter elements and associated chromatin structure. However, some recent studies have emerged showing that post-transcriptional influences on gene expression mediated by changes in non-coding microRNA (miRNA) expression are also associated with schizophrenia.7 We identified an increase in miR-181b expression in the superior temporal gyrus in schizophrenia and analyzed its activity with respect to candidate genes suppressed in the same tissue.8 We now present evidence suggesting that this represented a mere fraction of the overall scale of schizophrenia-associated alteration in miRNA expression. This striking deviation in global miRNA expression was observed in post-mortem tissue from both the superior temporal gyrus (STG) and the dorsolateral prefrontal cortex (DLPFC) and seemed to involve an increase in miRNA biogenesis.
Materials and methods
Tissue collection and dissection
Fresh frozen post-mortem STG gray matter tissue from 21 subjects with schizophrenia and 21 non-psychiatric controls and DLPFC gray matter from 15 subjects with schizophrenia and non-psychiatric controls was obtained through the NSW Tissue Resource Centre, The University of Sydney, Australia. The gray matter tissue was taken from the outer edge of blocks of STG tissue that was obtained from the most caudal coronal brain slice containing the STG (Brodmann's Area 22) or DLPFC (Brodmann's Area 9). Dissections were performed blind on coded tissue blocks such that disease status was not identifiable during this procedure. In all cases, a diagnosis of schizophrenia in accordance with DSM-IV criteria was confirmed by medical file review using the Item Group Checklist of the Schedules for Clinical Assessment in Neuropsychiatry and the Diagnostic Instrument for Brain Studies. Consent was obtained from the next of kin, and subjects with a significant history of drug or alcohol abuse, or other condition or gross neuropathology that might have influenced the agonal state were excluded. In addition, control subjects were excluded if there was a history of alcoholism or suicide. All subjects were of Caucasian descent. Subjects with schizophrenia were matched for gender, age, brain hemisphere, postmortem interval (PMI) and pH (Supplementary Table 1). The tissue was dissected and RNA extracted as described earlier.8
MicroRNA expression profiling
The microarray procedure was carried out as described earlier,9 with some alterations.8 miRNAs were labeled directly by a ligation approach using 3 μg total RNA. A synthetic reference library consisting of oligonucleotides (representing the entirety of miRBase version 7.1) was labeled with Ulysis platinum-conjugated AlexaFluor 647 (equivalent to Cy5) for detection in the control channel, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The unconjugated label was then removed by gel filtration through a Sephadex G-25 spin column (GE Healthcare, Piscataway, NJ). The labeled reference library was used at 1/700 dilution, alongside the Cy3-labeled miRNAs in each array hybridization. Microarrays were prepared using anti-sense Locked Nucleic Acid (LNA) oligonucleotides (Exiqon, Vedbaek, Denmark) corresponding to the miRBase Version 7.1, containing 322 human miRNA sequences. The oligonucleotide probes were printed in quadruplicate onto GAPS-2 glass slides (Corning, Lowell, MA) by the Australian Genome Research Facility. Feature deposition consistency was analyzed before hybridization by examining spot autofluorescence and after hybridization by comparing the reference intensity (AlexaFluor 647) from probe replicates. Single- and double-mismatch probes for a number of miRNAs were also printed in quadruplicate to assess the hybridization specificity. The slides were prepared as described earlier8 and hybridized for 2 h at 55 °C in a rotary hybridization oven. Arrays were then scanned and raw image extracted with a Genepix 4000B Scanner and Genepix Pro 3.0 software (Axon Instruments, Sunnyvale, CA). Features with a median Cy3 pixel intensity value more than 200% background had the background subtracted and were normalized with respect to U6 small nuclear RNA (snRNA) expression. Differential miRNA expression was analyzed using Significance Analysis of Microarrays(SAM) version 2.23 (Stanford University)10 (available from http://www-stat.stanford.edu/~tibs/SAM/). SAM uses permutations of the data to determine whether multiple differences are significantly related to the disorder. The threshold for significance was determined with respect to the false-discovery rate. Any miRNA showing a false-discovery rate <5% was considered as significantly altered. The SAM analysis parameters were: two-class unpaired analysis, t-test statistic and 5000 permutations of the data.
Total RNA analysis
Total RNA from the STG was quantified using a RNA Quant-it assay according to the manufacturer's instructions (Invitrogen). Individual and pooled RNA samples (schizophrenia and control groups) (100 ng μl−1) were dephosphorylated in 1 × SAP buffer and 1 unit of shrimp alkaline phosphatase (Fermentas, Vilnius, Lithuania) at 37 °C for 30 min. After heat inactivation, the dephosphorylated RNA was then rephosphorylated in the presence of [32P-γ] adenosine triphosphate in 1 × polynucleotide kinase forward reaction buffer and 1 unit of polynucleotide kinase (Fermentas). Labeled RNA was then combined with an equal volume of formamide/bromophenol blue/25 m EDTA loading dye and denatured at 95 °C before electrophoresis on a 16% denaturing (TBE/Urea) sequencing gel. Images were generated and analyzed from the radiolabeled gel using a Typhoon phosphorimager and ImageQuant software (GE Bioscience, Piscataway, NJ).
Quantitative reverse transcription-PCR (Q-PCR)
Reverse transcription and Q-PCR was conducted as described earlier8 with some adaption. Reactions were carried out in a final volume of 12.5 μl using a 7500 Real Time PCR system (Applied Biosystems, Foster City, CA). For miRNA expression, the delta Ct was calculated by subtracting the Ct of the endogenous controls (geometric mean of U6 snRNA, U44 small nucleolar RNA (snoRNA) and U49 snoRNA) from the Ct of the miRNA. For mRNA expression, the delta Ct was calculated by subtracting the Ct of the endogenous controls (geometric mean of β-glucuronidase and hydroxymethylbilane synthase) from the Ct of the mRNA. Primary miRNA (pri-miRNA) transcripts and processed precursor miRNA (pre-miRNA) hairpins were reverse transcribed using random primers as described for mRNA.8 Primers specific for the pri-miRNAs were designed to amplify a segment of the primary transcript upstream of the precursor sequence. The pre-miRNA primers were designed to hybridize to partially single-stranded segments within the precursor hairpin in both the precursor miRNA and the primary miRNA transcript. A dissociation curve analysis was performed for every Q-PCR reaction. A single peak was observed for all reactions to confirm the specificity of the reaction. In some cases, further confirmation was achieved by gel electrophoresis, in which products of the correct size were observed on the gel.
Statistical analyses
The demographic characteristics for each cohort were compared using Student's t-tests to verify matching for age, PMI and tissue pH. PMI was higher in the DLPFC schizophrenia group (29.9±11.0 h) when compared with the control group (21.9±9.9 h; P=0.045). The Kolmogorov–Smirnov test for normality was used to determine whether each data set was normally distributed to satisfy the conditions for parametric statistics (Prism 4.00, GraphPad Software, Inc., La Jolla, CA). To determine the significance of differential miRNA expression between the two cohorts, an un-paired one-tailed t-test was applied. Differential gene expression (mRNA) was determined by un-paired two-tailed Student's t-tests, in which significance was considered as P<0.05. In addition, Pearson's product moment correlations were performed for miRNA/mRNA expression levels and the demographic variables (age, PMI and pH) using control data. Where expression showed correlation, the significance was further tested for analysis of covariance (ANCOVA) using the demographic variable as a covariate (SPSS Statistics 17.0, SPSS Inc., Chicago, IL).
Target gene and pathway analyses
Putative target genes were identified using the publically available database, TargetCombo (which combines information gathered from multiple databases such as Diana-microT, PicTar, TargetScanS and miRanda; available at http://www.diana.pcbi.upenn.edu/cgi-bin/TargetCombo.cgi). Pathway analyses of the target gene lists were carried out using the Database for Annotation, Visualization, and Integrated Discovery bioinformatics resource (available at http://david.abcc.ncifcrf.gov/).
Cell culture, transfection and target gene reporter assay
Human embryonic kidney cells (HEK)-293 cell cultures were maintained as confluent monolayers at 37 °C with 5% CO2 and 90% humidity in Dulbecco's modified Eagles's medium (DMEM) with 10% (vol vol−1) fetal calf serum, 20 m HEPES (4-(2-hydroxyethyl)-1piperazineethanesulfonic acid), 0.15% (wt vol−1) sodium bicarbonate and 2 m-glutamine. Cells were seeded into 24-well plates and transfected 24 h later using Lipofectamine 2000 (Invitrogen). In each case transfections were performed according to the manufacturer's instructions, with 100 n synthetic miRNA or anti-miR oligonucleotide (Table 1). Validation of predicted target genes was accomplished by co-transfecting HEK293 cells with synthetic miRNA or an LNA-modified antisense inhibitor and recombinant firefly luciferase reporter gene constructs containing 3′-UTR sequences substituted from the target gene. Oligonucleotides encoding target gene miRNA recognition elements were annealed to form SpeI and HindIII restricted overhangs of a ligatable cassette compatible with SpeI and HindIII digested pMIR-REPORT vector (Ambion, Austin, TX) (Table 1). Reporter gene silencing in response to miRNA co-transfection was monitored with respect to a control plasmid expressing renilla luciferase using the dual-luciferase reporter assay (Promega, Madison, WI). To control for the nonspecific effects associated with siRNA transfection, the controls were co-transfected with mutant miRNAs or mutant anti-miRs. The miR-15 family miRNAs (that share an identical seed pairing region) were all controlled with a mutant form of miR-16 containing a scrambled seed-pairing region. Transfection of miR-107 was controlled by a mutant control version of miR-107 containing a scrambled seed pairing sequence. Although these reporter gene constructs, consisting of the miRNA recognition elements (MREs) and a small amount of 3′-UTR (Table 1), are useful for evaluating target–miRNA interactions, the reporter genes' mRNA secondary structure will be different from that of the native transcript and could positively or negatively influence the extent of gene silencing observed.
Table 1. Oligonucleotide sequences.
| Type | Name | Sequence | Target |
|---|---|---|---|
| Primersa | U6-probe | 5′-GCCATGCTAATCTTCTCTGTATC-3′ | U6 snRNA |
| U6-F339 | 5′-CGGCAGCACATATACTAAAATTGG-3′ | U6 snRNA | |
| U49-F | 5′-ATCACTAATAGGAAGTGCCGTC-3′ | U49 snoRNA | |
| U49-R | 5′-ACAGGAGTAGTCTTCGTCAGT-3′ | U49 snoRNA | |
| U44-F | 5′-TGATAGCAAATGCTGACTGA-3′ | U44 snoRNA | |
| U44-R | 5′-CAGTTAGAGCTAATTAAGACC-3′T | U44 snoRNA | |
| 107-F | 5′-AGCAGCATTGTACAG-3′ | miR-107 | |
| 107-R | 5′-GTAAAACGACGGCCAGTTGATAGCC-3′ | miR-107 | |
| 15a-Fb | 5′-T+AG+CAGCACATAA-3′ | miR-15a | |
| 15a-R | 5′-GTAAAACGACGGCCAGTCACAAACCA-3′ | miR-15a | |
| 15b-F | 5′-TAGCAGCACATCAT-3′ | miR-15b | |
| 15b-R | 5′-GTAAAACGACGGCCAGTTGTAAACC-3′ | miR-15b | |
| 16-F | 5′-TAGCAGCACATCAT-3′ | miR-16 | |
| 16-R | 5′-GTAAAACGACGGCCAGTTGTAAACC-3′ | miR-16 | |
| 128a-F | 5′-TCACAGTGAACCG-3′ | miR-128a | |
| 128a-R | 5′-GTAAAACGACGGCCAGTAAAAGAGAC-3′ | miR-128a | |
| 181a-F | 5′-AACATTCAACGCTG-3′ | miR-181a | |
| 181a-R | 5′-GTAAAACGACGGCCAGTACTCACCGA-3′ | miR-181a | |
| 181b-F | 5′-TTTCTAACATTCATTGCT-3′ | miR-181b | |
| 181b-R | 5′-CAACCTTCTCCCACCGAC-3′ | miR-181b | |
| 195-F | 5′-T+AGCAGCACAGA-3′ | miR-195 | |
| 195-R | 5′-GTAAAACGACGGCCAGTGCCAATATT-3′ | miR-195 | |
| 19a-F | 5′-TGTGCAAATCTATGC-3′ | miR-19a | |
| 19a-R | 5′-GTAAAACGACGGCCAGTTCAGTTTT-3′ | miR-19a | |
| 20a-F | 5′-T+AA+AGTGCTTATAGTG-3′ | miR-20a | |
| 20a-R | 5′-GTAAAACGACGGCCAGTCTACCTG-3′ | miR-20a | |
| 219-F | 5′-T+GAT+TGTCCAAAC-3′ | miR-219 | |
| 219-R | 5′-GTAAAACGACGGCCAGTAGAATTGC-3′ | miR-219 | |
| 26b-F | 5′-TT+CA+AGTAATTCAGG-3′ | miR-26b | |
| 26b-R | 5′-GTAAAACGACGGCCAGTAACCTAT-3′ | miR-26b | |
| 27a-F | 5′-TT+CACAGTGGCTA-3′ | miR-27a | |
| 27a-R | 5′-GTAAAACGACGGCCAGTGCGGAACT-3′ | miR-27a | |
| 29b-F | 5′-T+AG+CACCATTTGAA-3′ | miR-29c | |
| 29c-R | 5′-GTAAAACGACGGCCAGTTAACCGAT-3′ | miR-29c | |
| 338-F | 5′-AA+CAATATCCTGGT-3′ | miR-338 | |
| 338-R | 5′-GTAAAACGACGGCCAGTCACTCAGC-3′ | miR-338 | |
| 7-F | 5′-T+GGAAGACTAGTGA-3′ | miR-7 | |
| 7-R | 5′-GTAAAACGACGGCCAGTACAACAAAA-3′ | miR-7 | |
| let-7d-F | 5′-AGA+GGTAGTAGGTT-3′ | let-7d | |
| let-7d-R | 5′-GTAAAACGACGGCCAGTAACTATGC-3′ | let-7d | |
| let-7e-F | 5′-TG+AGGTAGGAGGT-3′ | let-7e | |
| let-7e-R | 5′-GTAAAACGACGGCCAGTACTATACA-3′ | let-7e | |
| M13-F | 5′-GTAAAACGACGGCCAGT-3′ | Rev primer for miRNA Q-PCR | |
| GUSB-F | 5′-GCCAATGAAACCAGGTATCCC-3′ | GUSB | |
| GUSB-R | 5′-GCTCAAGTAAACAGGCTGTTTTCC-3′ | GUSB | |
| HMBS-F | 5′-GAGAGTGATTCGCGTGGGTA-3′ | HMBS | |
| HMBS-R | 5′-CAGGGTACGAGGCTTTCAAT-3′ | HMBS | |
| FXR2-F | 5′-ACCGCCAGCCAGTGACTGTG-3′ | FXR2 | |
| FXR2-R | 5′-AGTCACCCTTCTGTCCTGAAA-3′ | FXR2 | |
| DICER1-F | 5′-CACATCAATAGATACTGTGCT-3′ | DICER | |
| DICER-R | 5′-TTGGTGGACCAACAATGGAGG-3′ | DICER | |
| DGCR8-F | 5′-GCTGAGGAAAGGGAGGAG-3′ | DGCR8 | |
| DGCR8-R | 5′-ACGTCCACGGTGCACAG-3′ | DGCR8 | |
| DROSHA-F | 5′-AAGCGTTAATAGGAGCTGTTTACT-3′ | DROSHA | |
| DROSHA-R | 5′-CGTCCAAATAACTGCTTGGCT-3′ | DROSHA | |
| XPO5-F | 5′-ATATATGAGGCACTGCGCC-3′ | EXP-5 | |
| XPO5-R | 5′-AAACTGGTCCAGTGAGTCCTT-3′ | EXP-5 | |
| DDX26-F2 | 5′-AGATCCGAAAGCCAGGAAGAAAA-3′ | DDX26 | |
| DDX26-R2 | 5′-TTTGTAAACTGCCTTGCACATGC-3′ | DDX26 | |
| DDX5-F | 5′-AAGGATGAAAAACTTATTCGT-3′ | DDX5 | |
| DDX5-R | 5′-TTTTCCATGTTTGAATTCATT-3′ | DDX5 | |
| DDX17-F | 5′-GTGAAAAAGACCACAAGTTGA-3′ | DDX17 | |
| DDX17-R | 5′-TACACATAGCTGGCCAACCAT-3′ | DDX17 | |
| FXR2-F | 5′-ACCGCCAGCCAGTGACTGTG-3′ | FXR2 | |
| FXR2-R | 5′-AGTCACCCTTCTGTCCTGAAA-3′ | FXR2 | |
| pri-181b-2-F1 | 5′-AAGAAGAGCCAGGAGTCAGC-3′ | pri-181b-2 | |
| pri-181b-2-R1 | 5′-TCAGTTGGTGGGGTTGCCTT-3′ | pri-181b-2 | |
| pre-181b-2-F | 5′-CTGATGGCTGCACTCAACAT-3′ | pre-181b | |
| pre-181b-2-R | 5′-TGATCAGTGAGTTGATTCAGACT-3′ | pre-181b | |
| pri-26b-F | 5′-CCGTGCTGTGCTCCCT-3′ | pri-26b | |
| pri-26b-R | 5′-CGAGCCAAGTAATGGAGAACAG-3′ | pri-26b | |
| pre-26b-F | 5′-GACCCAGTTCAAGTAATTCAGGA-3′ | pre-26b | |
| pre-26b-R | 5′-CGAGCCAAGTAATGGAGAACAG-3′ | pre-26b | |
| Cassettesc | VSNL1-107-T | 5′-CTAGTTCCTCCAAAGCCTGGGCAGAAATGTGCTGCAAA-3′ | VSNL1 |
| VSNL1-107-B | 5′-AGCTTTTGCAGCACATTTCTGCCCAGGCTTTGGAGGAA-3′ | VSNL1 | |
| RELN-107-T | 5′-CTAGTTTACTTGTTATGTTGTAATATTTTGCTGCTGAATTT-3′ | RELN | |
| RELN-107-B | 5′-AGCTAAATTCAGCAGCAAAATATTACAACATAACAAGTAAA-3′ | RELN | |
| HTR2A-107-T | 5′-CTAGCTATTTTCAAGTGGAAACCTTGCTGCTATGCTGTTCA-3′ | HTR2A | |
| HTR2A-107-B | 5′-AGCTTGAACAGCATAGCAGCAAGGTTTCCACTTGAAAATAG-3′ | HTR2A | |
| GRIN3A-107-T | 5′-CTAGGCACAAACCCTATCAAGAGCTGCTGCTTCCCT-3′ | GRIN3A | |
| GRIN3A-107-B | 5′-AGCTAGGGAAGCAGCAGCTCTTGATAGGGTTTGTGC-3′ | GRIN3A | |
| PLEXNA2-107-T | 5′-CTAGGACAGTTCTGCCTCTGTGACTGCTGCTTTGCATG-3′ | PLEXNA2 | |
| PLEXNA2-107-B | 5′-AGCTCATGCAAAGCAGCAGTCACAGAGGCAGAACTGTC-3′ | PLEXNA2 | |
| DLG4-107-T | 5′-CTAGGTCCGGGAGCCAGGGAAGACTGGAAATGCTGCCG-3′ | DLG4 | |
| DLG4-107-B | 5′-AGCTCGGCAGCATTTCCAGTCTTCCCTGGCTCCCGGAC-3′ | DLG4 | |
| DRD1-107-T | 5′-CTAGAATTTACGATCTTAGGTGGTAATGAAAAGTATATGCTGCTTTG-3′ | DRD1 | |
| DRD1-107-B | 5′-AGCTCAAAGCAGCATATACTTTTCATTACCACCTAAGATCGTAAATT-3′ | DRD1 | |
| GRM7-107-T | 5′-CTAGGTTTGTAATAAGTACTTTCGTTAATCTTGCTGCTTATGTG-3′ | GRM7 | |
| GRM7-107-B | 5′-AGCTCACATAAGCAGCAAGATTAACGAAAGTACTTATTACAAAC-3′ | GRM7 | |
| RGS4-107-T | 5′-AATGCACTAGTCCACATTGTAGCCTAATATTCATGCTGCCTGCCA TGAAGCTTAATGC-3′ | RGS4 | |
| RGS4-107-B | 5′GCATTAAGCTTCATGGCAGGCAGCATGAATATTAGGCTACAATGTGGA CTAGTGCATT-3′ | RGS4 | |
| miRNAd | miR-107+ | 5′-AGCAGCAUUGUACAGGGCUAUCA-3′ | miR-107 |
| miR-107− | 5′-AUAGCCCUGUACAAUGCUGUAUU-3′ | miR-107 | |
| miR-15a+ | 5′-UAGCAGCACAUAAUGGUUUGUG-3′ | miR-15a | |
| miR-15a− | 5′-CAAACCAUUAUGUGCUGUUAUU-3′ | miR-15a | |
| miR-15b+ | 5′-UAGCAGCACAUCAUGGUUUACA-3′ | miR-15b | |
| miR-15b− | 5′-UAAACCAUGAUGUGCUGUUAUU-3′ | miR-15b | |
| miR-16+ | 5′-UAGCAGCACGUAAAUAUUGGCG-3′ | miR-16 | |
| miR-16− | 5′-CCAAUAUUUACGUGCUGUUAUU-3′ | miR-16 | |
| miR-195+ | 5′-UAGCAGCACAGAAAUAUUGGC-3′ | miR-195 | |
| miR-195− | 5′-CAAUAUUUCUGUGCUGUUAUU-3′ | miR-195 | |
| control-miRNA-1+ | 5′-AUCCACCACGUAAAUAUUGGCG-3′ | miR-15 family | |
| control-miRNA-1− | 5′-CCAAUAUUUACGUGGUGGAUCG-3′ | miR-15 family | |
| control-miRNA-2+ | 5′-UCCACCAAUGUACAGGGCUAUCA-3′ | miR-107 | |
| control-miRNA-2− | 5′-AUAGCCCUGUACAUUGGUGAAUU-3′ | miR-107 | |
| Anti-miRse | Anti-miR-107 | 5′-T+GAT+AGC+CCT+GTA+CAA+TGC+TG-3′ | miR-107 |
| Anti-miR-15a | 5′-C+ACA+AAC+CAT+TAT+GTG+CTG+CTA-3′ | miR-15a | |
| Anti-miR-15b | 5′-T+GTA+AAC+CAT+GAT+GTG+CTG+CTA-3′ | miR-15b | |
| Anti-miR-16 | 5′-C+GCC+AAT+ATT+TAC+GTG+CTG+CTA-3′ | miR-16 | |
| Anti-miR-195 | 5′-G+CCA+ATA+TTT+CTG+TGC+TGC+TA-3′ | miR-195 | |
| Control anti-miR-1 | 5′-C+GCC+AAT+ATT+TAC+GTG+GTG+GAT-3′ | miR-15 family | |
| Control anti-miR-2 | 5′-T+GAT+AGC+CCT+GTA+CAT+TGG+TG-3′ | miR-107 |
The direction of primers with respect to the target sequence was denoted as either F or R for forward and reverse, respectively. The underlined sequence is not gene specific and was used to provide a primer recognition sequence. For miRNA Q-PCR, the Rev primer is used for reverse transcription, and the For primer is used in the Q-PCR with M13-F as the reverse primer.
The positions of LNA-modified bases are preceded by a ‘+' symbol.
SpeI/HindIII cassettes containing putative target recognition elements were used to generate recombinant luciferase reporter gene constructs. ‘T' indicates top strand and ‘B' indicates bottom strand.
Synthetic miRNAs are used to over-express microRNA. ‘+' indicates top strand and ‘–‘ indicates bottom strand.
Anti-miRs are used to suppress endogenous microRNA.
Results
Elevation of miRNA expression in the STG in schizophrenia
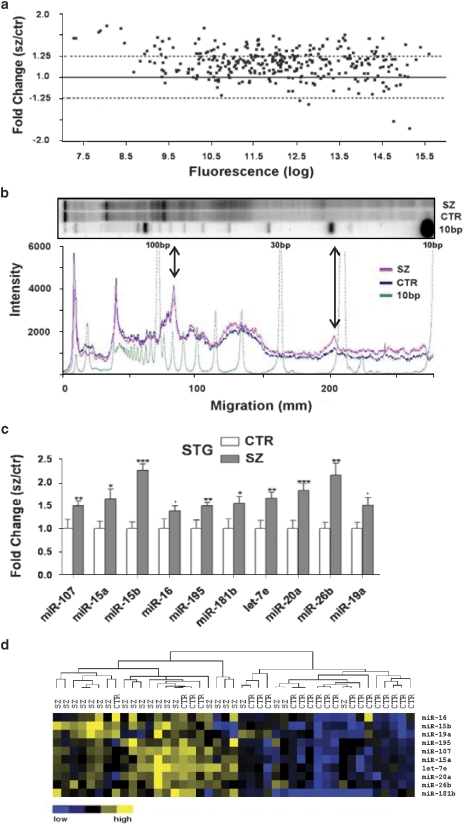
Changes in miRNA expression have broad implications for disease, as each miRNA molecule is capable of influencing the expression of hundreds of target genes. The expression of miRNA has been shown to be important during development, particularly in the mammalian brain;11 therefore, it is plausible that these molecules have great significance in neurodevelopmental disorders such as schizophrenia. In this study, we analyzed miRNA expression in the STG (Brodmann's Area 22, 17 matched pairs of schizophrenia and non-psychiatric controls) and the DLPFC (Brodmann's Area 9, 15 matched pairs), using a microarray printed with LNA-modified capture probes corresponding to miRBase version 7.1 (Exiqon).9 In contrast to our earlier study, which identified two differentially expressed miRNAs in a relatively small cohort of STG tissue normalized by global miRNA expression,8 these arrays were also furnished with two probes specific for different sites in the U6 snRNA that enabled external or miRNA-independent normalization of miRNA expression between samples. In this new analysis, miR-181b (earlier found to be upregulated in the STG) represented only one of many significantly elevated miRNAs in the schizophrenia group. This observation, apparent in scatter plots of the average expression between schizophrenia and controls for each miRNA (Figure 1a), implied there was a schizophrenia-associated global elevation of miRNA expression in the STG. The significance of these changes was supported by SAM analysis, which reported that 59 miRNAs (or 21% of the miRNAs expressed in the STG) were upregulated (Supplementary Table 2). With the apparent scope of this alteration in small RNA expression, we considered that it might be possible to directly visualize this in the small RNA fraction of 32P-labeled total RNA separated on a sequencing gel. On pooled samples this experiment revealed that the ∼22-nt band (corresponding to the miRNAs) was 1.5 times more intense for the schizophrenia sample than that of the controls (normalized to the ∼75-nt tRNA band) (Figure 1b). In individual samples, the schizophrenia-associated increase in the 22/75-mer ratio was 1.26-fold (P<0.05), indicative of a general increase in small RNA expression in schizophrenia (Supplementary Table 3).
Figure 1.
Schizophrenia-associated miRNA expression in the superior temporal gyrus (STG). (a) Average fold change of miRNA expression (schizophrenia to control) was plotted against log2-transformed fluorescence values (n=17 matched pairs). A global increase in miRNA expression in the STG in schizophrenia is indicated by the majority of miRNA showing a ratio >1.0. (b) Electrophoresis of dephosphorylated total RNA (pooled samples) labeled with polynucleotide kinase (PNK). Whole lane densitometry of the phosphor image indicated an increase in small RNA in the schizophrenia cohort (pink trace) compared with the controls (blue trace), particularly in the small RNA fraction (20–24 nt) region corresponding to most miRNAs (indicated by long arrow). Densitometry was normalized to the ∼75-nt band in each lane (indicated by short arrow). (c) Increased miRNA expression in the STG was validated using quantitative real-time reverse transcription-PCR (Q-PCR) (n=21 matched pairs). Level of expression for controls was set at 1. Bars are mean+s.e.m. *P<0.05; **P<0.01; ***P<0.001;+miR-16 and miR-19a were significant by t-test but were found to correlate with pH (r=−0.459 and −0.443, respectively) and fell short of significance by analysis of covariance. (d) Q-PCR expression data hierarchically clustered (correlation uncentered, average linkage; Cluster 3.0). Blue indicates low expression and yellow indicates high expression (Java Treeview; http://www.sourceforge.net/projects/jtreeview/files).
For more specific evidence of this phenomenon, Q-PCR assays for 10 miRNAs shown to be among the most significantly upregulated by microarray analysis were established. The relative expression values for each miRNA across an extended cohort, consisting of 21 schizophrenia samples and 21 matched controls of post-mortem cortical gray matter from the STG, were normalized with respect to the geometric mean of three constitutively expressed small RNAs (including U6 snRNA, U44 and U49 snoRNA) (Figure 1c). In each case the level of concordance with the microarray and Q-PCR was very high, and in many cases the average schizophrenia-associated increase was even greater by Q-PCR than that observed by microarray. This trend was also highly visible in individual samples clustered by expression and visualized by heat map (Figure 1d). Unsupervised clustering of these differentially expressed miRNAs was characterized by a high degree of segregation between the schizophrenia and control groups. Prominent among this group of miRNAs associated with schizophrenia was the apparent upregulation of the miR-15 family miRNAs such as miR-15a, miR-15b and miR-195, all of which share the same functionally important seed-pairing region and consequently a large proportion of target genes. In addition, miR-107 was among the most significantly upregulated miRNA in the STG and also shared a high degree of seed region homology with the miR-15 family (Figure 1c). Collectively they are predicted to target a wide array of target genes, with many of them implicated in schizophrenia, such as brain BDNF, NRG1, RELN, DRD1, HTR4, GABR1, GRIN1, GRM7, CHRM1 and ATXN2.
Elevation of miRNA expression in the DLPFC in schizophrenia
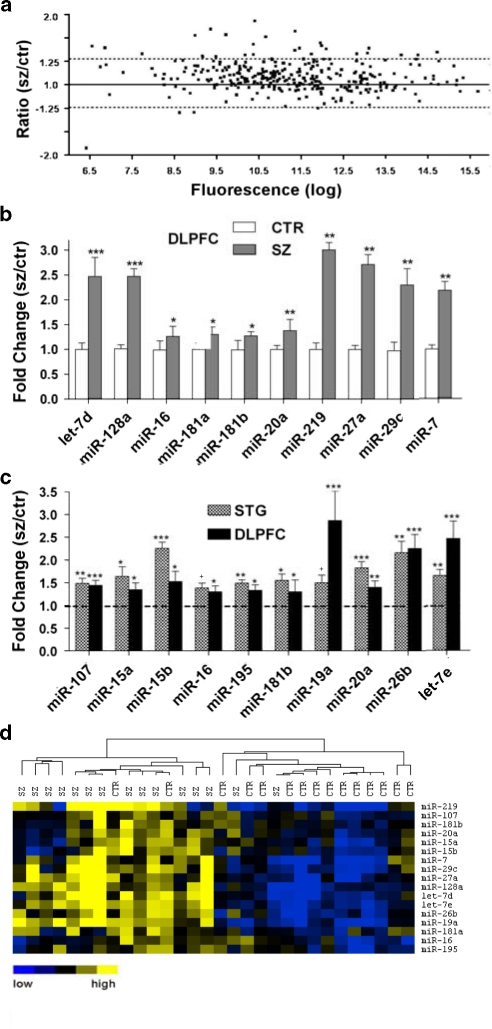
In view of the possibility that these changes in miRNA expression were merely STG-related phenomena, we initiated a similar investigation of the DLPFC (BA9), a region most frequently identified in the neuropathology of schizophrenia. Total RNA from post-mortem gray matter from a cohort of 15 cases with a history of schizophrenia and 15 matched controls with no record of psychiatric illness was extracted and subjected to microarray analysis as described for the STG. The miRNA expression profile in this tissue was similar to that in the STG, with 274 expressed miRNAs (compared with 280 in the STG). Importantly, the DLPFC showed a schizophrenia-associated global increase in miRNA expression that was broadly consistent with the observations in the STG (Figure 2a). According to SAM analysis, 26 (9.5%) of the miRNAs expressed in the DLPFC were significantly upregulated, such as miR-181b, miR-16 and miR-20a, which were also increased in the STG.
Figure 2.
Schizophrenia-associated miRNA expression in the dorsolateral prefrontal cortex (DLPFC). (a) miRNA expression in the DLPFC in schizophrenia was characterized by the global upregulation illustrated in this scatter plot (see Figure 1a for description), with the majority of individual miRNAs showing a ratio >1.0. (b) Increased miRNA expression in the DLPFC was validated using quantitative real-time reverse transcription-PCR (Q-PCR) (n=15 matched pairs). Level of expression for controls was set at 1. (c) Further Q-PCR expression analysis indicated that 11 miRNAs with altered expression showed an upregulation in both the superior temporal gyrus and DLPFC. Bars indicate mean fold change+s.e.m. *P<0.05; **P<0.01; ***P<0.001. (d) Q-PCR expression data were subjected to hierarchical clustering and heat map, shown as described in Figure 1d.
Again, to validate the microarray results, Q-PCR assays were performed on a subset of 10 differentially expressed miRNAs as described for the STG, using the expression of three constitutively expressed small RNAs as a reference. This analysis supported the array findings and in some cases exceeded the expectation by showing an even stronger schizophrenia-associated upregulation in miRNAs (Figure 2b). To determine whether the differentially expressed miRNAs in common extended beyond the scope identified by SAM analysis of the DLPFC microarray experiment, the DLPFC samples were also examined for the remaining schizophrenia-associated miRNAs validated for the STG cohort. These miRNAs, including let-7e, miR-19a, miR-26b, miR-107 and the remaining members of the miR-15 family, were all found to be significantly upregulated in the DLPFC by Q-PCR (Figure 2c). In a manner similar to the STG cohort, unsupervised hierarchical clustering of miRNA expression in individual DLPFC samples also produced very good segregation between the schizophrenia and control groups (Figure 2d).
Altered miRNA biogenesis in the STG and DLPFC
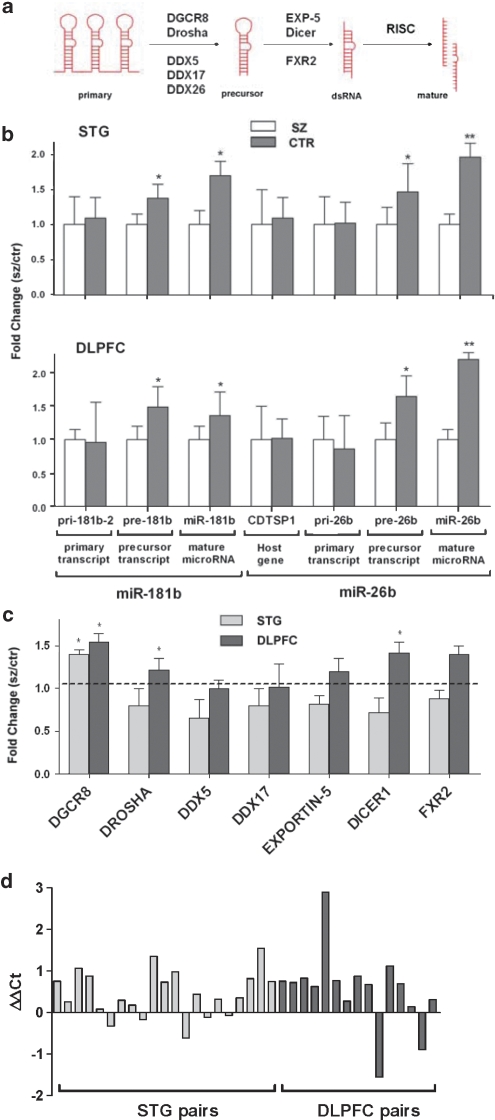
The scope and consistency of the schizophrenia-associated increase in miRNA expression led us to consider both miRNA processing and the activity of genes in the miRNA biogenesis pathway in this context. For this purpose, we analyzed the relative expression of pri-miRNA and pre-miRNA in addition to the mature miRNA transcripts for miR-181b and miR-26b. Interestingly, although there was a significant increase in pre-miRNA species (consistent with the mature miR-181b and miR-26b), there was no difference in transcription of the pri-miRNA, or the host gene mRNA (CDTSP1) for the intronic miR-26b (Figure 3b). This supported the hypothesis that there was a schizophrenia-associated increase in miRNA biogenesis rather than any change at the level of miRNA transcription. To further support this assertion, we examined the expression of the microprocessor constituents Drosha and DGCR8 involved in primary miRNA processing.12 The mRNA for both of these microprocessor components was found to be significantly upregulated in the DLPFC, and DGCR8 was also upregulated in the STG (Figure 3c). Importantly, DGCR8 was shown to be upregulated in 13 of the 15 matched pairs of DLPFC tissue, and in 16 of the 21 matched pairs of STG when analyzed as matched pairs rather than by their cohort-wide average (Figure 3d). These microprocessor components are thought to be rate limiting in the miRNA biogenesis pathway,13 and, as a consequence, their elevation in schizophrenia represents a highly plausible explanation for the corresponding increase in both pre-miRNA and mature miRNA expression. The expression of additional genes implicated in primary miRNA processing, such as the deadbox helicases DDX5 and DDX17, were also examined. Neither of these genes, however, was significantly altered in either part of the cerebral cortex. The difference in magnitude observed in differential miRNA and pre-miRNA expression (Figure 3b) was possibly due to some dilution of the pre-miRNA by pri-miRNA template as the pre-miRNA primer set has the capacity to amplify both of these sequences. However, it is also conceivable that other influences downstream of the microprocessor could further elevate mature miRNA expression and contribute to this difference. In this regard, we examined the expression of Exportin-5, Dicer and FXR2 by Q-PCR and found that Dicer was also significantly upregulated in schizophrenia in the DLPFC (Figure 3c). Dicer remained significant after ANCOVA using brain pH as a covariate, despite showing some correlation with pH in the DLPFC (r=−0.553; F=4.26; d.f.=1, 27; P=0.048).
Figure 3.
Alterations in miRNA processing in schizophrenia. (a) Simplified schematic of miRNA biogenesis showing genes involved in key enzymatic steps. (b) Primary, precursor and mature transcripts for miR-181b were analyzed by Q-PCR in the superior temporal gyrus (STG). The primary transcript was not altered in schizophrenia; however, the precursor and mature transcripts were both upregulated 1.4-fold (P=0.048) and 1.7-fold (P=0.039), respectively. The host gene of miR-26b (CDTSP1) and primary transcript were not altered. The precursor and mature miR-26b transcript were both upregulated in schizophrenia (1.5-fold (P=0.023) and 1.9-fold (P=0.001), respectively). In the dorsolateral prefrontal cortex (DLPFC), a similar trend followed. Host gene and primary transcripts were not altered in schizophrenia. For miR-181b, the precursor and mature were upregulated 1.5-fold (P=0.043) and 1.4-fold (P=0.039), respectively. For miR-26b, the precursor and mature were upregulated 1.6-fold (P=0.046) and 2.2-fold (P=0.001), respectively. (c) Expression of miRNA biogenesis genes was analyzed in the STG (n=21 matched pairs) and the DLPFC (n=15 matched pairs). DGCR8 was significantly upregulated in the STG and DLPFC, whereas Drosha and Dicer were significantly upregulated in the DLPFC only. Bars indicate mean fold change (schizophrenia to control)+s.e.m. *P<0.05; **P<0.01 unpaired Student's t-test. (d) DGCR8 expression was determined by Q-PCR in matched paired samples (SZ vs CTR). DGCR8 was upregulated in 16 out of 21 matched pairs of STG tissue and in 13 out of 15 matched pairs of DLPFC tissue.
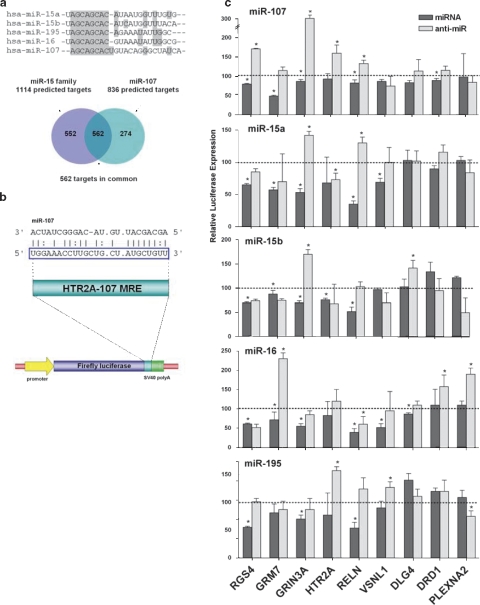
Target genes and pathways associated with miR-107 and miR-15 family microRNAs
To gain some appreciation of the biological implications of changes in miRNA expression observed in schizophrenia, we examined predicted miRNA targets and their associated pathways to see whether any patterns emerged. A conspicuous aspect of miRNA expression analyses in the STG and DLPFC was the prominence of all members of the miR-15 family and miR-107, all of which share a similar seed region (Figure 4a). To ascertain an overall perspective of this influence, a collection of predicted target genes derived using a range of search algorithms (collated on the TargetCombo web service http://www.diana.pcbi.upenn.edu/cgi-bin/TargetCombo.cgi)14 was subjected to pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery bioinformatics resource (http://david.abcc.ncifcrf.gov/tools.jsp)15 Predicted target genes common to the miR-15 family and miR-107 were highly enriched in pathways involved in neural connectivity and synaptic plasticity, such as axon guidance, long-term potentiation, Wingless/int (Wnt), epidermal growth factor receptor family (ErbB) and Mitogen-activated protein (MAP) kinase signaling (Supplementary Table 4). These processes are repeatedly implicated in the pathophysiology of schizophrenia and a number of individual genes have been shown to be associated with schizophrenia.
Figure 4.
Regulation of schizophrenia-associated reporter gene constructs by miRNA. (a) Sequence alignment showing miR-107 and the miR-15 family seed region homology (gray highlight). Together, the two groups were predicted to have many target genes in common (Venn diagram). (b) The pMIR-REPORT miRNA expression reporter system contains a firefly luciferase gene under the control of the cytomegalovirus (CMV) promoter. Putative miRNA recognition elements for various schizophrenia candidate genes were inserted into the multiple cloning site in the 3′-UTR of the firefly luciferase gene (HTR2A shown as an example). (c) A matrix chart showing the relative activity of reporter gene constructs (x-axis) in response to co-transfected miRNA (dark bars) or their cognate anti-miR (light bars). Relative luciferase activity for each reporter/miRNA/anti-miR combination was expressed as a percentage of the response to scrambled controls (+s.d.; *P<0.05).
Target gene silencing in vitro
To substantiate a link between these schizophrenia-associated target genes and altered expression in this group of miRNAs, the respective MREs from nine target genes, such as RGS4, GRM7, GRIN3A, HTR2A, RELN, VSNL1, DLG4, DRD1 and PLXNA2, were cloned into the 3′-UTR region of a luciferase reporter gene construct and co-transfected into a recipient cell line with miRNA or anti-miRs (miRNA antagonists). The extent of reporter gene activity and the influence of miRNAs were then determined by measuring the relative luciferase activity (Figure 4c).16 Many of these constructs behaved in accordance with the expectation and were significantly repressed in the presence of synthetic miRNA, and significantly de-repressed (increased luciferase) in the presence of the corresponding anti-miR (Figure 4c, Supplementary Table 5). The most consistently responsive targets were derived from the 3′-UTR of RGS4, GRM7, GRIN3A and RELN, whereas the least responsive was PLXNA2. With respect to the miRNA, miR-107 appeared to have the greatest overall effect, whereas miR-195 had the least effect on these target gene constructs. Collectively, these reporter assays showed a potential relationship between genes reported to be associated with schizophrenia and a large functionally related group of upregulated miRNAs.
Discussion
Analysis of the post-mortem brain tissue from two regions of the cerebral cortex in schizophrenia revealed a substantial alteration of the post-transcriptional regulatory environment, characterized by a global increase in miRNA expression. This change could have profound implications for the development and ongoing pathophysiology of the disorder, as each miRNA has the capacity to regulate the expression of hundreds of target genes. Studies in model systems have highlighted the significance of miRNA expression in brain development and a host of more specific neurobiological functions, such as regulation of the left–right asymmetry, long-term potentiation and establishment or maintenance of dendrites.17, 18, 19, 20, 21 In this work, we focused on the implications of miRNAs in the miR-15 family and closely related miR-107 as these functionally convergent miRNAs were consistently upregulated in both regions of the cerebral cortex in schizophrenia, and could collectively contribute a significant biological influence.
The miR-15 family of miRNAs has already been shown to have an important role in chronic lymphocytic leukemia, with a well-characterized association between a reduction of miR-15a/miR-16 concentration and increased expression of the anti-apoptosis gene BCL2.22 The relationship between miR-15 family expression and this gene may also have the opposite implications in schizophrenia, which has been associated with a downregulation of BCL2 expression. Reduced BCL2 expression in schizophrenia, perhaps in response to increased miR-15 family expression, is thought to contribute to elevated cortical apoptosis, cerebral atrophy and even a reduction in the risk of some forms of cancer.23, 24, 25
Pathway analysis of predicted target genes suggested that there are probably many other ways of influence of this group of miRNAs that are of significance to schizophrenia, such as axon guidance, long-term potentiation, WNT, ErbB and MAP kinase signaling (Supplementary Table 4). Many of these predicted target genes, such as RGS4, GRM7, GRIN3A, HTR2A, RELN, VSNL1, DLG4, DRD1 and PLXNA2, have been shown to be associated with schizophrenia.26, 27, 28, 29, 30, 31, 32, 33 In relation to the current study, RGS4 and VSNL1 were reported to be downregulated in the same STG tissue;8, 34 however, the expression of these and other candidate genes has not been analyzed at the protein level in these cohorts. To further examine the potential for a functional relationship between MREs in these candidate genes and the miR-15-related miRNAs, we established luciferase reporter constructs and measured the degree of silencing from individual miRNA. Regulation of 3′-UTR elements from the metabotropic glutamate receptor GRM7 and the N-methyl--aspartic acid (NMDA) receptor subunit GRIN3A was particularly strong and, along with DLG4 (PSD95; scaffold protein that supports these and other receptors in the post-synaptic density), provides a post-transcriptional mechanism that could underlie the many accounts of schizophrenia-associated glutamatergic hypofunction.35 It may also explain the apparent conflict between the schizophrenia-associated reduction of region-specific protein expression in the absence of change or even paradoxical increase in corresponding mRNA.36 Another target gene element that showed a consistent response to miR-107 and the miR-15 family miRNAs was one derived from the Reelin (RELN) 3′-UTR. RELN is a secreted glycoprotein involved in neuronal migration and synaptogenesis during development. It is also important for the establishment of long-term memory in the adult brain because of its role in the modulation of synaptic activity and dendritic spine development.37 RELN is a highly plausible candidate gene and its expression has been shown to be altered in schizophrenia.30, 38 Although this alteration has been associated with epigenetic regulation though promoter hypermethylation,39, 40 it is now conceivable that post-transcriptional gene silencing is also contributing to RELN dysregulation in schizophrenia.
Collectively, these experiments were broadly supportive of a role for this group of miRNAs in the regulation of schizophrenia-associated target genes; however, the response was quite variable for the individual miRNAs, with miR-107 showing the most consistent activity, whereas miR-195 appeared to have the least activity against the elements tested here. In contrast, a recent study has found that miR-195 (among others) was capable of regulating BDNF expression in vitro.41 The temporal and spatial expression pattern of this miR-15 family member in the DLPFC was inversely correlated with BDNF and may be important for the developmental regulation of this schizophrenia candidate gene.
Experiments in animal systems may also provide important insight into the behavioral consequences of altered cortical miRNA expression. In a recent study, mice treated with the NMDA receptor antagonist MK801 and hypomorphic GRIN1 (NR1) mutants showed a marked decrease in miR-219 expression.42 CaMKIIγ, a predicted target gene for this miRNA involved in NMDA signaling, was shown to be sensitive to miR-219 concentration in vitro. Moreover, suppression of miR-219 expression via intraventricular delivery of the corresponding LNA-modified anti-miR restored MK801 induced neurobehavioral dysfunction back to levels approaching that of the controls.42 Interestingly, in our study miR-219 was the most highly upregulated miRNA in the DLPFC and, in addition to the miR-15 family-related miRNA, could also be mediating a schizophrenia-associated reduction in NMDA signaling. These observations add support to the idea that these altered miRNAs are influential in the regulation of schizophrenia-associated genes and provide the basis of a model for the influence of disease-related miRNAs on genes involved in synaptic structure and function (Figure 5).
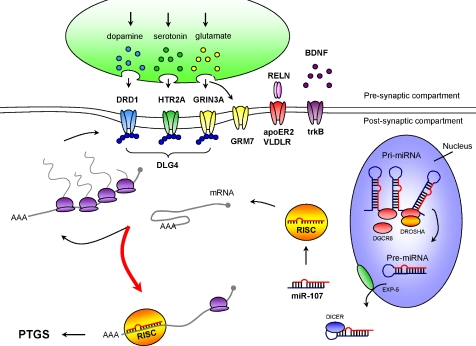
Figure 5.
Model for miRNA-associated dysregulation of synaptic structure and function in schizophrenia. The microprocessor activity is elevated in cortical nuclei as a consequence of a schizophrenia-associated increase in DGCR8 expression. The increase in pri-miRNA processing results in an increase in pre-miRNAs, which are exported from the nucleus and processed without delay by Exportin-5 (XPO5) and Dicer, respectively. Mature miRNAs are recruited into the RNA-induced silencing complex (RISC) and associate with the 3′-UTR of their target transcripts encoding synaptic components (among other proteins), such as neurotropins/ligands (BDNF, Reelin), neurotransmitter receptors (GRM7, GRIN3A, HTR2A, DRD1) and structural components of the post-synaptic density (DLG4). This association reduces the stability of the transcript and reduces its ability to undergo translation. Inappropriate levels of mature miRNA and gene silencing (red arrow) result in the reduction of synaptic proteins and consequently a loss of synaptic structure and function.
Although the examples of gene–miRNA interactions mentioned above and modeled in Figure 5 provide a conceptual framework for the mechanisms that may take place in the context of cortical miRNA dysregulation, they may only touch the surface of the broader ramifications for gene regulation in this altered environment. In this regard, it is worth noting that gene expression profiles in the same STG cohort (albeit smaller than the one examined in this study) showed more than twice as many downregulated genes in schizophrenia compared with those upregulated.5 This observation at the mRNA level has been observed in other studies as well,4, 43, 44 and may be reflective of an elevation in global gene silencing mediated by increased miRNA expression in these tissues.
The question of why such an extensive and consistent change in cortical miRNA expression was seen in the schizophrenia group led to the consideration of key components of the miRNA biogenesis pathway. Significantly, we identified a corresponding upregulation of the microprocessor component DGCR8 mRNA in both the STG and DLPFC. This alteration was consistent with an increase of both mature miRNA and precursor forms of miR-181b and miR-26b in the absence of a change in their level of transcription. Although the mechanism behind this apparent increase in DGCR8 expression at the mRNA level is currently unknown, the gene is situated within a region of the genome that is prone to spontaneous structural variation associated with schizophrenia and other neurodevelopmental disorders.45, 46, 47 Microdeletion at this locus (22q11.2) is responsible for the DiGeorge/Velocardiofacial syndrome, which is also strongly associated with schizophrenia.48 In a neurodevelopmental model, mice with a specific deficiency in DGCR8 and miRNA biogenesis showed similar behavioral deficits to the larger hemizygous deletion of a larger region of chromosome 16 syntenic to that of the 22q11 locus in humans.49 Although the deletion model and human syndrome, involving haploinsufficiency, does not accord with the increased DGCR8 activity observed in this study, the low copy repeats that give rise to deletion can also induce microduplication. Interestingly, the syndrome associated with duplication at this locus also appears to be associated with behavioral abnormalities and cognitive deficits akin to those seen in the deletion syndrome, but, like schizophrenia, shows fewer dysmorphic features.50 As a consequence of this more subtle phenotype, the frequency of this poorly characterized syndrome may be underrepresented through misdiagnosis.50 Microduplications are also more difficult to identify by classical cytogenetic approaches than microdeletions, which may have historically masked their relative abundance. In theory, however, they are just as abundant as microdeletions, a view supported in a recent molecular analysis of copy number variation, which found that microduplications outnumbered microdeletions and were more highly associated with schizophrenia.47 In view of these observations, it is possible that changes in DGCR8 expression in some individuals could be due to increased gene dosage through chromosomal microduplication.
Alternatively, the increase in DGCR8 may be due to transcriptional or post-transcriptional dysregulation. A recent study has shown that DGCR8 is post-transcriptionally autoregulated by its own microprocessor complex.51 Contrary to expectation, this is probably not miRNA-mediated as the 3′-UTR for DGCR8 mRNA is almost devoid of predicted MREs, an exception being an MRE for the microprocessor-independent miRNA-intron or mirtron hsa-miR-1227.52 The mechanism is instead related to the presence of primary miRNA-like hairpin structures in the DGCR8 mRNA, which are themselves substrates for cleavage by the microprocessor. Cleavage results in destabilization of the mRNA and reduction in DGCR8 expression.51 Polymorphisms with a capacity to destabilize these secondary structures in the mRNA could hinder feedback inhibition and result in DGCR8 elevation.
In conclusion, our data suggest that schizophrenia is associated with a global increase in miRNA biogenesis and expression in the cerebral cortex. This could have profound neurodevelopmental and broader neurological implications in the context of schizophrenia by influencing genes involved in cortical structure and neural plasticity. It also has significance for our understanding of the mechanism underlying patterns of cortical gene expression associated with the disorder.
Acknowledgments
This study was supported by the Schizophrenia Research Institute, using funding from NSW Health and the Henderson Foundation; a NARSAD Young Investigator Award (MC); a University of Newcastle pilot grant; a Hunter Medical Research Institute project grant; and the MC Ainsworth Research Fellowship in Epigenetics (MC). Tissues were received from the Australian Brain Donor Program's NSW Tissue Resource Centre, which is supported by The University of Sydney, National Health and Medical Research Council of Australia, Schizophrenia Research Institute, National Institute of Alcohol Abuse and Alcoholism, Neurobehavioural Genetics Unit and NSW Department of Health. We thank Professor Vaughan Carr for his critical reading of the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Harrison PJ. Schizophrenia: a disorder of neurodevelopment. Curr Opin Neurobiol. 1997;7:285–289. doi: 10.1016/s0959-4388(97)80018-9. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenhofer J, Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the amygdala in schizophrenia: up-regulation of genes located in the cytomatrix active zone. Mol Cell Neurosci. 2006;31:243–250. doi: 10.1016/j.mcn.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics. 2008;9:199. doi: 10.1186/1471-2164-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Webster MJ.Correlation analysis between genome-wide expression profiles and cytoarchitectural abnormalities in the prefrontal cortex of psychiatric disorders Mol Psychiatry 2008(E-pub ahead of print). [DOI] [PubMed]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarskog LF, Gilmore JH, Selinger ES, Lieberman JA. Cortical bcl-2 protein expression and apoptotic regulation in schizophrenia. Biol Psychiatry. 2000;48:641–650. doi: 10.1016/s0006-3223(00)00988-4. [DOI] [PubMed] [Google Scholar]
- Jarskog LF, Selinger ES, Lieberman JA, Gilmore JH. Apoptotic proteins in the temporal cortex in schizophrenia: high Bax/Bcl-2 ratio without caspase-3 activation. Am J Psychiatry. 2004;161:109–115. doi: 10.1176/appi.ajp.161.1.109. [DOI] [PubMed] [Google Scholar]
- Catts VS, Catts SV. Apoptosis and schizophrenia: is the tumour suppressor gene, p53, a candidate susceptibility gene. Schizophr Res. 2000;41:405–415. doi: 10.1016/s0920-9964(99)00077-8. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Koga M, Ishiguro H, Horiuchi Y, Arai M, Niizato K, et al. A polymorphism of the metabotropic glutamate receptor mGluR7 (GRM7) gene is associated with schizophrenia. Schizophr Res. 2008;101:9–16. doi: 10.1016/j.schres.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Mueller HT, Meador-Woodruff JH. NR3A NMDA receptor subunit mRNA expression in schizophrenia, depression and bipolar disorder. Schizophr Res. 2004;71:361–370. doi: 10.1016/j.schres.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Burnet PW, Eastwood SL, Harrison PJ. 5-HT1A and 5-HT2A receptor mRNAs and binding site densities are differentially altered in schizophrenia. Neuropsychopharmacology. 1996;15:442–455. doi: 10.1016/S0893-133X(96)00053-X. [DOI] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma T, Kato H, Arai H, Faull RL, McKenna PJ, Emson PC. Gene expression of PSD95 in prefrontal cortex and hippocampus in schizophrenia. Neuroreport. 2000;11:3133–3137. doi: 10.1097/00001756-200009280-00019. [DOI] [PubMed] [Google Scholar]
- Okubo Y, Suhara T, Suzuki K, Kobayashi K, Inoue O, Terasaki O, et al. Decreased prefrontal dopamine D1 receptors in schizophrenia revealed by PET [see comments] Nature. 1997;385:634–636. doi: 10.1038/385634a0. [DOI] [PubMed] [Google Scholar]
- Mah S, Nelson MR, Delisi LE, Reneland RH, Markward N, James MR, et al. Identification of the semaphorin receptor PLXNA2 as a candidate for susceptibility to schizophrenia. Mol Psychiatry. 2006;11:471–478. doi: 10.1038/sj.mp.4001785. [DOI] [PubMed] [Google Scholar]
- Bowden NA, Scott RJ, Tooney PA. Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia. Schizophr Res. 2007;89:165–168. doi: 10.1016/j.schres.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Curr Opin Pharmacol. 2007;7:48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006;11:737–747. doi: 10.1038/sj.mp.4001844. [DOI] [PubMed] [Google Scholar]
- Fatemi SH. Reelin glycoprotein: structure, biology and roles in health and disease. Mol Psychiatry. 2005;10:251–257. doi: 10.1038/sj.mp.4001613. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci USA. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Gorman JM, Haroutunian V. Variations in differential gene expression patterns across multiple brain regions in schizophrenia. Schizophr Res. 2005;77:241–252. doi: 10.1016/j.schres.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, et al. Microduplication and triplication of 22q11 2 a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.