Abstract
The separation of the first two lineages – trophectoderm (TE) and inner cell mass (ICM) – is a crucial event in the development of the early embryo. The ICM, which constitutes the pluripotent founder cell population, develops into the embryo proper, whereas the TE, which comprises the surrounding outer layer, supports the development of the ICM before and after implantation. Cdx2, the first transcription factor expressed specifically in the developing TE, is crucial for the differentiation of cells into the TE, as lack of zygotic Cdx2 expression leads to a failure of embryos to hatch and implant into the uterus. However, speculation exists as to whether maternal Cdx2 is required for initiation of TE lineage separation. Here, we show that effective elimination of both maternal and zygotic Cdx2 transcripts by an RNA interference approach resulted in failure of embryo hatching and implantation, but the developing blastocysts exhibited normal gross morphology, indicating that TE differentiation had been initiated. Expression of keratin 8, a marker for differentiated TE, further confirmed the identity of the TE lineage in Cdx2-deficient embryos. However, these embryos exhibited low mitochondrial activity and abnormal ultrastructure, indicating that Cdx2 plays a key role in the regulation of TE function. Furthermore, we found that embryonic compaction does not act as a `switch' regulator to turn on Cdx2 expression. Our results clearly demonstrate that neither maternal nor zygotic Cdx2 transcripts direct the initiation of ICM/TE lineage separation.
Keywords: Cdx2, Trophectoderm, Lineage specification, Preimplantation mouse embryos
INTRODUCTION
The first visible cell lineage split in the development of the mammalian embryo occurs during blastulation. The cells of the inner part of the blastocyst, called the inner cell mass (ICM), are pluripotent and eventually give rise to the embryo proper and to the extra-embryonic tissues. By contrast, the cells of the outer layer differentiate into an epithelium, called the trophectoderm (TE), which subsequently develops into the placenta. To date, a full understanding has been lacking of the molecular mechanisms that underlie the fate decision for the initial totipotent cells of the embryo to become either the TE or ICM lineage.
Cdx2 is a class I homeobox transcription factor that belongs to the caudal-related homeobox gene family, which comprises mammalian homologs of Drosophila caudal (James et al., 1994; Suh et al., 1994). Cdx family members function as upstream positive regulators of Hox genes (Lorentz et al., 1997), which are purported to confer positional identity to cells along the anteroposterior body axis and are sequentially activated in a temporal and spatial manner (Davidson et al., 2003). Loss of Cdx function has been shown to be associated with inactivation of Hox gene expression (Davidson et al., 2003). Cdx1 and Cdx2, which are regulated by p38 MAPK (Mapk14) (Houde et al., 2001), are involved in defining the anteroposterior body axis (Chawengsaksophak et al., 1997; Chawengsaksophak et al., 2004) and in establishing anteroposterior patterning of the intestine (Silberg et al., 2000). Dysregulation of Cdx2 expression has been found in intestinal metaplasia and carcinomas (da Costa et al., 1999; Bai et al., 2002; Eda et al., 2003). In colon cancer cells, for example, oncogenic ras downregulates Cdx2 expression by activating protein kinase C (PKC) pathway signaling and by reducing activity of the Cdx2 promoter AP-1 site through changes in the relative expression of c-June/c-Fos (Lorentz et al., 1999); by contrast, in the embryo, ras-MAPK signaling activates Cdx2 expression (Lu et al., 2008). Most importantly, one study found that although Cdx2 expression appears to be required for TE cell fate specification in the early mouse embryo, Cdx2 zygotic mutants can still initiate TE differentiation and blastulation (Strumpf et al., 2005). Similarly, another study found that Cdx2-deficient embryos, generated from Cdx2 short hairpin (sh) RNA-expressing somatic cells using nuclear transfer, failed to implant into the uterus, just like Cdx2–/– embryos (Meissner and Jaenisch, 2006). However, neither study eliminated maternal Cdx2 expression. More recent studies have found that Tead4 acts upstream of Cdx2 and is essential for TE specification and blastocyst formation (Yagi et al., 2007; Nishioka et al., 2008), although a subsequent study contradicted these findings by reporting that maternal Cdx2 is responsible for compaction and TE lineage initiation (Jedrusik et al., 2010). Therefore, although it is well established that Cdx2 is indispensable for the maintenance and proper functioning of the TE lineage, its role in the initiation of TE lineage specification remains obscure. It is therefore imperative to study the full effect of Cdx2 expression in early mammalian embryonic development by eliminating both maternal and zygotic Cdx2 transcripts.
In the present study, we microinjected a robust Cdx2 small interfering (si) RNA duplex into zygotes and metaphase II (MII) oocytes and determined the effects of eliminating both maternal and zygotic expression of Cdx2 on the initiation of ICM/TE lineage separation and cellular differentiation. As Cdx2 acts downstream of Tead4, we also targeted Tead4 with siRNA to further confirm these results. The findings of this study help to better define the roles played by Cdx2 during early mammalian development.
MATERIALS AND METHODS
Embryo culture and microinjection of siCdx2 duplex
Fertilized oocytes were collected from the oviducts of primed B6C3F1 female mice after mating with CD1 male mice 18 hours post-hCG in M2 medium. Oocytes were cultured in KSOMAA (potassium simplex optimized medium plus 19 natural amino acids) (Ho et al., 1995) at 37°C and 5% CO2 in air until microinjection. For MII oocyte microinjection, mature oocytes were collected from the oviducts of primed B6C3F1 female mice 14 hours post-hCG in M2 medium, injected with siCdx2, and then fertilized in vitro in modified KSOM (Summers et al., 2000) with epididymal spermatozoa from adult OG2 male mice.
We used the online tool BLOCK-iT RNAi Designer to select specific target sequences for siRNA (https://rnaidesigner.invitrogen.com/rnaiexpress/). The lyophilized siRNA duplexes (Invitrogen, Karlsruhe, Germany) were resuspended in 1 ml DEPC-treated water according to the manufacturer's instructions and stored in single-use aliquots at –20°C. We tested three regular oligonucleotides and three Stealth RNAi oligonucleotides containing the coding region of Cdx2 (target sequence 5′-GCAGTCCCTAGGAAGCCAA-3′) in zygotes and selected the most effective duplex (siRNA3) for use in all our experiments. Unless otherwise specified, a scrambled siRNA duplex was used as control (target sequence 5′-GCACCCGATAAGCGGTCAA-3′).
siRNAs were microinjected using an Eppendorf FemtoJet microinjector and Narishige micromanipulators. Microinjection pipettes were pulled with a Sutter P-97 pipette puller. siRNA solution (5 μl of 8 μM) was loaded into the pipette and ∼2 pl was injected into the cytoplasm of each oocyte. A relatively consistent amount was carefully injected each time. Specifically, from 73 separate experiments, 3894 of 4093 zygotes (95.1%) were successfully injected with Cdx2 siRNA, in comparison with 3846 of 4065 zygotes (94.6%) successfully injected with scrambled siRNA.
In addition, from 18 separate experiments, 345 of 1202 MII oocytes successfully injected with Cdx2 siRNA were fertilized in vitro, in comparison with 207 of 743 MII oocytes successfully injected with scrambled siRNA and fertilized in vitro and with 327 of 415 uninjected MII oocytes that were fertilized in vitro. Unsuccessfully injected oocytes or lysed oocytes were excluded from all subsequent experiments.
Following microinjection, oocytes were washed and cultured in KSOMAA at 37°C and 5% CO2 in air and evaluated for cleavage twice daily. The implantation capability of early-stage embryos was evaluated by transfer of embryonic day (E) 3.5 mouse embryos into the uterus of a 2.5 days post-coitum (dpc) pseudopregnant CD1 female mouse.
Animal care was in accordance with the institutional guidelines of the Max Planck Institute.
RNA extraction, cDNA synthesis and real-time reverse transcription PCR
For real-time analysis of gene expression, embryos were harvested in RLT buffer (Qiagen, Hilden, Germany) at different stages of development and processed as previously described (Boiani et al., 2005). Briefly, total RNA was extracted from individual blastocysts using the MicroRNeasy Kit (Qiagen) and cDNA synthesis was performed with the High-Capacity cDNA Archive Kit (Applied BioSystems, Darmstadt, Germany) according to the manufacturer's instructions.
Real-time PCR was carried out on the ABI PRISM 7900HT Sequence Detection System (Applied BioSystems) using TaqMan probes (Applied Biosystems) for the housekeeping gene Hprt1 (Mm00446968_m1) as a control and the following genes of interest: Cdx2 (Mm00432449_m1), Eomes (Mm01351984_m1), Efs (Mm00468700_m1), Atpaf1 (Mm00619286_g1), Atp1b1 (Mm00437612_m1), Akt3 (Mm00442194_m1), Fgfr2 (Mm00438941_m1), Rab13 (Mm00503311_m1), Cdh1 (Mm00486906_m1), Hand1 (Mm00433931_m1), Grn (Mm00433848_m1), Oct4 (Mm00658129_gH), Gata6 (Mm00802636_m1), Sox17 (Mm00488363_m1). Oligos for Nanog amplification were custom designed: PF, 5′-AACCAGTGGTTGAATACTAGCAATG-3′; PR, 5′-CTGCAATGGATGCTGGGATACT-3′; and probe, 5′-6FAM-TTCAGAAGGGCTCAGCAC-MGB-3′. Three to six biological replicates were used and each sample was run with three technical replicates; negative controls lacked reverse transcriptase or template. The cycle threshold (CT) values were collected using Applied Biosystems SDS v2.0 software and transferred to a Microsoft Excel spreadsheet for further relative quantification analysis using the ΔΔCT method (User Bulletin #2, ABI Prism 7700 Sequence Detection System, 1997). Some of the data were presented using the percentage of peak method, which is an adaptation of the ΔΔCT method (Wang et al., 2004).
Immunofluorescent staining of embryos
Immunocytochemical staining was performed as described previously with minor modifications (Strumpf et al., 2005). Briefly, samples were fixed in 4% paraformaldehyde for 20 minutes, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St Louis, MO, USA) for 1 hour, and blocked with 3% BSA in PBS for 1 hour. Control and Cdx2-deficient embryos were processed and examined in parallel. Samples were incubated with the following antibodies: monoclonal mouse anti-Cdx2 IgG (1:100; BioGenex, San Ramon, CA, USA), rabbit polyclonal anti-Cdx2 [1:2000; gift from Felix Beck (Victoria, Australia) and Alexey Tomilin, (Freiburg, Germany)], rabbit anti-Nanog IgG (1:500; Cosmo Bio Company, Tokyo, Japan), monoclonal mouse anti-Oct4 IgG (1:200; SC-5279; Santa Cruz Biotech., Santa Cruz, CA, USA), rabbit anti-Oct4 IgG [1:2500; generated in our laboratory (Palmieri et al., 1994)], rat anti-E-cadherin IgG (1:100; ECCD-2; Calbiochem, Darmstadt, Germany), Troma-1 (1:100; DSHB, Iowa City, IA, USA) or rabbit anti-ISP1 IgG [1:100; kindly provided by Dr D. Rancourt (O'Sullivan et al., 2001)], all at 4°C overnight, or with rabbit anti-ZO-1 IgG (1:150; Zytomed, Berlin, Germany) at 37°C for 1 hour.
Embryo samples were then washed several times with PBS and stained with Alexa 488-conjugated goat anti-rabbit IgG, Alexa 488-conjugated goat anti-mouse IgG (Invitrogen), Cy3-conjugated goat anti-rat IgG, or Cy3-conjugated goat anti-mouse IgG (Jackson Laboratory, Bar Harbor, ME, USA) at 1:400 dilution for 1 hour at room temperature. After further washes and counterstaining with 5 μM DRAQ5 (Biostatus, Shepshed, UK) for 30 minutes, samples were examined under a laser-scanning confocal microscope (UltraVIEW; PerkinElmer Life Sciences, Jügesheim, Germany) with 488, 568 and 647 nm lasers. ECCD-2 immunostaining was quantified with ImageJ 1.42 software (http://rsbweb.nih.gov/ij/download.html). Troma-1 antibody and DRAQ5 staining were used to determine the ICM:TE cell number ratio by comparing confocal images of neighboring sections using a 5 μm scanning distance.
Transmission electron microscopy
Cells were fixed in 2.5% glutaraldehyde (Merck, Darmstadt, Germany) in 0.1 M sodium cacodylate buffer (pH 7.4), post-fixed in 1% aqueous osmium tetroxide, dehydrated in ethanol and embedded in Epon 812 (Fluka, Buchs, Switzerland). Ultrathin sections (50 nm) were prepared with an EM UC6 ultramicrotome (Leica Microsystems, Wetzlar, Germany), stained with 1% uranyl acetate and then 3% lead citrate, and subsequently examined under a Zeiss EM 109 electron microscope (Carl Zeiss, Oberkochen, Germany).
Measurement of mitochondrial membrane potential and ATP content
Mitochondrial membrane potential was measured by staining with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl carbocyanide iodide (JC-1; Molecular Probes, Eugene, OR, USA) (Cossarizza et al., 1994). Cells were incubated in KSOMAA containing 5.0 μg/ml JC-1 for 15 minutes at 37°C in the dark. Cells were washed with KSOMAA, excited at 488 nm, and then observed at either 510 nm (green mitochondria) or 590 nm (red-to-orange mitochondria) with the confocal imaging system. The ATP content of each embryo was measured as previously described (Boiani et al., 2004).
Induction and inhibition of embryonic compaction
To induce compaction, 4-cell stage embryos were transferred into KSOMAA containing 200 μM DiC8 (Sigma-Aldrich) and incubated overnight at 37°C. To prevent compaction, precompacted 8-cell stage embryos were cultured in KSOMAA containing 200 μg/ml anti-E-cadherin antibody (ECCD-1; Calbiochem). Embryo samples were collected after 18 and 24 hours of treatment and subjected to immunocytochemical analysis for Cdx2 expression.
Measurement of DNA fragmentation by TUNEL assay
The degree of apoptosis in embryo samples was determined using terminal deoxynucleotidyl transferase (TdT)-mediated dUTP-FITC nick end labeling (TUNEL) with an in situ cell death detection kit (DeadEnd Fluorimetric TUNEL System; Promega, Madison, WI, USA) according to the manufacturer's recommendations. Cells were then viewed/counted under a confocal microscope.
Statistical analysis
The Jarque-Bera test was performed on the ICM:TE cell number ratio and apoptotic cell numbers, revealing that the cell number did not follow a normal, or Gaussian, distribution with a significance level α=0.05. Therefore, the Wilcoxon rank-sum test was performed as an appropriate non-parametric statistical test with significance level α=0.05 to compare differences in cell number.
RESULTS
Maternal and zygotic expression of Cdx2
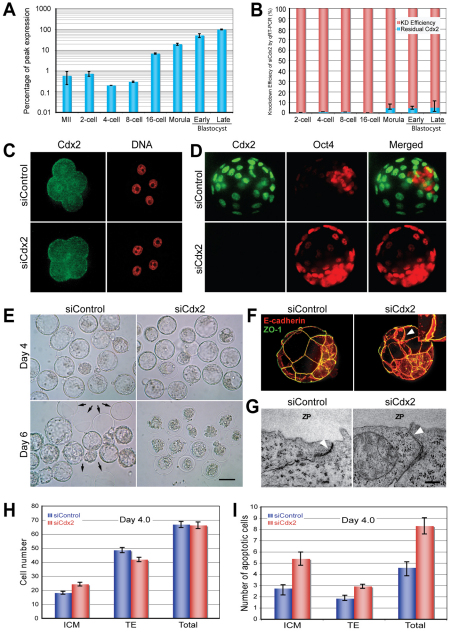
We examined Cdx2 mRNA and protein levels in developing early mouse embryos by quantitative real-time PCR (qRT-PCR) and immunocytochemistry. Maternal Cdx2 mRNA was readily detectable in oocytes (CT value of 36.5 from pools of ten MII oocytes) and 2-cell stage embryos, but levels were reduced by 73% in 4-cell stage embryos and increased by 23-fold upon Cdx2 activation in 8- to 16-cell stage embryos (Fig. 1A). As previously reported (Niwa, H. et al., 2005; Strumpf et al., 2005; Dietrich and Hiiragi, 2007; Yagi et al., 2007), Cdx2 protein levels could only be detected as a very weak nuclear signal in 8-cell stage embryos (Fig. 1C); however, a stronger Cdx2 protein signal was detected in some blastomeres during the next cell cycle, gradually extending to all outer blastomeres (see Fig. S1 in the supplementary material). This expression profile provides evidence for the presence of maternal Cdx2 transcripts in early mouse embryos and indicates that activation of zygotic Cdx2 expression is concomitant with the first lineage differentiation event during the asymmetric fourth division after compaction in 8-cell stage mouse embryos (Johnson and McConnell, 2004).
Fig. 1.
Efficient reduction of Cdx2 mRNA and protein levels in different preimplantation stage mouse embryos by siCdx2 results in phenotypes similar to Cdx2 knockout. (A) Relative Cdx2 mRNA expression levels as a percentage of peak expression in mouse embryos at different preimplantation stages, as assessed by qRT-PCR. The maternal mRNA decreased at the 4-cell stage, followed by upregulation at the 8-cell stage due to the zygotic activation of Cdx2. Embryo collection times (post-hCG): 2-cell stage, 40 hours; 4-cell stage, 52 hours; 8-cell stage, 62 hours; 16-cell stage, 72 hours; morula, 85 hours; early blastocyst, 96 hours; late blastocyst, 115 hours. (B) A comparison of Cdx2 mRNA levels in siControl- and siCdx2-treated zygotes at different preimplantation stages demonstrated robust downregulation of Cdx2 by siCdx2 treatment. KD, knockdown. (C) Confocal images of 8-cell stage embryos immunolabeled with anti-Cdx2 monoclonal antibody illustrate the reduction of Cdx2 protein (green) by siCdx2 treatment. Red, nuclear counterstaining with DRAQ5. (D) Confocal images of blastocyst stage embryos immunolabeled with anti-Cdx2 (green) and anti-Oct4 (red) antibodies. Following siCdx2 treatment, Cdx2 was eliminated, but Oct4 was ectopically expressed in the trophectoderm (TE). (E) DIC images of E4.0 and E6.0 embryos. All siCdx2-treated embryos failed to hatch. Arrows indicate empty zonae pellucidae left by hatched control embryos. Scale bar: 100 μm. (F) Confocal images of ZO-1 (green) and E-cadherin (red) immunohistochemistry show normal E-cadherin distribution, which marks the lateral-basal cell boundary, but disoriented cell polarity of the TE, as indicated by the presence of ZO-1 at both the apical and basal sides (arrowhead and inset). (G) Defective tight junction (arrowhead) in the TE of a Cdx2-deficient blastocyst as shown by electron microscopy. Scale bar: 0.2 μM. (H) Effect of Cdx2 depletion on TE and inner cell mass (ICM) cell numbers. The number of ICM cells was significantly increased (P<0.01), but the total cell number remained unchanged (P=0.64) upon Cdx2 depletion. (I) Quantitation of apoptotic cells shows an increase in apoptotic activity in a Cdx2-deficient E4.0 blastocyst,. MII, metaphase II oocyte; ZP, zona pellucida. Error bars indicate standard deviation.
Effective reduction in maternal and zygotic Cdx2 by RNAi has profound effects on TE cell differentiation, function and embryo developmental potential
Of the six Cdx2-specific siRNA duplexes tested, siRNA3 was selected for subsequent experiments owing to its unique sequence and high efficiency (see Fig. S2A in the supplementary material); siRNA3 is hereafter referred to as siCdx2. A scrambled siRNA duplex (siControl) with the same composition as siCdx2 but no specific target was used as control. A single injection of siCdx2 into MII oocytes or zygotes was found to reduce Cdx2 mRNA levels by at least 95%, as determined by qRT-PCR (Fig. 1B). Furthermore, siCdx2 injection also led to an efficient reduction in Cdx2 protein to undetectable levels in each of the 8-cell (siControl, n=26; siCdx2, n=28), morula (siControl, n=26; siCdx2, n=25) and blastocyst stage (siControl, n=71; siCdx2, n=67) embryos, as determined by three independent immunocytochemistry experiments (Fig. 1C,D; see Fig. S2B in the supplementary material) and a western blot with replicates (see Fig. S2C in the supplementary material). The robust elimination of both maternal and zygotic Cdx2 expression did not affect the rate of blastocyst formation (see Table S1 in the supplementary material). As shown in our time-lapse observations (see Fig. S2D in the supplementary material), compaction and the onset of blastocyst formation were unaffected in siCdx2-treated embryos. However, most likely owing to a defect in tight junctions, Cdx2-deficient blastocysts could not initially maintain the integrity of the blastocyst cavity, collapsed three times and were delayed by ∼7.5 hours in starting cavity expansion, but were of similar size and gross morphology to siControl embryos at 4.0 dpc. Then, after 5-8 pulses of collapse and expansion, all Cdx2-deficient embryos eventually collapsed in the zonae pellucidae between 5.5 and 6.0 dpc, rather than hatching like control blastocysts.
To exclude the possibility that any residual maternal Cdx2 protein could have driven TE lineage separation and blastocyst formation in siCdx2-treated embryos, we co-injected siCdx2 with a rabbit polyclonal Cdx2 antibody at 100-fold higher concentration than the effective concentration for immunostaining, but still observed no phenotype prior to the blastocyst stage (see Table S2 in the supplementary material).
In addition, an increase in the ICM:TE cell number ratio was found (0.58 versus 0.37; siControl, n=40; siCdx2, n=41; P=1.05×10–5), but the total number of cells in Cdx2-deficient E4.0 blastocysts was not different from that of control blastocysts (66.3 versus 66.9; P=0.64; Fig. 1H). TUNEL analysis revealed the presence of slightly more apoptotic cells within both the inner (5.4 versus 2.7; P=7.35×10–5) and outer (2.9 versus 1.8; P=3.48×10–4; siControl, n=19; siCdx2, n=13) cells of Cdx2-deficient blastocysts (Fig. 1I). However, transfer of Cdx2-deficient blastocysts into the uterus of pseudopregnant foster mice did not lead to any implantation (0 implanted out of 109 transferred in the siCdx2-treated group versus 60 out of 108 in the siControl, from three separate experiments), constituting another specific phenotype of the zygotic Cdx2–/– embryos.
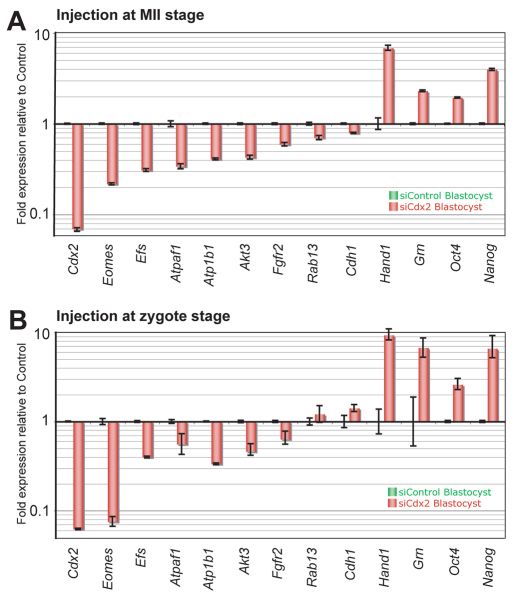
Gene expression analysis by qRT-PCR revealed that knockdown of maternal Cdx2 led to a 77.5% reduction in the transcript levels of the TE-associated gene Eomes in 8-cell stage embryos, whereas a reduction in the Fgfr2 transcript and an increase in the transcripts of the pluripotency gene Oct4 (Pou5f1 – Mouse Genome Informatics) became significant only after blastulation (Fig. 2A,B; Fig. 4; see Fig. S3A in the supplementary material). Notably, Hand1, which is expressed in the TE and regulates trophoblast cell differentiation into trophoblast giant cells (Cross et al., 1995), and Grn, which is an important factor for blastocyst hatching, adhesion and outgrowth (Qin et al., 2005), were overexpressed in Cdx2-deficient blastocysts (Fig. 4).
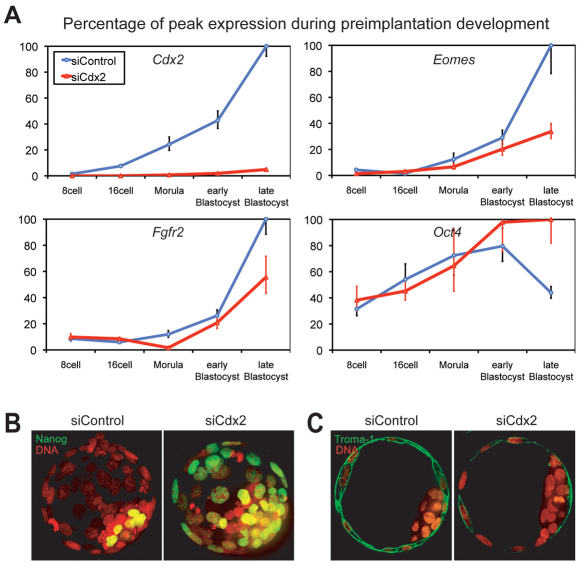
Fig. 2.
Significant effect of Cdx2 reduction on gene expression at various developmental stages but no blockage of expression of the differentiated TE marker Krt8. (A) Relative gene expression levels as a percentage of peak expression of Cdx2, Eomes, Fgfr2 and Oct4 in mouse preimplantation embryos after siControl and siCdx2 injection into zygotes. The downstream transcription factor Eomes was reduced to a basal level. Fgfr2 reduction is shown at the early and expanded blastocyst stages. Oct4 expression was increased ∼2-fold in both the early and expanded blastocyst stages (P<0.0001). Error bars indicate standard deviation. (B) Ectopic expression of Nanog in the TE of Cdx2-deficient blastocysts after microinjection of siCdx2 at the zygote stage. The embryonic stem cell marker Nanog was ectopically expressed in TE, as shown in this z-stack confocal image of an immunostained siCdx2-treated blastocyst. (C) Sectional confocal image showing the presence of Krt8 (as detected by the Troma-1 antibody) in an interrupted pattern in Cdx2 knockdown blastocysts.
Fig. 4.
Relative gene expression levels in E4.0 blastocysts after microinjection of siCdx2 into MII oocytes and zygotes. Elimination of Cdx2 RNA by microinjection of siCdx2 into mouse (A) MII oocytes and (B) zygotes had similar effects on gene expression levels, as assessed by qRT-PCR. Cdx2 knockdown reduced expression of genes crucial in TE lineage differentiation, with overexpression of pluripotency-related genes. Error bars indicate standard deviation.
To determine why the normally shaped Cdx2-deficient blastocysts failed to hatch from the zonae pellucidae, we performed gene expression studies with qRT-PCR or immunocytochemistry on E4.0 Cdx2-deficient blastocysts. We found that Grn (an autocrine growth factor) and ISP1 (a tryptase; Prss28 – Mouse Genome Informatics), which have been reported to play crucial roles in the hatching process (O'Sullivan et al., 2001; Qin et al., 2005), were not reduced in Cdx2-deficient blastocysts (Fig. 4; see Fig. S3B in the supplementary material). However, further examination revealed the cause of this hatching failure. Immunocytochemical analysis of embryos using the tight junction-associated protein zona occludens 1 (ZO-1; Tjp1 – Mouse Genome Informatics) showed that the apical zonular structure of the tight junctions was discontinuous in siCdx2-treated embryos (Fig. 1F), indicating compromised intercellular sealing and epithelial integrity. This specific phenotype was previously observed in experiments with mouse Cdx2–/– embryos (Strumpf et al., 2005). Moreover, we also observed the presence of the zonular form of ZO-1 at both the apical and basal sides of the epithelium, suggesting that some TE cells exhibited disorientation in cellular apicobasal polarity. Examination of Cdx2-deficient blastocysts under the electron microscope (Fig. 1G) showed that the tight junction structure was actually missing, although ZO-1 protein was detected by immunocytochemistry. However, the ultrastructure of lateral-basal adherens junctions was present as in control embryos (Fig. 1G). Immunocytochemical analysis of embryos revealed a stronger intensity of E-cadherin, a major component of adherens junctions, at the basolateral border of TE cells in Cdx2-deficient embryos than in control embryos (Fig. 1F; P<0.01), without an increase in mRNA levels (Fig. 4). Therefore, although adherens junctions could compensate for the lack of tight junctions and support delayed blastocoele formation and expansion in the mutated embryos, they failed to maintain intercellular sealing for the more demanding process of hatching.
Finally, to further confirm the identity of the outer cells, we performed immunocytochemical analysis of blastocysts for keratin 8 (Krt8; also known as EndoA), a widely used marker for differentiated TE, with the monoclonal antibody Troma-1 (Brulet et al., 1980; Ralston and Rossant, 2008). We found that Krt8 was present in the outer cells of Cdx2-deficient blastocysts, albeit with an uneven distribution, clearly showing that establishment of the TE lineage was independent of Cdx2 at the molecular level (Fig. 2C). Our time-lapse analysis revealed that Cdx2-deficient embryos compacted and initiated cavitation at the correct time and only failed to hatch afterwards. In addition, the zonular form of ZO-1 was found to be present at the apical sides of the outer cells and certain TE-associated genes (Hand1 and Grn) were actually overexpressed in Cdx2-depleted embryos. Taking all these data together, we conclude that elimination of both maternal and zygotic Cdx2 expression did not block the initiation of ICM/TE lineage separation, but it did impact the continuation of TE differentiation and function.
Effect of Cdx2 knockdown on mitochondrial activity
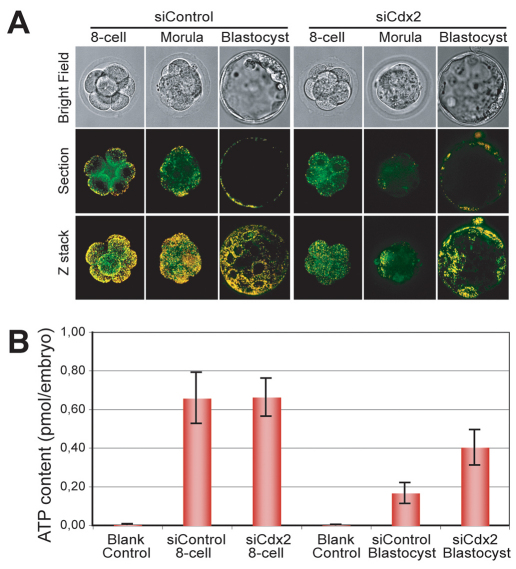
Upregulation of mitochondrial activity is a feature of TE differentiation. The JC-1 assay was performed to explore the impact of Cdx2 expression on mitochondrial function and revealed a significant reduction in mitochondrial activity at the apical cortex of 8-cell embryos and TE cells of blastocysts, indicating the presence of a functionally defective TE (Fig. 3A). This finding was corroborated by the underexpression of genes involved in both ATP synthesis (Atpaf1) and consumption (Atp1b1) (Fig. 4A,B) in Cdx2-deficient embryos, as determined by qRT-PCR. However, an increase in cytoplasmic ATP content was found in blastocyst stage embryos that had Cdx2 knockdown-induced mitochondrial dysfunction, as compared with wild-type blastocysts (0.41 versus 0.17 pmol per embryo; Fig. 3B).
Fig. 3.
Significant effect of Cdx2 knockdown on mitochondrial activity. (A) Confocal images showing mitochondrial activity as assessed by JC-1 staining at the 8-cell, morula and blastocyst stages. siCdx2 treatment significantly reduced mitochondrial activity, as indicated by the accumulation of the red aggregated form of JC-1 in the mitochondria (appears yellow, as all mitochondria were stained with the green form of JC-1). (B) Measurement of ATP content in individual embryos, showing a significant increase in ATP from 0.17 pmol to 0.41 pmol in Cdx2-deficient blastocysts. Error bars indicate standard deviation.
siCdx2 microinjection into MII oocytes produces a phenotype identical to that of zygotes
Our results showed that microinjection of siCdx2 into MII oocytes also led to an efficient reduction in Cdx2 mRNA expression levels (Fig. 4A), with no protein detectable in blastocyst stage embryos (siControl, n=61; siCdx2, n=52) by immunostaining. Changes in the gene expression profile at E4.0 were similar to those observed after injection of siCdx2 into zygotes (Fig. 4B), as determined by qRT-PCR. Injection of siCdx2 into MII oocytes led to ectopic expression of Oct4 and Nanog proteins in the outer cells of Cdx2-deficient blastocysts (see Fig. S3C in the supplementary material), comparable to that observed after siCdx2 injection into zygotes (Fig. 1D; Fig. 2B). Injection of siCdx2 at an earlier time point resulted in the same phenotype and gene expression changes as at the zygote stage, further supporting the conclusion that the initiation of compaction and separation of ICM/TE in Cdx2-deficient embryos was not due to Cdx2 protein produced from maternal mRNA before the mRNA was knocked down.
Embryonic compaction does not lead to activation of Cdx2 expression
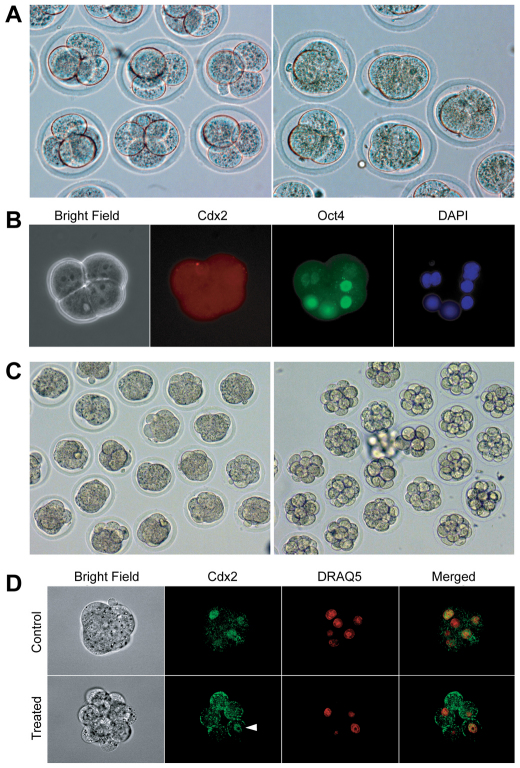
To evaluate the effect of embryonic compaction on the activation of Cdx2 expression, we used two complementary approaches. First, we induced compaction in 4-cell stage embryos with the natural PKC agonist DiC8 (Pauken and Capco, 1999). Embryos at this stage of normal development do not initiate compaction. However, upon treatment with DiC8, 4-cell stage embryos rapidly initiated compaction, as seen by the flattening and adherence of blastomeres, which became well compacted within 30 minutes (Fig. 5A). Cdx2 expression could not be detected after 18 hours of treatment by immunocytochemistry (Fig. 5B; DiC8, n=21; Control, n=19). Using the second approach, we blocked the compaction of 8-cell stage embryos (Fig. 5C) with ECCD-1, an antibody directed against the extracellular domain of E-cadherin (Yoshida-Noro et al., 1984), and examined Cdx2 expression after 24 hours in 12- to 16-cell stage embryos (Fig. 5D; ECCD-1, n=20; Control, n=21). Although there was no compaction, Cdx2 expression could still be detected in 12-cell stage embryos. These results clearly demonstrate that the compaction process does not lead to activation of Cdx2 expression in mouse embryos.
Fig. 5.
Neither induction nor blockage of embryonic compaction affects the activation of Cdx2 expression. (A) Premature compaction induced within 30 minutes of treatment with the PKC agonist DiC8 (right). Normal 4-cell stage mouse embryos are shown on the left, with obvious, individual blastomeres. (B) Cdx2 expression (red) could not be detected by immunocytochemistry 18 hours after DiC8 treatment. Nuclei are blue (DAPI) and Oct4 (a positive control for transcriptional activity of the treated embryos) green. (C) Compaction was inhibited by anti-E-cadherin (ECCD-1) antibody treatment for 24 hours (right). Normally compacted 14-cell stage embryos are shown on the left. (D) Cdx2 was detected (arrowhead) in uncompacted 12-cell stage embryos. Heavy cytoplasmic background (especially around the cell surface) was due to cross-reaction of ECCD-1 (rat IgG monoclonal), which was used at a very high concentration (200 μg/ml) to block embryo compaction, with the Cdx2 antibody (mouse IgG monoclonal).
Reduction in Tead4, which is upstream of Cdx2, prevents the cavitation and separation of the ICM and TE but does not prevent compaction
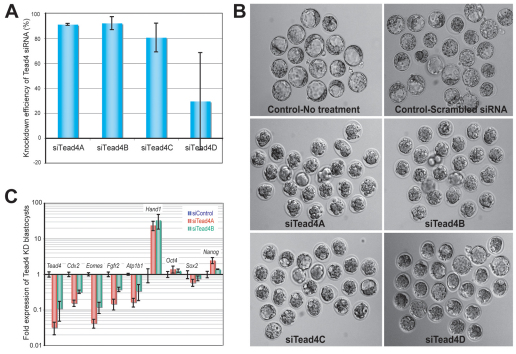
We tested the efficiency of four Tead4-specific siRNA duplexes to knock down Tead4 mRNA expression levels by the same microinjection approach that we used for Cdx2 knockdown. Injected oocytes were cultured for 72 hours to reach the morula/blastocyst stage and collected individually or in groups for qRT-PCR. Two siRNA duplexes were found to result in a ∼90% knockdown efficiency of Tead4 (siTead4A and siTead4B; Fig. 6A). Consistent with the zygotic Tead4 knockout phenotype (Yagi et al., 2007; Nishioka et al., 2008), these two duplexes were found to effectively prevent embryo cavitation and clear separation of the ICM and TE (Fig. 6B), but had no effect upon compaction at 2.5 dpc (see Fig. S4 in the supplementary material), just as with Cdx2 knockdown embryos. Significant underexpression of TE marker genes (Cdx2, Eomes, Fgfr2 and Atp1b1), but not pluripotency-related genes (Oct4, Nanog and Sox2), was observed by qRT-PCR upon Tead4 knockdown (Fig. 6C).
Fig. 6.
Effective knockdown of Tead4 in preimplantation mouse embryos by microinjection of siRNA into zygotes reproduces the Tead4–/– embryo phenotype. (A) Tead4 knockdown efficiency (%) determined by qRT-PCR in a single E4.5 mouse blastocyst in comparison to the scrambled siRNA control. Error bars indicate standard deviation. (B) Failure of blastulation 72 hours (E3.5) after microinjection of siTead4A and siTead4B into mouse zygotes. In this particular experiment, 25 zygotes per group were successfully injected. Note that the phenotype is consistent and highly reproducible within the siTead4A and siTead4B groups (except for one embryo in each group that was blocked at the 2-cell stage), but was variable in groups with lower Tead4 knockdown efficiency. (C) Tead4 knockdown reduced the expression of genes crucial in TE lineage differentiation but did not reduce the expression of pluripotency-related genes. Shown are qRT-PCR results from pools of six embryos, with biological and technical triplicates.
DISCUSSION
In the present study, we defined the expression profile of Cdx2 in mouse oocytes and embryos by the sensitive and specific technique of qRT-PCR and systematically investigated the impact of knocking down both maternal and zygotic mRNA levels by an siRNA approach on the development of preimplantation embryos. The consistency in gene expression profile and phenotype of the siCdx2-treated embryos with those of the zygotic Cdx2-null embryos (Strumpf et al., 2005), such as the failure of embryos to hatch from the zonae pellucidae and implant into the uterus, the underexpression of Eomes, and the ectopic expression of Oct4 and Nanog in the TE, demonstrates the efficacy and specificity of our siCdx2 treatment.
Developmental biologists have sought for years to understand how mammalian embryos establish their developmental pattern and initiate their first differentiation event – the separation of the TE from the ICM (Hiiragi et al., 2006). Cdx2, the first TE marker gene to be expressed in mouse embryos, was reported in a study by Strumpf and colleagues to be required for correct TE fate specification (Strumpf et al., 2005). The zygotic Cdx2–/– embryos failed to downregulate Oct4 and Nanog levels in the outer cells of the blastocyst, resulting in loss of epithelial integrity and in a subsequent failure to hatch and implant. However, these Cdx2-null embryos were still capable of forming a TE, as evidenced by cavitation and blastocyst formation, a finding that contradicted the main conclusion of that study and the authors hypothesized that the expression of maternal Cdx2 in these Cdx2–/– embryos might be responsible for the initiation of TE lineage differentiation. However, the same group later proposed that Cdx2 does not lead the TE transcription factor hierarchy (Rossant and Tam, 2009), as a loss-of-function Cdx2 mutation had no effect on the initiation of blastocyst formation (Strumpf et al., 2005), and that Cdx2-mutant cells were not excluded from the TE layer in chimeric blastocysts (Ralston and Rossant, 2008). These cells, however, do not undergo further trophoblast differentiation and thus are not functional (Strumpf et al., 2005).
In the present study, we used an siRNA approach to robustly knock down both maternal and zygotic Cdx2 transcripts, and are the first group to provide unequivocal and direct evidence that maternal Cdx2 is not a determinant of TE lineage commitment. In these maternal and zygotic Cdx2-deficient embryos, compaction and cavitation initiation were not interrupted, as clearly shown by our time-lapse observations (see Fig. S2D in the supplementary material), even though the morula-to-blastocyst transformation was delayed as a consequence of a defect in ZO-1/tight junctions (Wang et al., 2008). The outer cells of Cdx2-deficient blastocysts expressed the TE marker Krt8, confirming their TE identity. However, disorientation of the TE, as indicated by the presence of linear ZO-1 at the base of the epithelium, also highlights the importance of Cdx2 in cell polarization at the preimplantation stages. An increased ICM:TE cell number ratio might reflect compromised TE proliferation and function in regulating the number of ICM cells. These embryos subsequently failed to hatch, implant into the uterus or survive, indicating that TE lineage separation is independent of Cdx2 but that TE function supporting embryogenesis is completely dependent on Cdx2.
A recent report from M. Zernicka-Goetz's group (Jedrusik et al., 2010) has provided results that are contradictory to those of the present study. In that study, the authors achieved a much lower knockdown efficiency of maternal Cdx2 transcripts, of ∼80-85% before the 8-cell stage, with Cdx2 protein levels still detectable in some of the treated embryos, but they reported a more severe phenotype and claimed that the maternal pool of Cdx2 mRNA played a crucial role in compaction and polarization at the 8- to 16-cell stages of embryonic development. In our experiments, we achieved a knockdown efficiency of maternal Cdx2 transcripts of greater than 99.0%, without any detectable Cdx2 proteins, and yet 88.4% of these embryos compacted and developed to the blastocyst stage, with the same morphology and developmental rate as those injected with scrambled siRNA (87%) or non-injected embryos (85.3%) (see Table S1 in the supplementary material). The failure of such embryos to hatch and implant and the formation of embryoid body (EB)-like structures on mouse embryonic fibroblasts (MEFs) (Wu et al., 2010) were reported to be phenotypes specific to zygotic Cdx2–/– embryos by Rossant's group (Strumpf et al., 2005), yet we did not observe any significant effect on compaction and blastocyst formation.
In the study by M. Zernicka-Goetz's group, heterogeneous phenotypes were observed as embryos arrested at various stages of development. Furthermore, their Cdx2-deficient embryos not only totally lacked expression of TE-associated genes, such as Krt8, Eomes, Ecad (Cdh1), aPKC (Prkcz), Par1 (Mark1) and Par3 (Pard3), but also that of the pluripotent marker gene Nanog, which contradicts the findings of other publications, as a reciprocal relationship between Nanog and Cdx2 has already been clearly established (Chen et al., 2009) and overexpression of Nanog has been observed in zygotic Cdx2 knockout experiments (Strumpf et al., 2005) as well as in the present study. In support of our observations, an independent group has reported that following injection of Cdx2-specific siRNA at the 1-cell stage, Cdx2-deficient embryos do not exhibit developmental arrest prior to the blastocyst stage and do overexpress the pluripotent gene Oct4 at the blastocyst stage (Wang et al., 2010). In a broader sense, research on primates has also indicated that CDX2 plays an essential role in functional TE formation and that the ICM lineage of CDX2-depleted embryos supports the isolation of functional embryonic stem cells (Sritanaudomchai et al., 2009), which is consistent with our observations in the mouse.
In the present study, we observed an increase in mitochondrial activity concomitant with TE differentiation. This is consistent with the involvement of Na+/K+-ATPase in mouse embryos preparing for cavitation and production of nascent blastocoele fluid (Wiley, 1984) and with the stage-specific translocation of active mitochondria during early porcine embryo development (Sun et al., 2001). Interestingly, TE cells have been reported to produce and consume most of the ATP of the blastocyst (Houghton, 2006). Functional analysis with JC-1 staining showed reduced activity in the mitochondria of Cdx2-deficient embryos. Further gene expression analysis demonstrated that the reduction in mitochondrial activity was mediated by the underexpression of genes involved in both ATP synthesis and consumption. Krt8 has been shown to modulate the shape, distribution and function of hepatocyte mitochondria and to protect cells from apoptosis (Tao et al., 2009), suggesting that some of the phenotypes of Cdx2-deficient embryos might be mediated by the reduced and uneven Krt8 expression pattern that we observed. Reduction in Krt8 expression has also been observed in zygotic Cdx2–/– embryos (Ralston and Rossant, 2008). Of note, an increase in ATP content was consistently found in Cdx2-deficient embryos, in duplicate experiments with triplicate tests, which was probably due to underexpression of the ATP-dependent Na+ pump of the TE, a main consumer of ATP at this developmental stage (Houghton, 2006). These data indicate that deficiency in Cdx2 has profound effects on mitochondrial activity and that Cdx2 is indeed required for proper functioning of the TE. Further study is needed to determine precisely how Cdx2 regulates mitochondrial activity and ATP levels in the embryo.
Observations from classical embryonic experiments have suggested that ICM/TE lineage separation is initiated by asymmetric cell divisions that act to establish the inner and outer cell populations following compaction and polarization in 8-cell stage mouse embryos (Johnson and Rossant, 1981). During compaction, changes at the cellular level include enhanced cell adhesion between blastomeres resulting in the formation of a smooth ball of blastomeres, and the establishment of cell polarity, as demonstrated by the formation of apical and basolateral surfaces on the blastomeres (Hyafil et al., 1980; Butz and Larue, 1995; Ohsugi et al., 1996). At the next cell division after compaction, the developmental fate of the embryo becomes restricted, as outer blastomeres become destined to form the first epithelial layer, the TE, and inner blastomeres are pluripotent and form the ICM (Tarkowski and Wroblewska, 1967; Johnson and Rossant, 1981). Par3, Par6 and aPKC are components of an apical polarity complex that has been shown to influence TE/ICM fate choice (Plusa et al., 2005). In contrast to the two studies by M. Zernicka-Goetz's group (Jedrusik et al., 2008; Jedrusik et al., 2010), other studies have shown that Cdx2 appears to act downstream of the first lineage decision in the early mouse embryo – i.e. following cell polarization – to promote TE cell fate determination in a cell-autonomous manner, suggesting that processes influencing lineage allocation or morphogenesis might regulate Cdx2 expression along the inside/outside axis of the embryo (Ralston and Rossant, 2008). However, it is not yet clear whether compaction induces Cdx2 expression. It is known that compaction can be induced prematurely in 4-cell stage embryos by PKC activators (Winkel et al., 1990; Ohsugi et al., 1993; Pauken and Capco, 1999). With this approach, we showed that induction of compaction does not activate Cdx2 expression. Moreover, blocking compaction with ECCD-1, a monoclonal antibody that specifically neutralizes E-cadherin-mediated adhesion (Johnson et al., 1986) and induces disorientation of cell polarity in mouse early embryos (Fleming et al., 1989), does not prevent Cdx2 expression. These results suggest that Cdx2 expression and compaction are two parallel processes in regulating TE differentiation. This conclusion is consistent with a very recent report from Rossant's group that demonstrated that the epithelial integrity mediated by E-cadherin is not required for Cdx2 expression (Stephenson et al., 2010).
We next sought to confirm that even though we effectively knocked down maternal Cdx2 transcripts and attempted to block the function of any residual proteins with a Cdx2 antibody, we still could not confer an essential role for maternal Cdx2 in embryo compaction and initiation of the TE lineage, contrary to M. Zernicka-Goetz's group (Jedrusik et al., 2010). Here, we add another piece of evidence to support our conclusions. Tead4 has been reported by two studies to act upstream of Cdx2 and to be essential for TE specification (Yagi et al., 2007; Nishioka et al., 2008). In these two studies, zygotic Tead4–/– embryos were found to be devoid of the TE lineage, and expression of Tead4 was activated at the 2-cell stage and was postulated to subsequently trigger differentiation of totipotent blastomeres into trophoblast by the differential activation of the protein through the Hippo signaling pathway (Nishioka et al., 2008; Nishioka et al., 2009). In the present study, we attempted to knock down Tead4 by the same siRNA approach that we used to knock down Cdx2. After specifically and efficiently knocking down Tead4 transcripts, we found that none of the treated embryos was capable of initiating cavitation, consistent with the results of the two previous Tead4 knockout studies. In these Tead4–/– embryos, the maternal pool of Cdx2, which was not at all affected, did not sustain the embryos through the early stages of development to support initiation of TE separation in the positive-feedback loop manner proposed by M. Zernicka-Goetz's group. Our Tead4 knockdown results provided another solid piece of evidence to support the validity of our Cdx2 knockdown approach and confirmed that maternal Cdx2 does not lead the cascade that regulates TE lineage specification.
Notably, Nishioka et al. (Nishioka et al., 2008) performed a trophoblast outgrowth assay and showed that all the TE-specific genes examined, including Cdx2, Eomes, Fgfr2 (Arman et al., 1998), integrin alpha 7 (Itga7) (Klaffky et al., 2001), placental cadherin (Cdh3) (Niwa, T. et al., 2005) and the giant-cell-specific genes Hand1 (Riley et al., 1998) and Prl3d1 (Faria et al., 1991), were not expressed in Tead4–/– embryos corresponding to the late blastocyst stage. Similarly, expression of Hand1 and Prl3d1 was detected in Cdx2+/+ and Cdx2+/– embryos but not in Cdx2–/– embryos after 72 hours of culture of 3.5 dpc embryos (Strumpf et al., 2005). On the contrary, another Tead4 knockout study (Yagi et al., 2007) concluded that Tead4 was required for expression of some, but not all, TE-specific genes, as expression of Fgfr2 was not affected in Tead4–/– embryos. Our qRT-PCR analysis (Fig. 6C) showed significant downregulation of TE-associated genes (Cdx2, Eomes, Atp1b1 and Fgfr2) and significant overexpression of Hand1. These results, together with previous observations that Tead4–/– embryos weakly and briefly express Cdx2 between the 8- and 18-cell stages, and that they appear to have normal adherens junctions, cell polarities and JNK and p38 MAPK signaling (Nishioka et al., 2008), lead us to propose the existence of additional, Cdx2- and Tead4-independent mechanisms in TE lineage specification, which might include the subcortical maternal complex (SCMC) and its component Floped (also known as Moep19 or Ooep) in the mouse oocyte cortex (Herr et al., 2008; Li et al., 2008). Although the SCMC does not appear to be sufficient for establishing the TE lineage, the persistence of the SCMC in the outer cells of E3.5 Tead4-null embryos indicates that it might be present in early mouse embryos as an intrinsic pre-patterning factor related to blastomere polarity and TE specification (Li et al., 2008).
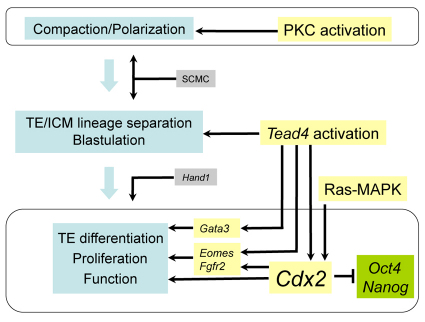
As illustrated in Fig. 7, based on the results of this study we confirm the findings of recent studies and clarify conflicting studies by concluding that although Cdx2 plays a crucial role in the maintenance of TE differentiation and function, neither maternal nor zygotic Cdx2 expression is essential for initiating the segregation of the ICM and TE lineages in the early stages of mouse embryonic development.
Fig. 7.
Postulated TE differentiation regulatory network. Cdx2 is not required for the TE lineage specification but it is critical for further differentiation of TE. Interactions among the processes of compaction and cellular polarization could affect the distribution of Cdx2 along the inside/outside axis and influence tight junction formation and localization. These interactions could therefore affect ICM/TE lineage allocation. SCMC, subcortical maternal complex.
Supplementary Material
Acknowledgments
We thank Jeanine Mueller-Keuker, Margit Preusser and Susanne Koelsch for assistance in preparing the manuscript and Dr D. Rancourt for providing the ISP1 antibody. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health. This research was supported by the Max Planck Society and grant NIH R01HD059946-01 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. This work was also supported in part by DFG grant Germ Cell Potential DFG FOR 1041 SCHO 340/7-1. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.056630/-/DC1
References
- Arman E., Haffner-Krausz R., Chen Y., Heath J. K., Lonai P. (1998). Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. USA 95, 5082-5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y. Q., Yamamoto H., Akiyama Y., Tanaka H., Takizawa T., Koike M., Kenji Yagi O., Saitoh K., Takeshita K., Iwai T., et al. (2002). Ectopic expression of homeodomain protein CDX2 in intestinal metaplasia and carcinomas of the stomach. Cancer Lett. 176, 47-55 [DOI] [PubMed] [Google Scholar]
- Boiani M., Gambles V., Schöler H. R. (2004). ATP levels in clone mouse embryos. Cytogenet. Genome Res. 105, 270-278 [Google Scholar]
- Boiani M., Gentile L., Gambles V. V., Cavaleri F., Redi C. A., Scholer H. R. (2005). Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: distinct developmental potentials in different culture environments. Stem Cells 23, 1089-1104 [DOI] [PubMed] [Google Scholar]
- Brulet P., Babinet C., Kemler R., Jacob F. (1980). Monoclonal antibodies against trophectoderm-specific markers during mouse blastocyst formation. Proc. Natl. Acad. Sci. USA 77, 4113-4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz S., Larue L. (1995). Expression of catenins during mouse embryonic development and in adult tissues. Cell Adhes. Commun. 3, 337-352 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., James R., Hammond V. E., Kontgen F., Beck F. (1997). Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84-87 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641-7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yabuuchi A., Eminli S., Takeuchi A., Lu C. W., Hochedlinger K., Daley G. Q. (2009). Cross-regulation of the Nanog and Cdx2 promoters. Cell Res. 19, 1052-1061 [DOI] [PubMed] [Google Scholar]
- Cossarizza A., Kalashnikova G., Grassilli E., Chiappelli F., Salvioli S., Capri M., Barbieri D., Troiano L., Monti D., Franceschi C. (1994). Mitochondrial modifications during rat thymocyte apoptosis: a study at the single cell level. Exp. Cell Res. 214, 323-330 [DOI] [PubMed] [Google Scholar]
- Cross J. C., Flannery M. L., Blanar M. A., Steingrimsson E., Jenkins N. A., Copeland N. G., Rutter W. J., Werb Z. (1995). Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121, 2513-2523 [DOI] [PubMed] [Google Scholar]
- da Costa L. T., He T. C., Yu J., Sparks A. B., Morin P. J., Polyak K., Laken S., Vogelstein B., Kinzler K. W. (1999). CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene 18, 5010-5014 [DOI] [PubMed] [Google Scholar]
- Davidson A. J., Ernst P., Wang Y., Dekens M. P., Kingsley P. D., Palis J., Korsmeyer S. J., Daley G. Q., Zon L. I. (2003). cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 425, 300-306 [DOI] [PubMed] [Google Scholar]
- Dietrich J. E., Hiiragi T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219-4231 [DOI] [PubMed] [Google Scholar]
- Eda A., Osawa H., Satoh K., Yanaka I., Kihira K., Ishino Y., Mutoh H., Sugano K. (2003). Aberrant expression of CDX2 in Barrett's epithelium and inflammatory esophageal mucosa. J. Gastroenterol. 38, 14-22 [DOI] [PubMed] [Google Scholar]
- Faria T. N., Ogren L., Talamantes F., Linzer D. I., Soares M. J. (1991). Localization of placental lactogen-I in trophoblast giant cells of the mouse placenta. Biol. Reprod. 44, 327-331 [DOI] [PubMed] [Google Scholar]
- Fleming T. P., McConnell J., Johnson M. H., Stevenson B. R. (1989). Development of tight junctions de novo in the mouse early embryo: control of assembly of the tight junction-specific protein, ZO-1. J. Cell Biol. 108, 1407-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr J. C., Chertihin O., Digilio L., Jha K. N., Vemuganti S., Flickinger C. J. (2008). Distribution of RNA binding protein MOEP19 in the oocyte cortex and early embryo indicates pre-patterning related to blastomere polarity and trophectoderm specification. Dev. Biol. 314, 300-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiiragi T., Louvet-Vallee S., Solter D., Maro B. (2006). Embryology: does prepatterning occur in the mouse egg? Nature 442, E3-E4 [DOI] [PubMed] [Google Scholar]
- Ho Y., Wigglesworth K., Eppig J. J., Schultz R. M. (1995). Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 41, 232-238 [DOI] [PubMed] [Google Scholar]
- Houde M., Laprise P., Jean D., Blais M., Asselin C., Rivard N. (2001). Intestinal epithelial cell differentiation involves activation of p38 mitogen-activated protein kinase that regulates the homeobox transcription factor CDX2. J. Biol. Chem. 276, 21885-21894 [DOI] [PubMed] [Google Scholar]
- Houghton F. D. (2006). Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 74, 11-18 [DOI] [PubMed] [Google Scholar]
- Hyafil F., Morello D., Babinet C., Jacob F. (1980). A cell surface glycoprotein involved in the compaction of embryonal carcinoma cells and cleavage stage embryos. Cell 21, 927-934 [DOI] [PubMed] [Google Scholar]
- James R., Erler T., Kazenwadel J. (1994). Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J. Biol. Chem. 269, 15229-15237 [PubMed] [Google Scholar]
- Jedrusik A., Parfitt D. E., Guo G., Skamagki M., Grabarek J. B., Johnson M. H., Robson P., Zernicka-Goetz M. (2008). Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 22, 2692-2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A., Bruce A. W., Tan M. H., Leong D. E., Skamagki M., Yao M., Zernicka-Goetz M. (2010). Maternally and zygotically provided Cdx2 have novel and critical roles for early development of the mouse embryo. Dev. Biol. 344, 66-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. H., Rossant J. (1981). Molecular studies on cells of the trophectodermal lineage of the postimplantation mouse embryo. J. Embryol. Exp. Morphol. 61, 103-116 [PubMed] [Google Scholar]
- Johnson M. H., McConnell J. M. (2004). Lineage allocation and cell polarity during mouse embryogenesis. Semin. Cell Dev. Biol. 15, 583-597 [DOI] [PubMed] [Google Scholar]
- Johnson M. H., Maro B., Takeichi M. (1986). The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J. Embryol. Exp. Morphol. 93, 239-255 [PubMed] [Google Scholar]
- Klaffky E., Williams R., Yao C. C., Ziober B., Kramer R., Sutherland A. (2001). Trophoblast-specific expression and function of the integrin alpha 7 subunit in the peri-implantation mouse embryo. Dev. Biol. 239, 161-175 [DOI] [PubMed] [Google Scholar]
- Li L., Baibakov B., Dean J. (2008). A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell 15, 416-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz O., Duluc I., Arcangelis A. D., Simon-Assmann P., Kedinger M., Freund J. N. (1997). Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J. Cell Biol. 139, 1553-1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz O., Cadoret A., Duluc I., Capeau J., Gespach C., Cherqui G., Freund J. N. (1999). Downregulation of the colon tumour-suppressor homeobox gene Cdx-2 by oncogenic ras. Oncogene 18, 87-92 [DOI] [PubMed] [Google Scholar]
- Lu C. W., Yabuuchi A., Chen L., Viswanathan S., Kim K., Daley G. Q. (2008). Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat. Genet. 40, 921-926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Jaenisch R. (2006). Generation of nuclear transfer-derived pluripotent ES cells from cloned Cdx2-deficient blastocysts. Nature 439, 212-215 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. (2008). Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270-283 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., et al. (2009). The hippo signaling pathway components lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410 [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917-929 [DOI] [PubMed] [Google Scholar]
- Niwa T., Yamashita S., Tsukamoto T., Kuramoto T., Nomoto T., Wakazono K., Fujita H., Matsushima T., Tatematsu M., Sugimura T., et al. (2005). Whole-genome analyses of loss of heterozygosity and methylation analysis of four tumor-suppressor genes in N-methyl-N′-nitro-N-nitrosoguanidine-induced rat stomach carcinomas. Cancer Sci. 96, 409-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M., Ohsawa T., Semba R. (1993). Similar responses to pharmacological agents of 1,2-OAG-induced compaction-like adhesion of two-cell mouse embryo to physiological compaction. J. Exp. Zool. 265, 604-608 [DOI] [PubMed] [Google Scholar]
- Ohsugi M., Hwang S. Y., Butz S., Knowles B. B., Solter D., Kemler R. (1996). Expression and cell membrane localization of catenins during mouse preimplantation development. Dev. Dyn. 206, 391-402 [DOI] [PubMed] [Google Scholar]
- O'Sullivan C. M., Rancourt S. L., Liu S. Y., Rancourt D. E. (2001). A novel murine tryptase involved in blastocyst hatching and outgrowth. Reproduction 122, 61-71 [DOI] [PubMed] [Google Scholar]
- Palmieri S. L., Peter W., Hess H., Scholer H. R. (1994). Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166, 259-267 [DOI] [PubMed] [Google Scholar]
- Pauken C. M., Capco D. G. (1999). Regulation of cell adhesion during embryonic compaction of mammalian embryos: roles for PKC and beta-catenin. Mol. Reprod. Dev. 54, 135-144 [DOI] [PubMed] [Google Scholar]
- Plusa B., Frankenberg S., Chalmers A., Hadjantonakis A. K., Moore C. A., Papalopulu N., Papaioannou V. E., Glover D. M., Zernicka-Goetz M. (2005). Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 118, 505-515 [DOI] [PubMed] [Google Scholar]
- Qin J., Diaz-Cueto L., Schwarze J. E., Takahashi Y., Imai M., Isuzugawa K., Yamamoto S., Chang K. T., Gerton G. L., Imakawa K. (2005). Effects of progranulin on blastocyst hatching and subsequent adhesion and outgrowth in the mouse. Biol. Reprod. 73, 434-442 [DOI] [PubMed] [Google Scholar]
- Ralston A., Rossant J. (2008). Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614-629 [DOI] [PubMed] [Google Scholar]
- Riley P., Anson-Cartwright L., Cross J. C. (1998). The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 18, 271-275 [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P. P. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701-713 [DOI] [PubMed] [Google Scholar]
- Silberg D. G., Swain G. P., Suh E. R., Traber P. G. (2000). Cdx1 and cdx2 expression during intestinal development. Gastroenterology 119, 961-971 [DOI] [PubMed] [Google Scholar]
- Sritanaudomchai H., Sparman M., Tachibana M., Clepper L., Woodward J., Gokhale S., Wolf D., Hennebold J., Hurlbut W., Grompe M., et al. (2009). CDX2 in the formation of the trophectoderm lineage in primate embryos. Dev. Biol. 335, 179-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R. O., Yamanaka Y., Rossant J. (2010). Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 137, 3383-3391 [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093-2102 [DOI] [PubMed] [Google Scholar]
- Suh E., Chen L., Taylor J., Traber P. G. (1994). A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 14, 7340-7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. C., McGinnis L. K., Lawitts J. A., Raffin M., Biggers J. D. (2000). IVF of mouse ova in a simplex optimized medium supplemented with amino acids. Hum. Reprod. 15, 1791-1801 [DOI] [PubMed] [Google Scholar]
- Sun Q. Y., Wu G. M., Lai L., Park K. W., Cabot R., Cheong H. T., Day B. N., Prather R. S., Schatten H. (2001). Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction 122, 155-163 [PubMed] [Google Scholar]
- Tao G. Z., Looi K. S., Toivola D. M., Strnad P., Zhou Q., Liao J., Wei Y., Habtezion A., Omary M. B. (2009). Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J. Cell Sci. 122, 3851-3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski A. K., Wroblewska J. (1967). Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155-180 [PubMed] [Google Scholar]
- Wang H., Ding T., Brown N., Yamamoto Y., Prince L. S., Reese J., Paria B. C. (2008). Zonula occludens-1 (ZO-1) is involved in morula to blastocyst transformation in the mouse. Dev. Biol. 318, 112-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C. A., Henry R. W., Knott J. G. (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One 5, e10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. T., Piotrowska K., Ciemerych M. A., Milenkovic L., Scott M. P., Davis R. W., Zernicka-Goetz M. (2004). A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6, 133-144 [DOI] [PubMed] [Google Scholar]
- Wiley L. M. (1984). Cavitation in the mouse preimplantation embryo: Na/K-ATPase and the origin of nascent blastocoele fluid. Dev. Biol. 105, 330-342 [DOI] [PubMed] [Google Scholar]
- Winkel G. K., Ferguson J. E., Takeichi M., Nuccitelli R. (1990). Activation of protein kinase C triggers premature compaction in the four-cell stage mouse embryo. Dev. Biol. 138, 1-15 [DOI] [PubMed] [Google Scholar]
- Wu G., Gentile L., Do J. T., Cantz T., Sutter J., Psathaki K., Arauzo-Bravo M. J., Ortmeier C., Scholer H. (2010). Efficient derivation of pluripotent stem cells from siRNA-mediated Cdx2-deficient mouse embryos. Stem Cells Dev. doi:10.1089/scd.2010.0128 [DOI] [PMC free article] [PubMed]
- Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., DePamphilis M. L., Buonanno A. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827-3836 [DOI] [PubMed] [Google Scholar]
- Yoshida-Noro C., Suzuki N., Takeichi M. (1984). Molecular nature of the calcium-dependent cell-cell adhesion system in mouse teratocarcinoma and embryonic cells studied with a monoclonal antibody. Dev. Biol. 101, 19-27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.