Abstract
Sonic hedgehog signalling is essential for the embryonic development of many tissues including the central nervous system, where it controls the pattern of cellular differentiation. A genome-wide screen of neural progenitor cells to evaluate the Shh signalling-regulated transcriptome identified the forkhead transcription factor Foxj1. In both chick and mouse Foxj1 is expressed in the ventral midline of the neural tube in cells that make up the floor plate. Consistent with the role of Foxj1 in the formation of long motile cilia, floor plate cells produce cilia that are longer than the primary cilia found elsewhere in the neural tube, and forced expression of Foxj1 in neuroepithelial cells is sufficient to increase cilia length. In addition, the expression of Foxj1 in the neural tube and in an Shh-responsive cell line attenuates intracellular signalling by decreasing the activity of Gli proteins, the transcriptional mediators of Shh signalling. We show that this function of Foxj1 depends on cilia. Nevertheless, floor plate identity and ciliogenesis are unaffected in mouse embryos lacking Foxj1 and we provide evidence that additional transcription factors expressed in the floor plate share overlapping functions with Foxj1. Together, these findings identify a novel mechanism that modifies the cellular response to Shh signalling and reveal morphological and functional features of the amniote floor plate that distinguish these cells from the rest of the neuroepithelium.
Keywords: Cilia, Neural tube, Sonic hedgehog signalling, Mouse, Chick
INTRODUCTION
Organising centres that coordinate the molecular, cellular and morphogenetic events underpinning tissue formation are of fundamental importance for embryonic development. In the central nervous system one such organising centre is the floor plate (FP). This morphologically distinct structure, first described in 1888 by Wilhelm His, is situated in the ventral midline of the neural tube (Kingsbury, 1930; Placzek and Briscoe, 2005). FP cells play an important role in the morphogenesis of the neural tube and are the source of positional cues that instruct cells to acquire distinct neuronal fates and guide axons as they establish functional connections (Strähle et al., 2004).
The secreted protein sonic hedgehog (Shh) performs the principal extrinsic organising function of the FP (Marti et al., 1995; Roelink et al., 1995). Moreover, the induction of the FP itself depends on Shh produced from the notochord that underlies the neural tube (Chiang et al., 1996). The intracellular transmission of Shh signalling requires two transmembrane proteins: patched 1 (Ptch1), the receptor that binds Shh, and smoothened (Smo), which initiates intracellular signalling. In the absence of Shh, Ptch1 inhibits Smo activity (Hooper and Scott, 2005; Huangfu and Anderson, 2006; Lum et al., 2003; Varjosalo and Taipale, 2008). Relief of this inhibition, which is achieved by Shh binding to Ptch1, results in changes in the activity of the Gli transcription factors, reducing their transcriptional inhibitory activity and increasing their transcriptional activator function (Bai et al., 2004; Huangfu and Anderson, 2006; Huangfu et al., 2003; Jacob and Briscoe, 2003; Ruiz i Altaba et al., 2003; Stamataki et al., 2005). The mechanism of signal transduction between Ptch1, Smo and Gli proteins is still poorly understood but, in vertebrates, it appears to depend on the primary cilium of a cell and on the integrity of intraflagellar transport (Goetz and Anderson, 2010; Huangfu and Anderson, 2006; Huangfu et al., 2003). Several components of the Shh pathway, including Ptch1, Smo and Gli proteins, accumulate in primary cilia, and primary cilia are necessary for the formation of both repressor and activator isoforms of Gli proteins (Corbit et al., 2005; Haycraft et al., 2005; Ko et al., 2010; Milenkovic et al., 2009; Rohatgi et al., 2007). Nevertheless, the exact role of the cilium in Shh signalling is unclear. It is notable that some cells possess cilia that differ from the primary cilia found on most cells: these include motile cilia found on cells of the node and in some epithelia and the non-motile specialised cilia of olfactory neurons and photoreceptors (Eggenschwiler and Anderson, 2007; Satir and Christensen, 2007). Whether the architecture and morphology of cilia affect the transduction of Shh signalling and whether alterations in cilia are exploited during normal development to modulate Shh signalling remain to be determined.
Within responding cells in the neural tube, Shh signalling controls a gene regulatory network (Dessaud et al., 2008). This comprises a set of transcription factors that are differentially expressed along the dorsal-ventral (DV) axis of the neural tube in response to a ventral-to-dorsal gradient of Shh signalling (Briscoe and Ericson, 2001; Ericson et al., 1997). For example, high concentrations of Shh induce the expression of FoxA2 and Nkx2.2, whereas lower Shh concentrations are sufficient to induce Olig2 (Dessaud et al., 2007; Ericson et al., 1997). Ultimately, the combination of transcription factors expressed in a progenitor cell determines its fate (Briscoe and Ericson, 2001; Dessaud et al., 2008). Despite the importance of the transcriptional network, our knowledge of the genes that are regulated by Shh signalling in neural cells is fragmentary. In the case of the FP, the Shh-dependent induction of the forkhead transcription factor FoxA2 has a prominent function (Ang and Rossant, 1994; Norton et al., 2005; Ribes et al., 2010; Ruiz i Altaba et al., 1993; Weinstein et al., 1994). Whether expression of FoxA2 is sufficient to explain all the features of FP cells has not been established.
To better understand the Shh-controlled gene regulatory network in the neural tube, we carried out a genome-wide expression screen in neural cells. This led us to focus on Foxj1, a forkhead family transcription factor that is upregulated by Shh signalling and expressed in the FP of both mouse and chick embryos. Foxj1 is associated with the production of motile cilia that are several times the length of primary cilia (Blatt et al., 1999; Chen et al., 1998; Stubbs et al., 2008; Tichelaar et al., 1999; Yu et al., 2008). Consistent with this, FP cells produce cilia that are two to three times the length of the primary cilia produced by other neural progenitors. Strikingly, we found that Foxj1 attenuates Shh signal transduction and we provide evidence that this relies on its ability to alter cilia structure and to modify the intracellular localisation of Gli2 protein. However, neither ciliogenesis nor the dynamics of Shh signalling appears to be affected in the FP of mice lacking Foxj1. We provide evidence that the expression of FoxA2 in FP cells is sufficient to block Shh signalling, whereas Rfx3, another transcription factor implicated in the generation of motile cilia (Bonnafe et al., 2004), is also expressed in the FP and sufficient to induce long cilia. Together, the data provide insight into the establishment of FP identity and reveal a novel mechanism involving changes in cilia architecture that modulates the response of cells to Shh signalling.
MATERIALS AND METHODS
Immunohistochemistry, in situ hybridisation and electron microscopy
Antibody reagents and protocols have been described previously (Briscoe et al., 2000; Dessaud et al., 2007; Stamataki et al., 2005). In addition, acetylated α-tubulin (Sigma), Arl13b (Caspary et al., 2007), ZO-1 (Tjp1) (Abcam), Arx (Poirier et al., 2004), Smo (Rohatgi et al., 2007) and Gli2 (Ko et al., 2010) antibodies were used. Immunofluorescence microscopy was carried out using a Leica TCS SP2 confocal microscope and images were processed with Photoshop 7.0 software (Adobe Systems, San Jose, CA, USA). In situ hybridisation was performed as described (Schaeren-Wiemers and Gerfin-Moser, 1993) using mouse probes to Foxj1 (BC082543), Shh (Echelard et al., 1993), Ptch1 (C. C. Hui, University of Toronto, ON, Canada), Rfx3 (BC017598) and Lrd (AF183144) and chick probes to Foxj1 (XM_001233326), Ptch1 (Persson et al., 2002) and Gli1 (C. Tabin, Harvard University, MA, USA). Scanning electron microscopy and transmission electron microscopy were performed as described previously (Hirst and Howard, 1992).
Mouse and chick lines and in ovo electroporation
Mice heterozygous for the Shh null allele (Chiang et al., 1996), Foxj1 null allele (Brody et al., 2000) and talpid3 heterozygous chicks (Davey et al., 2006) were used to generate homozygous mutant embryos. Electroporation constructs were based on the pCAGGS expression vector (Niwa et al., 1991) engineered to bi-cistronically express nuclear-targeted GFP (pCAGGS-IRES NLS-GFP). Gli3AHIGH (Stamataki et al., 2005), Ptc1Δloop2 (Briscoe et al., 2001), SmoM2 (Hynes et al., 2000) and FoxA2 (Jacob et al., 2007) were described previously. cDNAs encoding Foxj1 (BC082543) and Rfx3 (BC017598) were cloned into the pCAGGS-IRES-GFP vector. HH stage 10-12 chick embryos were electroporated and incubated in ovo before dissection and processing for immunohistochemistry, in situ hybridisation or FACS.
FACS and RNA extraction
Briefly, HH stage 10-12 chick embryos were electroporated in ovo and embryos collected at the indicated time points. Cells from electroporated embryos were dissociated and GFP-expressing cells purified by FACS. RNA was extracted using Trizol (Invitrogen) and the quality assessed with a Bioanalyser 2100 (Agilent).
Acquisition and analysis of microarray data
Hybridisation to microarrays and array processing were carried out according to the manufacturer's instructions (Affymetrix). Two-cycle cDNA synthesis was performed from 35-50 ng of total RNA and hybridised to the GeneChip Chicken Genome Array (Affymetrix). Analysis of microarray data was performed using GeneSpring 7.2 and Bioconductor (Gentleman et al., 2004). Signal intensity measurements from individual arrays were obtained using the Affymetrix Mas5.0 algorithm. For statistical analysis, data from three biological replicates of each experiment were averaged. Data were filtered to remove probes with a signal intensity that was not significantly above background. The significance analysis of microarrays (SAM) algorithm was used to identify significant differences in expression by pairwise comparisons between data sets and a false discovery rate (FDR) of below 15% was used (Tusher et al., 2001). Data from this analysis were then subjected to hierarchical and k-means clustering. Mammalian orthologues of chick genes were identified using BioMart (www.ensembl.org). Gene ontology annotation was assigned using FatiGO (Al-Shahrour et al., 2007). Microarray data are available from ArrayExpress with accession E-MEXP-2212.
Cell culture
For immunohistochemistry and luciferase reporter assays in NIH 3T3 cells, 24 hours after seeding, cells were transfected using FuGENE HD Transfection Reagent (Roche) or Lipofectamine (Sigma). After reaching confluency (24-48 hours), cells were switched to medium containing 0.5% NBCS (newborn calf serum; Hyclone) and 12 hours later the medium was supplemented with recombinant Shh protein (464-SH, R&D Systems) or vehicle control for 24-48 hours.
Luciferase reporter assays
Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega). Foxj1, SmoM2, Gli3AHIGH expression plasmids or pCAGGS empty vector were transfected into NIH 3T3 or HH stage 10-12 chick embryos together with the GBS-Luc firefly luciferase reporter (Sasaki et al., 1997) as previously described (Stamataki et al., 2005).
RESULTS
Transcriptional profiling of neural cells identifies the transcription factor Foxj1
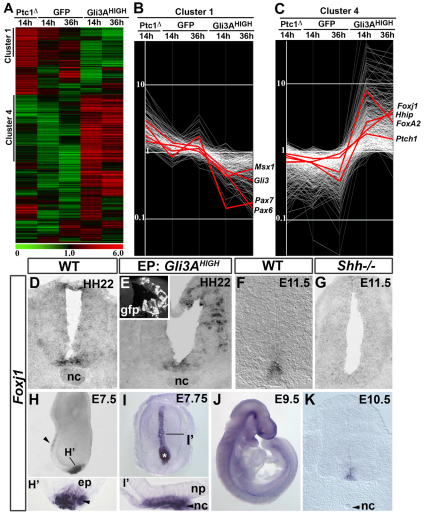
We systematically compared gene expression in chick neural progenitors in which Shh signalling had been cell-autonomously activated (Gli3AHIGH) or blocked (Ptc1Δloop2) (Briscoe et al., 2001; Stamataki et al., 2005). Hamburger and Hamilton (HH) stage 11 chick embryos electroporated with these constructs were allowed to develop in ovo for 14 or 36 hours. Bi-cistronically encoded nuclear-targeted GFP was then used to FACS purify dissociated transfected cells. RNA was extracted and gene expression assayed with Affymetrix GeneChips. Clustering the data identified seven groups (Fig. 1A; see Tables S1 and S2 in the supplementary material), two of which were of immediate interest. Cluster 1 contained 182 transcripts including several previously identified as dorsally expressed in the neural tube and downregulated by Shh signalling, e.g. Gli3, Msx1, Pax6 and Pax7 (Fig. 1B). By contrast, genes in cluster 4 displayed the opposite behaviour. This cluster comprised 464 transcripts (Fig. 1C) and included 12 genes previously characterised as being induced by Shh signalling and/or restricted to the ventral neural tube. The expression pattern and regulation by Shh signalling of selected genes from these clusters were validated (see Fig. S1 in the supplementary material), confirming that the screening strategy accurately predicted the expression profile of a series of DV restricted genes.
Fig. 1.
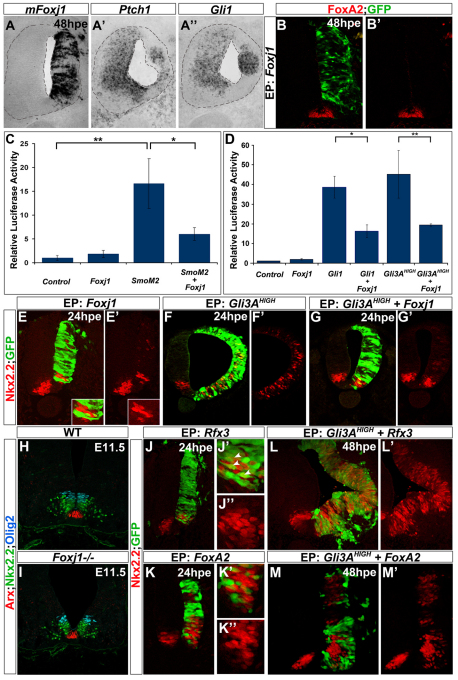
Transcriptional profiling identifies Foxj1 as differentially regulated by Shh signalling in the neural tube. (A) Hierarchical clustering of genes up- and downregulated in chick neural tube in each of the datasets. Columns represent the mean expression of three biological replicates for each dataset and rows represent individual probes (red, increased expression; green, decreased expression). Two clusters (1 and 4) of co-regulated genes have opposite modes of regulation by positive and negative Shh signalling. (B,C) Plots of normalised expression values for genes in cluster 1 (B) and cluster 4 (C). (B) Cluster 1 comprises genes downregulated by Shh signalling and includes Msx1, Gli3, Pax6 and Pax7. (C) Cluster 4 includes FoxA2, Ptch1, Hhip and Foxj1, which are induced by Shh signalling. (D-K) In situ hybridisation of Foxj1 in chick (D,E) and mouse (F-K) embryos. Foxj1 is expressed within floor plate (FP) cells of HH stage 22 chick neural tube and E11.5 mouse embryos (D,F). Gli3AHIGH induces Foxj1 expression at 48 hours post-electroporation (hpe) (E). By contrast, Foxj1 expression is extinguished in the neural tube of Shh–/– embryos (G). At E7.5 and E7.75, axial mesoderm cells and ventral cells of the node, but not neural cells, express Foxj1 (H,I and transverse section in H′,I′). At E9.5 and E10.5, Foxj1 is expressed in the midline of the neural tube (J,K). ep, epiblast; np, neural plate; nc, notochord; Ptc1Δ, Ptc1Δloop2.
We initially focused on the forkhead family transcription factor Foxj1 in the cluster of genes upregulated by Shh signalling (Clevidence et al., 1993; Hackett et al., 1995). In agreement with the microarray analysis (Fig. 1C), Foxj1 was expressed prominently in the ventral neural tube within FP cells in both mouse and chick (Fig. 1D,F,K). Moreover, Foxj1 expression was ectopically induced in the dorsal neural tube by Gli3AHIGH (Fig. 1E) and was lost in the spinal cord of Shh mutant embryos (Fig. 1F,G). In chick, the neural expression of Foxj1 was first observed at around HH stage 18 [∼30-35 somite stage (ss)] (see Fig. S2E-H in the supplementary material). In mouse, Foxj1 is induced within the ventral midline of the neural tube at embryonic day (E) 9.0 (16-20 ss), similar to Shh (Jeong and McMahon, 2005) (Fig. 1J). Before these stages, whole-mount in situ hybridisation confirmed Foxj1 expression in the node of E7.5 mouse embryos (Fig. 1H,H′) (Brody et al., 2000) and within all axial mesoderm cells and ventral cells of the node at E7.75 (Fig. 1I,I′). This mesodermal expression declined from E8.5 onwards, corresponding to the time at which Foxj1 was induced in the anterior neural tube (Fig. 1K; see Fig. 3A, inset).
Fig. 3.
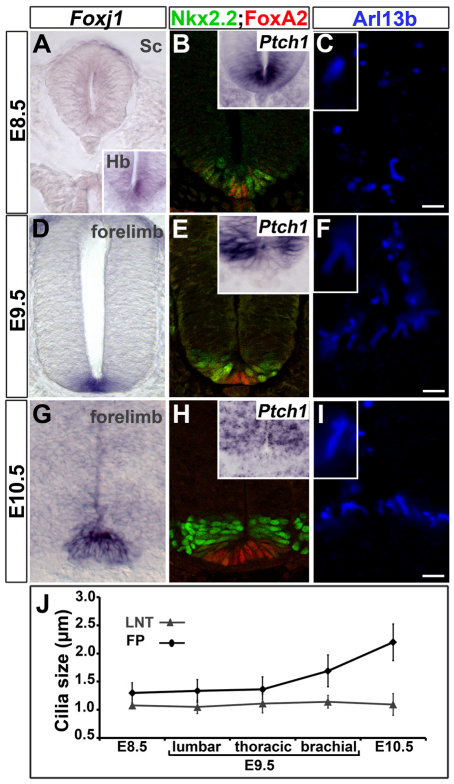
Dynamics of floor plate gene expression and generation of long cilia. (A-I) In situ hybridisation of Foxj1 (A,D,G) and Ptch1 (insets in B,E,H) and immunostaining for Nkx2.2 (green) and FoxA2 (red) (B,E,H) and Arl13b (white; C,F,I) in transverse sections of the forelimb region of mouse embryos at the indicated stages. At E8.5, Foxj1 is detected within the ventral midline of the anterior (inset in A) but not caudal neural tube (main panel in A). Nkx2.2, FoxA2 and Ptch1 (B) are expressed in ventral midline cells. At E9.5 and E10.5, FP expresses Foxj1 (D,G) and FoxA2, whereas Nkx2.2 and Ptch1 (E,H) are no longer expressed in the midline. Insets (C,F,I) show a ventral midline cilium at higher magnification (2× zoom). (J) Quantification of cilia length in the lateral neural tube (LNT) and ventral midline (FP) at the indicated stages (mean ± s.d.). At E8.5, and in the lumbar and thoracic regions of E9.5 mouse embryos, all neuroepithelial cells have primary cilia of similar lengths (C). At brachial regions of E9.5 and E10.5 embryos, FP cells have longer cilia than do neural progenitors (F,I). Scale bars: ∼2 μm.
Foxj1 expression is linked to a distinct ciliogenesis programme in the floor plate
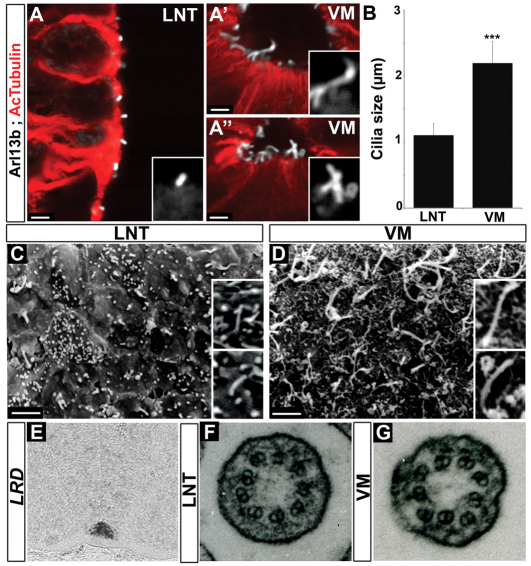
Foxj1 is associated with the production of motile cilia in several tissues (Blatt et al., 1999; Brody et al., 2000; Chen et al., 1998; Stubbs et al., 2008; Tichelaar et al., 1999; Yu et al., 2008; Yu et al., 2008). We therefore characterised neural tube cilia in chick and mouse. We first examined the distribution of Arl13b, a small GTPase that localises to cilia (Caspary et al., 2007), and of acetylated α-tubulin (AcTubulin), which marks stable microtubules found in cilia (Muresan et al., 1993). Analysis of sections of mouse E10.5 and chick HH stage 20 neural tube revealed the presence of long cilia protruding from the surface of the FP into the ventricle (Fig. 2A-B; see Fig. S2A-B′ in the supplementary material). These were ∼2 μm in length, twice as long as the primary cilia found on adjacent non-FP neuroepithelial cells (Fig. 2B; see Fig. 4D). Scanning electron microscopy (SEM) confirmed the presence of primary cilia ∼1 μm in length in the centre of the apical surface of non-FP neuroepithelial cells of both mouse and chick (Fig. 2C; see Fig. S2D in the supplementary material). By contrast, the cilia on the apical surface of FP cells of both species were much longer (Fig. 2D; see Fig. S2C in the supplementary material), and in both cases the density of these cilia appeared consistent with FP cells producing monocilia (Satir and Christensen, 2007). Thus, the amniote FP, like its zebrafish counterpart (Yu et al., 2008), produces morphologically distinct cilia that are longer than the primary cilia generated by other neuroepithelial cells.
Fig. 2.
Floor plate cells generate long cilia. (A-A′) Acetylated α-tubulin (AcTubulin; red) and Arl13b (white) staining of transverse sections of the brachial region of E10.5 mouse embryos. Insets (A,A′) show a cilium at higher magnification (2× zoom). (B) Quantification of cilia length in ventral midline (VM) and lateral neural tube (LNT) cells (mean ± s.d.). Cilia averaging 2 μm in length are detected at the ventral midline (A′,A′,B), whereas cells from the lateral neural tube exhibit cilia of 1 μm (A,B). ***, P=3.2×10–13; Student's t-test. (C,D) SEM analysis of the ventricular surface of the neural tube of E11.5 mouse embryos. The apical surface of cells of the lateral neural tube exhibits short cilia (C), whereas ventral midline cells have long cilia (D). (E) In situ hybridisation of Lrd in transverse sections of the thoracic region of E11.5 mouse embryos indicates expression in the ventral midline. (F,G) TEM of cilia from the ventricular surface of the lateral neural tube (F) and ventral midline (G) of E11.5 mouse embryos. Cilia from both locations have nine peripheral doublets microtubule. Scale bars: ∼2 μm.
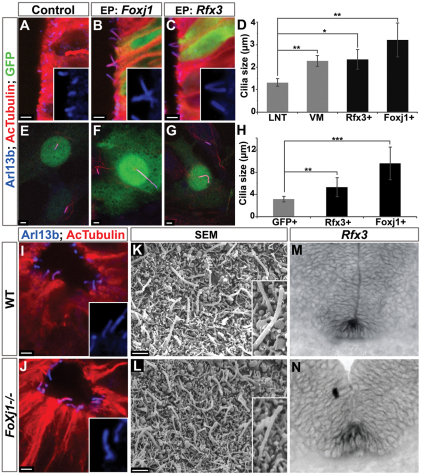
Fig. 4.
Foxj1 induces the formation of long cilia but is not required for floor plate ciliogenesis. (A-C) HH stage 10-12 chick embryos electroporated (EP) in ovo with Foxj1 (B) and Rfx3 (C) (GFP, green) and analysed at 48 hpe for the distribution of AcTubulin (red) and Arl13b (blue) versus control (A). (D) Quantification of cilia length in lateral neural tube (LNT), ventral midline (VM), and in Rfx3- and Foxj1-transfected cells (mean ± s.d.). *, P<10–3; **, P<10–9; Student's t-test. (E-G) NIH 3T3 cells transfected with GFP, Foxj1 or Rfx3 (transfected cells are green) and analysed 48 hours later for AcTubulin (red) and Arl13b (blue). (H) Quantification of cilia length in transfected NIH 3T3 cells (mean ± s.d.). **, P<10–9; ***, P<10–22; Student's t-test. Foxj1 and Rfx3 are sufficient to lengthen the monocilia of lateral neural tube and NIH 3T3 cells. (I,J) AcTubulin (red) and Arl13b (blue) staining of transverse sections at the forelimb level of E10.5 wild-type (WT, I) and Foxj1 mutant (J) mouse neural tube. (K,L) SEM analysis of cilia in the ventral midline of the neural tube of E11.5 Foxj1–/– (L) and control (K) mouse embryos. (M,N) Rfx3 expression in sections of the thoracic region of E11.5 Foxj1–/– (N) or control (M) mouse embryos. Foxj1 is not required for the generation of long cilia or for Rfx3 expression in the FP. All insets show a cilium at higher magnification (2× zoom). Scale bars: ∼2 μm.
To characterise FP cilia further, we examined the expression of Dnahc11 (left-right dynein; Lrd) and of the transcription factor Rfx3, both of which are implicated in the production of motile cilia (Bonnafe et al., 2004; Supp et al., 1999). Similar to Foxj1, both genes were expressed in the FP of mouse embryos (Fig. 2E; see Fig. 4M) (Cohen and Meininger, 1987; Supp et al., 1997). We next examined the ultrastructure of FP cilia by transmission electron microscopy (TEM). Motile cilia usually display an extra central pair of microtubules, although motile cilia with a 9+0 structure are observed in the node (Cohen and Meininger, 1987; Nonaka et al., 1998; Sulik et al., 1994; Takeda et al., 1999). TEM of E11.5 mouse embryos indicated that cilia from both FP and non-FP regions of the neural tube exhibit a 9+0 arrangement with nine peripheral doublet microtubules located around the circumference (n=15 individual cilia for ventral midline sections) (Fig. 2F,G). Thus, the molecular and ultrastructure data (Fig. 2D,F,G) suggested that FP cilia are similar to the long, motile 9+0 cilia found in the node. To assay for movement directly, we attempted video microscopy of FP cilia, but this failed to provide conclusive evidence of motility (data not shown). In part, this is likely to be because accessing the ventral neural tube at the requisite developmental stages is difficult and technical improvements will be required before definitive data can be obtained. Nevertheless, the molecular data are consistent with FP cilia being motile, in line with observations in zebrafish (Stubbs et al., 2008; Yu et al., 2008).
The timing of Foxj1 expression corresponds to the appearance of FP identity
In order to investigate the function of Foxj1 in the development of the FP, we first established the dynamics of its expression in the neural tube with respect to other hallmarks of FP identity. In E8.5 mouse embryos, definitive FP identity has yet to be established (Ribes et al., 2010) and direct targets of Shh signalling, including Ptch1, FoxA2 and Nkx2.2 (Vokes et al., 2007), were expressed in ventral midline cells (Fig. 3B). At this time, Foxj1 was not expressed in the spinal cord (Fig. 3A, main panel) and all neuroepithelial cells had primary cilia of similar lengths (Fig. 3C,J). By E9.5, presumptive FP cells are identifiable by their downregulation of Nkx2.2 and their decreased level of Shh signalling, indicated by the decreased expression of Ptch1 compared with cells in adjacent regions of the neural tube (Fig. 3E) (Ribes et al., 2010). Measurement of cilia length revealed that in anterior regions, which are developmentally more advanced, ventral midline cells had longer cilia than adjacent neural progenitors (Fig. 3F,J). This correlated with the induction of Foxj1 in the ventral midline of the neural tube (Fig. 3D). Finally, at E10.5, the FP robustly expressed Foxj1 and FoxA2 (Fig. 3G,H), whereas Nkx2.2 and Ptch1 were no longer expressed in these cells (Fig. 3H). At this stage, FP cilia were approximately twice the length of the cilia in other regions of the neural tube (Fig. 3I,J). Together, these data indicate that the onset of Foxj1 expression in the ventral midline of the neural tube correlates with these cells acquiring characteristic features of FP identity, including the generation of long cilia and the downregulation of Shh signalling and Nkx2.2 expression. This prompted us to examine whether Foxj1 is responsible for the acquisition of these FP attributes.
Foxj1 is sufficient but not necessary for the induction of long cilia in the FP
To test whether Foxj1 was sufficient to induce the long cilia characteristic of the FP, we assayed chick embryos transfected with Foxj1 (Fig. 4A,B,D). Long cilia protruding into the lumen of the neural tube were observed in regions ectopically expressing Foxj1 (Fig. 4B), but not on the control untransfected side of the neural tube (Fig. 4A). These cilia averaged up to three times the length of the primary cilia observed on untransfected neural progenitors (Fig. 4D). SEM analysis of transfected regions of the neural tube confirmed that Foxj1 was sufficient to increase the length of the primary cilium normally observed on each neural progenitor (see Fig. S2I,J in the supplementary material). Consistent with this observation, Foxj1 transfection in NIH 3T3 fibroblasts also increased the length of the primary cilia found in these cells (Fig. 4E,F,H). Together, these data suggest that Foxj1 induces the growth of a primary cilium, converting it into a long cilium.
To address whether Foxj1 is required for the generation of long cilia in the FP, we analysed mouse embryos containing a targeted deletion of Foxj1 (Brody et al., 2000). Immunofluorescence and SEM comparisons of cilia in the neural tube of E11.5 mouse embryos did not reveal marked defects in the length or distribution of cilia between control and Foxj1 mutant cells (Fig. 4I-L), although the gross morphology of the neural tube appeared abnormal in many mutants (see Fig. S3A,B in the supplementary material). Thus, Foxj1 does not appear to be necessary for FP cells to generate long cilia. We therefore analysed Shh-null embryos to determine whether Shh signalling was necessary for the generation of the long FP cilia. SEM indicated that the ventral midline of Shh–/– mouse embryos lacked long cilia; in their place, short cilia, similar to the primary cilia found on non-FP neuroepithelial cells, were present (see Fig. S3I,J in the supplementary material). These data suggested that other factors expressed in FP cells might compensate for the loss of Foxj1. Consistent with this, the expression of Dnahc11 (see Fig. S3G,H in the supplementary material) and Rfx3 (Fig. 4M,N) was unaffected by the loss of Foxj1. Moreover, in ovo electroporation of Rfx3 was sufficient to induce long monocilia in neural progenitors and NIH 3T3 cells (Fig. 4C,G). These cilia were similar in length to those normally found on FP cells (Fig. 4D,H). Together, these data suggest that Rfx3 might act redundantly with Foxj1 to induce the ciliogenesis programme that results in the lengthening of cilia in the FP.
Foxj1 alters the response of cells to Shh signalling
We next turned our attention to the decrease in sensitivity to Shh signalling that accompanies the induction and elaboration of FP identity (Ribes et al., 2010). We noticed that the forced expression of Foxj1 in the neural tube often resulted in a decrease in Ptch1 and Gli1 expression (Fig. 5A-A′). The inhibition of these direct targets of Shh signalling prompted us to assay Shh signalling using a reporter of Gli transcriptional activity (GBS-Luc) (Sasaki et al., 1997; Stamataki et al., 2005) in HH stage 10-12 chick embryos transfected with either activated Smo (SmoM2) (Xie et al., 1998) or Foxj1, or SmoM2 and Foxj1 together (Fig. 5C). Twenty-four hours post-electroporation (hpe), both SmoM2 alone and SmoM2 together with Foxj1 increased GBS-Luc reporter activity compared with controls. In the presence of Foxj1, however, the ability of SmoM2 to induce Gli activity was reduced by ∼50%. To test whether Foxj1 also attenuates Shh signalling in other cell types, we assessed the effect of Foxj1 expression in NIH 3T3 cells (see Fig. S4A in the supplementary material) (Taipale et al., 2000). NIH 3T3 cells were transfected with GBS-Luc, either alone or together with Foxj1, and their response to recombinant Shh protein assayed. Similar to neural cells, the expression of Foxj1 reduced the response of NIH 3T3 cells to Shh by ∼50%. Together, these data indicate that Foxj1 expression attenuates, but does not completely block, Shh signal transduction.
Fig. 5.
Foxj1 decreases the sensitivity of cells to Shh signalling at the level of Gli proteins. (A-A′) Transverse sections of chick neural tubes electroporated with Foxj1 and analysed 48 hpe for the expression of Foxj1 (A), Ptch1 (A′) and Gli1 (A′). Transfected regions are identified by Foxj1 expression (A). Reduced expression of Ptch1 and Gli1 was observed on the Foxj1-transfected side of the neural tube. (B,B′) HH stage 10-12 chick embryos were electroporated in ovo with Foxj1 and analysed 48 hpe for the expression of FoxA2. Foxj1 does not induce FoxA2 expression. (C) Relative luciferase activities in HH stage 10-12 chick embryos 24 hpe, electroporated with GFP (control), Foxj1, SmoM2 or SmoM2 and Foxj1, together with a Gli binding site firefly luciferase (GBS-Luc) reporter (Sasaki et al., 1997) and a normalisation plasmid. SmoM2 significantly induced Gli transcriptional activity compared with the control. Co-transfection of Foxj1 and SmoM2 resulted in ∼50% lower levels of Gli transcriptional activity compared with SmoM2 alone. Luciferase activity is shown relative to the control ± s.e.m. *, P=3×10–3; ** P<10–4; Student's t-test. (D) Relative GBS-Luc activities of HH stage 10-12 chick embryos electroporated with GFP (control), Foxj1, Gli1, Gli1 and Foxj1, Gli3AHIGH, Gli3AHIGH and Foxj1, assayed 24 hours after transfection. Gli1 generated a ∼35-fold and Gli3AHIGH a ∼45-fold increase in GBS luciferase activity compared with the control. Co-transfection of Foxj1 with Gli1 or Gli3AHIGH resulted in a ∼50% reduction in the induced Gli activity levels compared with Gli1 or Gli3AHIGH alone. Luciferase activity is shown relative to the control ± s.e.m. *, P=3×10–3; **, P<10–4; Student's t-test. (E,E′) HH stage 10-12 chick embryos were electroporated with Foxj1 and analysed 24 hpe for the expression of Nkx2.2 (red). Foxj1 is sufficient to repress Nkx2.2 in a cell-autonomous manner. (F-G′) HH stage 10-12 chick embryos electroporated with Gli3AHIGH (F,F′) or Gli3AHIGH together with Foxj1 (G,G′) analysed 24 hpe for the expression of Nkx2.2. Foxj1 represses the ectopic expression of Nkx2.2 (red) induced by Gli3AHIGH overexpression. (H,I) Arx (red), Nkx2.2 (green) and Olig2 (blue) expression in sections of the thoracic region of E11.5 Foxj1–/– (I) or wild-type (WT) control littermate (H) mouse embryos. Neural tube patterning is not affected in embryos lacking Foxj1. (J-M′) HH stage 10-12 chick embryos electroporated with Rfx3 (J-J′), FoxA2 (K-K′), Rfx3 together with Gli3AHIGH (L,L′) or FoxA2 together with Gli3AHIGH (M,M′) and analysed 24 hpe for the expression of Nkx2.2 (red). FoxA2, in contrast to Rfx3, blocks the endogenous as well as the Gli3AHIGH-induced expression of Nkx2.2 (red). Transfected cells are marked by GFP (green). EP, electroporation.
We next asked whether Foxj1 affects signal transduction at the level of Gli proteins. In ovo reporter assays were performed using Gli1 and Gli3AHIGH, which both generate high levels of Gli transcriptional activity (Fig. 5D) (Stamataki et al., 2005). Reporter activity was reduced by ∼50% in embryos co-transfected with Foxj1 and Gli1 or with Foxj1 and Gli3AHIGH, as compared with embryos transfected with Gli1 or Gli3AHIGH alone (Fig. 5D). This indicates that Foxj1 attenuates Shh signalling by inhibiting the ability of cells to produce the highest levels of Gli activity.
The inhibitory activity of Foxj1 on Shh signalling raised the possibility that Foxj1 represses the expression of Nkx2.2, which is downregulated in FP cells at a similar time to Ptch1 (Fig. 3B,E). Forced expression of Foxj1 was sufficient to block the expression of Nkx2.2 cell-autonomously (Fig. 5E). Importantly, this phenotype was not a consequence of cells acquiring an FP identity in response to Foxj1 expression, as expression of neither FoxA2 nor Arx was markedly altered (Fig. 5B; data not shown). To verify these observations, we examined whether Foxj1 was sufficient to inhibit SmoM2- or Gli3AHIGH-induced expression of Nkx2.2 (Stamataki et al., 2005). Both cell-autonomous and non-autonomous ectopic induction of Nkx2.2 was evident in embryos transfected with SmoM2 or Gli3AHIGH (Fig. 5F,F′; see Fig. S4B,B′ in the supplementary material), consistent with the ability of Shh signalling to induce Shh expression (data not shown). By contrast, in embryos that had been co-electroporated with Foxj1 and SmoM2 or Gli3AHIGH, the ability to induce Nkx2.2 was markedly reduced (Fig. 5G,G′; see Fig. S4C,C′ in the supplementary material). These data are consistent with Foxj1 decreasing the sensitivity of cells to Shh signalling and suggest that a consequence of the Foxj1 attenuation of Gli activity is the inhibition of Nkx2.2 expression.
To test whether Foxj1 is necessary for the changes in Shh sensitivity and gene expression normally observed during FP development, we analysed Foxj1–/– mouse embryos. Examination of molecular markers that identify FP cells, including Shh, Arx and FoxA2, indicated that the molecular identity of the FP was unchanged in Foxj1 mutants (Fig. 5H,I; see Fig. S3A-F in the supplementary material; data not shown), in accordance with the data on cilia length. Moreover, analysis of a range of neural progenitor markers, including Nkx2.2, Olig2 and Ptch1 (Fig. 5H,I; see Fig. S3C,D in the supplementary material and data not shown), did not reveal any marked alterations in the number or distribution of these cell types in Foxj1 mutants. The proliferation and differentiation of neural progenitors also appeared normal and the production of motoneurons was not significantly affected in the absence of Foxj1 (data not shown). Together, these data indicate that Foxj1 is not necessary for the establishment of FP identity, nor for DV patterning of the neural tube.
We asked whether Rfx3 could substitute for these functions of Foxj1. However, in cells ectopically expressing Rfx3, expression of Nkx2.2 was detected and the ability of SmoM2 or Gli3AHIGH to activate Nkx2.2 was not inhibited by Rfx3 (Fig. 5J,L; data not shown). These results suggest that (1) increasing the length of cilia is not sufficient, on its own, to modify Shh signalling and (2) other factors might compensate for Foxj1 loss of function to decrease Shh signalling in FP cells. Our attention turned to another member of the forkhead transcription factor family, FoxA2. We have previously shown that the ectopic expression of FoxA2 is able to downregulate the sensitivity of cells to Shh signalling (Ribes et al., 2010). Accordingly, in ovo electroporation of FoxA2 was sufficient to block the endogenous, as well as SmoM2- or Gli3AHIGH-induced, expression of Nkx2.2 (Fig. 5K,M). These data suggest that FoxA2 could share overlapping functions with Foxj1 in this aspect of FP development.
Cilia are required for Foxj1 to modify the sensitivity of cells to Shh
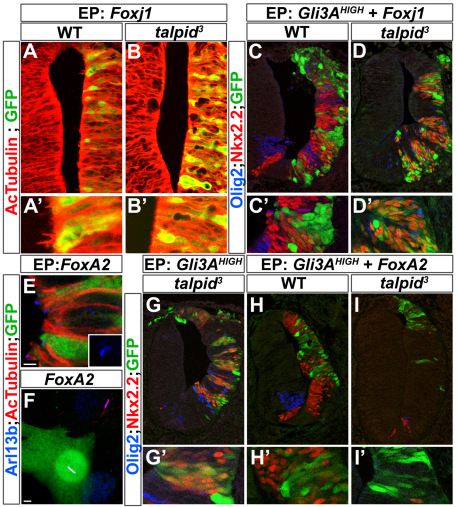
Finally, we asked whether a change in cilia structure is required for either Foxj1 or FoxA2 to repress Nkx2.2 expression. We compared the activity of Foxj1 and FoxA2 in talpid3 mutant chick embryos, which harbour a recessive mutation in a centrosomal protein and consequently lack all cilia (Davey et al., 2006; Yin et al., 2009). Consistent with the requirement for primary cilia in Shh signal transduction, embryos homozygous for the talpid3 mutation have defects in neural tube patterning (Davey et al., 2006). The expression of neither Foxj1 nor FoxA2 in talpid3 embryos restored ciliogenesis (Fig. 6A,B; data not shown). Moreover, FoxA2 did not appear to change the morphology of cilia in either wild-type neural progenitors or NIH 3T3 cells (Fig. 6E,F). In talpid3 mutant neural tube, the forced expression of Gli3AHIGH was sufficient to induce Nkx2.2 expression in a cell-autonomous manner, indicating that the absence of primary cilia did not prevent activated Gli proteins from upregulating Shh target gene expression (Fig. 6G,G′) (Davey et al., 2006). We therefore assayed Nkx2.2 induction in talpid3 embryos transfected with Gli3AHIGH and Foxj1 or FoxA2. In wild-type embryos, both transcription factors cell-autonomously inhibited Gli3AHIGH induction of Nkx2.2 (Fig. 6C,C′). Non-cell-autonomous induction was evident, as a result of non-transfected cells responding to the Shh produced from the transfected cells. In talpid3 mutant embryos, there was no non-cell-autonomous Nkx2.2 induction, consistent with the defective Shh response in talpid3 mutants (Fig. 6D,D′). By contrast, cell-autonomous induction of Nkx2.2 was observed in embryos transfected with Foxj1 and Gli3AHIGH (Fig. 6D,D′). Thus, in the absence of cilia, Foxj1 was unable to block the activity of Gli3AHIGH. However, FoxA2 continued to inhibit Gli3AHIGH induction of Nkx2.2 in talpid3 mutant embryos (Fig. 6H-I′), indicating that FoxA2 inhibits Nkx2.2 induction independently of cilia. By contrast, the activity of Foxj1 depends on its ability to alter cilia architecture.
Fig. 6.
Foxj1 does not repress Nkx2.2 induction in the absence of cilia. (A-B′) HH stage 10-12 wild-type (WT) and talpid3 chick embryos were electroporated with Foxj1 and analysed 48 hpe for AcTubulin (red). Foxj1 does not induce long cilia in talpid3 mutant embryos. (C-D′) HH stage 10-12 wild-type and talpid3 chick embryos were electroporated with Foxj1 together with Gli3AHIGH and analysed 48 hpe for the expression of Nkx2.2 (red) and Olig2 (blue). In wild-type embryos, Foxj1 cell-autonomously represses ectopic expression of Nkx2.2 and Olig2 induced by Gli3AHIGH. In talpid3 embryos, Foxj1 does not repress the induction of Nkx2.2 by Gli3AHIGH. (E,F) HH stage 10-12 chick embryos (E) and NIH 3T3 cells (F) electroporated with FoxA2 and analysed 48 hpe for the expression of AcTubulin (red) and Arlb13b (blue). FoxA2 is not sufficient to induce long cilia in the neural tube or in NIH 3T3 cells. (G-I′) HH stage 10-12 talpid3 chick embryos electroporated with Gli3AHIGH (G) or with Gli3AHIGH together with FoxA2 (H-I′) and analysed 48 hpe for the expression of Nkx2.2 (red) and Olig2 (blue). Gli3AHIGH overexpression is sufficient to induce Nkx2.2 and Olig2 expression in a cell-autonomous manner in talpid3 embryos. FoxA2 is sufficient to block the ectopic expression of Nkx2.2 and Olig2 induced by Gli3AHIGH in wild-type and talpid3 embryos. Transfected cells are marked by GFP (green). EP, electroporation.
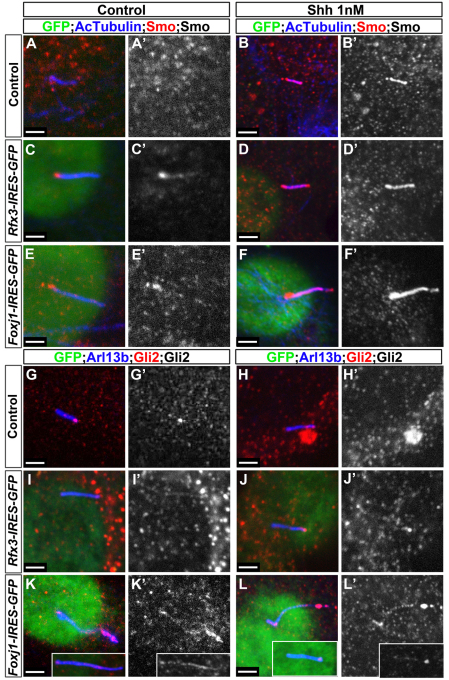
To investigate the mechanism by which Foxj1 modulates Gli activity, we analysed the subcellular localisation of Smo and Gli2, the Gli protein that provides the main activating function in vertebrates (Bai et al., 2004). Consistent with published data (Rohatgi et al., 2007), Smo translocated from the basal portion of the primary cilium to the entire shaft of the cilium of NIH 3T3 cells when exposed to Shh (Fig. 7A-B′), whereas Gli2 was observed at the tip of the cilium in the presence or absence of Shh (Fig. 7G-H′) (Haycraft et al., 2005). The localisation of Smo was unaffected in cells in which the ciliogenesis programme had been altered by expression of Foxj1 or Rfx3 (Fig. 7C-F′). By contrast, in many of the cells transfected with Foxj1, the concentration of Gli2 at the tip of the lengthened cilium was greatly increased compared with control or Rfx3-transfected cells (compare Fig. 7K-L′ with 7G-J′). In the remaining cells, lower levels of Gli2 were seen along the shaft of the cilia without accumulation at the tip (Fig. 7K-L′, insets). These data support the idea that the Foxj1-mediated attenuation of Gli activity is caused by defects in the ciliary localisation of Gli2 protein. Together, our findings suggest a novel mechanism for refining Shh signalling.
Fig. 7.
Gli2 protein accumulates in elongated cilia induced by Foxj1. (A-L′) Untransfected NIH 3T3 cells (A-B′,G-H′) or those transfected with Rfx3 (C-D′,I-J′) or with Foxj1 (E-F′,K-L′) were analysed 48 hours later for either AcTubulin (blue) and Smo (red) (A-F′) or Arl13b (blue) and Gli2 (red) (G-L′). Transfected cells are marked by GFP (green). In control cells, Smo is observed in the basal portion of the cilium in the absence of Shh (A,A′), whereas Smo is detected all along the shaft of the cilium in cells incubated with Shh (B,B′). The subcellular localisation of Smo is not affected by Foxj1 or Rfx3 transfection (C-F′). Gli2 is mainly detected at the tip of cilia in untransfected cells treated or otherwise with Shh (G-H′). This localisation is not affected in cells transfected with Rfx3 (I-J′). By contrast, in many Foxj1-transfected cells, Gli2 accumulation at the tip of the cilium is increased (main panels in K-L′). In the remaining Foxj1-transfected cells, Gli2 is detected along the shaft of the cilia (insets in K-L′). Scale bars: ∼2 μm.
DISCUSSION
A genome-wide screen identifies Shh signalling-regulated genes in the neural tube
We developed a systematic approach to profile in vivo gene expression in neural progenitors in which Shh signalling had been manipulated (Fig. 1). This genome-wide analysis identified two clusters of genes that responded in opposite ways to Shh signalling (see Tables S1 and S2 in the supplementary material). Alongside the well-studied genes in these two clusters, there were many genes that had not previously been implicated in neural development. In situ hybridisation with a subset of genes from the two clusters confirmed that their expression patterns were consistent with the microarray results (see Fig. S1 in the supplementary material). Functional annotation revealed a wide variety of attributes for genes in the two clusters and, moreover, there was an overlap between these genes and direct Gli target genes previously identified in mouse neural cells (Vokes et al., 2007) (see Tables S1 and S2 in the supplementary material). Together, the data confirm the efficacy of the screening strategy and highlight the pleiotropic activity of Shh signalling during neural development.
A distinct ciliogenesis programme associated with floor plate identity
The transcriptome analysis led us to the transcription factor Foxj1. The expression of Foxj1 in presumptive FP cells of the neural tube, but not in cells of the node or notochord, depends on Shh signalling (Fig. 1; data not shown). Whether Foxj1 is a direct target of Shh-induced Gli activity remains to be determined, but the observation in zebrafish as well as in amniotes that FP expression of Foxj1 is regulated by Shh signalling suggests a conserved regulatory mechanism (Yu et al., 2008).
At many sites of expression, Foxj1 is associated with the production and function of motile cilia (Blatt et al., 1999; Brody et al., 2000; Chen et al., 1998; Dziegielewska et al., 2001; Lim et al., 1997). Consistent with this, the FP in mouse and chick revealed the presence of long cilia. However, whether FP cilia are motile remains unclear. The molecular data are suggestive of motility because, in addition to Foxj1, FP cells express Dnahc11 (Supp et al., 1999), Pkd2 (McGrath et al., 2003; Pennekamp et al., 2002) and Rfx3 (Bonnafe et al., 2004), all of which are associated with motile cilia in the node. Furthermore, the cilia of the zebrafish FP appear to be motile (Yu et al., 2008). Nevertheless, it has proved impossible to visualise motility in the mouse or chick and the 9+0 structure of the FP cilia leaves open the possibility that FP cilia are long but immotile.
Foxj1 is necessary for the motile ciliogenesis programme of cells in several tissues (Brody et al., 2000; Chen et al., 1998), including the FP of zebrafish (Stubbs et al., 2008; Yu et al., 2008). Surprisingly, however, long cilia were still present in the FP of mouse embryos that lack Foxj1 (Fig. 4). This suggests that Foxj1 is not unique in its ability to alter the ciliogenesis programme of the mammalian FP. Consistent with this, Rfx3 expression is unaffected in mouse embryos lacking Foxj1 and ectopic expression of Rfx3 is sufficient to promote the generation of long cilia similar to those observed in the FP. Thus, Foxj1 and Rfx3 appear to have a similar capacity to alter the morphological features of cilia in neuroepithelial cells (Fig. 5). This suggests a parallel between the cilia found in the FP and those in the node. Analogous to the FP, cells in the mammalian node co-express Foxj1 and Rfx3 and generate long 9+0 cilia (Bonnafe et al., 2004; Brody et al., 2000; Takeda et al., 1999). Moreover, Foxj1 is dispensable for the formation of the long cilia in the node (Brody et al., 2000). This raises the possibility that the FP cilia are node-like in both generation and architecture.
Foxj1 alters the sensitivity of cells to Shh
Expression of Foxj1 alters the response of cells to Shh signalling. The attenuation of Shh signalling depends on the presence of cilia, suggesting that the change in the ciliogenesis programme elicited by Foxj1 affects Shh signal transduction. This is consistent with the observation that cilia are vital for the intracellular transmission of Shh signal (Goetz and Anderson, 2010; Huangfu et al., 2003; Caspary et al., 2007). In this context, our data raise the possibility that physiological changes to cilia during normal development are important for the quantitative interpretation of Shh signalling. However, such changes are unlikely to be simply in the length of cilia, as Rfx3 is sufficient to lengthen cilia but does not appear to diminish Shh signalling. Instead, the data suggest that specific alterations in the composition or architecture of cilia are responsible for the alteration in the intracellular transmission of Shh signal.
The effect on Shh signal transduction appears to be at the level of Gli protein activity. Gli proteins, along with other components of the signal transduction pathway, are present in cilia (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007), providing an opportunity for cilia structure to influence signal transmission to Gli proteins. Accordingly, there is a marked increase in the accumulation of Gli2 at the tip of the elongated cilia induced by ectopic expression of Foxj1, even though trafficking of Smo to the cilia appears unaffected (Fig. 7). Strikingly, accumulation of Gli2 has also been observed in some mouse mutants with shorter cilia, in which Shh signalling is impeded (Ko et al., 2010). However, an explanation of the changes in Gli activity that occur in cells expressing Foxj1 will require a better understanding of the molecular mechanisms of Shh signalling.
The in vivo significance of the Foxj1-dependent modulation of Shh signal transduction is unclear. A decrease in sensitivity of FP cells to Shh signalling is essential for the elaboration of their identity (Ribes et al., 2010). Although Foxj1 has the potential to mediate this activity, the analysis of embryos that lack Foxj1 indicates that it is not required for this. Instead, FoxA2 is likely to perform this function (Ribes et al., 2010) (Fig. 5K,M) and to act, at least in part, by inhibiting the expression of key components of the Shh signalling pathway. Whether the functional overlap in the roles of Foxj1 and FoxA2 is a fail-safe mechanism that guarantees downregulation of Shh signalling, is a by-product of other essential functions of Foxj1, or an evolutionary vestige that was important in the ancestors of amniotes, remains to be determined. Indeed, together with the evidence indicating that Rfx3 functions redundantly with Foxj1 in the induction of long cilia in the FP, it adds a further complication to the challenge of dissecting the developmental network that underlies FP development. It raises the question of whether Foxj1 has any unique role in the FP. In this context, it is notable that in many Foxj1 mutant embryos the neural tube is misshapen (see Fig. S3 in the supplementary material). Thus, it is possible that Foxj1 has an essential role in regulating the morphology of the neural tube, perhaps by controlling the shape of FP cells. The molecular and cellular bases for this require further analysis.
Overall, the identification of structurally distinct cilia in the amniote FP provides an easily recognisable cellular feature that separates this region from the adjoining neuroepithelium and emphasises the morphological distinctness of the FP. Moreover, the data provide new insight into the development of the FP. Finally, the ability of Foxj1 to attenuate Shh signalling suggests a novel mechanism by which changes in cilia configuration can be deployed by cells to refine and modulate their sensitivity to a hedgehog signal during tissue development.
Supplementary Material
Acknowledgments
We thank Cheryll Tickle, Rick Livesey and Suganthi Suren for advice and discussions; Nicola Powles-Glover for assistance with video microscopy; T. Caspary, J. Chelly, M. Scott and J. Eggenschwiler for providing antibodies; members of the laboratory, particularly Natascha Bushati and David Wilkinson, for helpful comments and discussions; and Chris Atkins, Bob Butler and Graham Preece for assistance with FACS and GeneChip hybridisations. C.C. was supported by Fundação para a Ciência e Tecnologia, Portugal and V.R. by an EMBO Long-Term Fellowship. Work in the laboratories of J.B. and D.N. is supported by the MRC (UK). Work in the E.M. laboratory is supported by the Spanish Ministry of Education grant BFU2004-00455/BMC. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.051714/-/DC1
References
- Al-Shahrour F., Minguez P., Tárraga J., Medina I., Alloza E., Montaner D., Dopazo J. (2007). FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35, W91-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang S. L., Rossant J. (1994). HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561-574 [DOI] [PubMed] [Google Scholar]
- Bai C. B., Stephen D., Joyner A. L. (2004). All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6, 103-115 [DOI] [PubMed] [Google Scholar]
- Blatt E. N., Yan X. H., Wuerffel M. K., Hamilos D. L., Brody S. L. (1999). Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am. J. Respir. Cell Mol. Biol. 21, 168-176 [DOI] [PubMed] [Google Scholar]
- Bonnafe E., Touka M., AitLounis A., Baas D., Barras E., Ucla C., Moreau A., Flamant F., Dubruille R., Couble P., et al. (2004). The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell. Biol. 24, 4417-4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. (2001). Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11, 43-49 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M., Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435-445 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Chen Y., Jessell T. M., Struhl G. (2001). A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol. Cell 7, 1279-1291 [DOI] [PubMed] [Google Scholar]
- Brody S. L., Yan X. H., Wuerffel M. K., Song S. K., Shapiro S. D. (2000). Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 23, 45-51 [DOI] [PubMed] [Google Scholar]
- Caspary T., Larkins C. E., Anderson K. V. (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell 12, 767-778 [DOI] [PubMed] [Google Scholar]
- Chen J., Knowles H. J., Hebert J. L., Hackett B. P. (1998). Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 102, 1077-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407-413 [DOI] [PubMed] [Google Scholar]
- Clevidence D. E., Overdier D. G., Tao W., Qian X., Pani L., Lai E., Costa R. H. (1993). Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. USA 90, 3948-3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Meininger V. (1987). Ultrastructural analysis of primary cilium in the embryonic nervous tissue of mouse. Int. J. Dev. Neurosci. 5, 43-51 [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005). Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018-1021 [DOI] [PubMed] [Google Scholar]
- Davey M. G., Paton I. R., Yin Y., Schmidt M., Bangs F. K., Morrice D. R., Smith T. G., Buxton P., Stamataki D., Tanaka M., et al. (2006). The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 20, 1365-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E., Yang L. L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B. G., Briscoe J. (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717-720 [DOI] [PubMed] [Google Scholar]
- Dessaud E., McMahon A. P., Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489-2503 [DOI] [PubMed] [Google Scholar]
- Dziegielewska K. M., Ek J., Habgood M. D., Saunders N. R. (2001). Development of the choroid plexus. Microsc. Res. Tech. 52, 5-20 [DOI] [PubMed] [Google Scholar]
- Echelard Y., Epstein D. J., St-Jacques B., Shen L., Mohler J., McMahon J. A., McMahon A. P. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417-1430 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J. T., Anderson K. V. (2007). Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J., Briscoe J., Rashbass P., van Heyningen V., Jessell T. M. (1997). Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harbor Symp. Quant. Biol. 62, 451-466 [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. C., Anderson K. V. (2010). The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett B. P., Brody S. L., Liang M., Zeitz I. D., Bruns L. A., Gitlin J. D. (1995). Primary structure of hepatocyte nuclear factor/forkhead homologue 4 and characterization of gene expression in the developing respiratory and reproductive epithelium. Proc. Natl. Acad. Sci. USA 92, 4249-4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst E. M., Howard J. E. (1992). SEM studies of surfaces hidden within bulk tissue: a simple technique to control the position and orientation of dry fracture planes. J. Microsc. 167, 239-244 [DOI] [PubMed] [Google Scholar]
- Hooper J. E., Scott M. P. (2005). Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6, 306-317 [DOI] [PubMed] [Google Scholar]
- Huangfu D., Anderson K. V. (2006). Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133, 3-14 [DOI] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83-87 [DOI] [PubMed] [Google Scholar]
- Hynes M., Ye W., Wang K., Stone D., Murone M., Sauvage F., Rosenthal A. (2000). The seven-transmembrane receptor smoothened cell-autonomously induces multiple ventral cell types. Nat. Neurosci. 3, 41-46 [DOI] [PubMed] [Google Scholar]
- Jacob J., Briscoe J. (2003). Gli proteins and the control of spinal-cord patterning. EMBO Rep. 4, 761-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Ferri A. L., Milton C., Prin F., Pla P., Lin W., Gavalas A., Ang S. L., Briscoe J. (2007). Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat. Neurosci. 10, 1433-1439 [DOI] [PubMed] [Google Scholar]
- Jeong J., McMahon A. P. (2005). Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development 132, 143-154 [DOI] [PubMed] [Google Scholar]
- Kingsbury B. F. (1930). The developmental significance of the floor plate of the brain and spinal cord. J. Comp. Neurol. 32, 113-135 [Google Scholar]
- Ko H. W., Norman R. X., Tran J., Fuller K. P., Fukuda M., Eggenschwiler J. T. (2010). Broad-minded links cell cycle-related kinase to cilia assembly and hedgehog signal transduction. Dev. Cell 16, 237-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L., Zhou H., Costa R. H. (1997). The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc. Natl. Acad. Sci. USA 94, 3094-3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. (2003). Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell 12, 1261-1274 [DOI] [PubMed] [Google Scholar]
- Marti E., Bumcrot D. A., Takada R., McMahon A. P. (1995). Requirement of 19K form of Sonic hedgehog for induction of distinct ventral cell types in CNS explants. Nature 375, 322-325 [DOI] [PubMed] [Google Scholar]
- McGrath J., Somlo S., Makova S., Tian X., Brueckner M. (2003). Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61-73 [DOI] [PubMed] [Google Scholar]
- Milenkovic L., Scott M. P., Rohatgi R. (2009). Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol. 187, 365-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V., Joshi H. C., Besharse J. C. (1993). Gamma-tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J. Cell Sci. 104, 1229-1237 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193-199 [DOI] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829-837 [DOI] [PubMed] [Google Scholar]
- Norton W. H., Mangoli M., Lele Z., Pogoda H. M., Diamond B., Mercurio S., Russell C., Teraoka H., Stickney H. L., Rauch G. J., et al. (2005). Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development 132, 645-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennekamp P., Karcher C., Fischer A., Schweickert A., Skryabin B., Horst J., Blum M., Dworniczak B. (2002). The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 12, 938-943 [DOI] [PubMed] [Google Scholar]
- Persson M., Stamataki D., te Welscher P., Andersson E., Bose J., Ruther U., Ericson J., Briscoe J. (2002). Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 16, 2865-2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek M., Briscoe J. (2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci. 6, 230-240 [DOI] [PubMed] [Google Scholar]
- Poirier K., Van Esch H., Friocourt G., Saillour Y., Bahi N., Backer S., Souil E., Castelnau-Ptakhine L., Beldjord C., Francis F., et al. (2004). Neuroanatomical distribution of ARX in brain and its localisation in GABAergic neurons. Brain Res. 122, 35-46 [DOI] [PubMed] [Google Scholar]
- Ribes V., Balaskas N., Sasai N., Cruz C., Dessaud E., Cayuso J., Tozer S., Yang L. L., Novitch B., Marti E., et al. (2010). Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 24, 1186-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelink H., Porter J. A., Chiang C., Tanabe Y., Chang D. T., Beachy P. A., Jessell T. M. (1995). Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445-455 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Scott M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372-376 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Cox C., Jessell T. M., Klar A. (1993). Ectopic neural expression of a floor plate marker in frog embryos injected with the midline transcription factor Pintallavis. Proc. Natl. Acad. Sci. USA 90, 8268-8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Nguyen V., Palma V. (2003). The emergent design of the neural tube: prepattern, SHH morphogen and GLI code. Curr. Opin. Genet. Dev. 13, 513-521 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M., Kondoh H. (1997). A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313-1322 [DOI] [PubMed] [Google Scholar]
- Satir P., Christensen S. T. (2007). Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69, 377-400 [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N., Gerfin-Moser A. (1993). A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431-440 [DOI] [PubMed] [Google Scholar]
- Stamataki D., Ulloa F., Tsoni S. V., Mynett A., Briscoe J. (2005). A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 19, 626-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strähle U., Lam C. S., Ertzer R., Rastegar S. (2004). Vertebrate floor-plate specification: variations on common themes. Trends Genet. 20, 155-162 [DOI] [PubMed] [Google Scholar]
- Stubbs J. L., Oishi I., Izpisua Belmonte J. C., Kintner C. (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40, 1454-1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik K., Dehart D. B., Iangaki T., Carson J. L., Vrablic T., Gesteland K., Schoenwolf G. C. (1994). Morphogenesis of the murine node and notochordal plate. Dev. Dyn. 201, 260-278 [DOI] [PubMed] [Google Scholar]
- Supp D. M., Witte D. P., Potter S. S., Brueckner M. (1997). Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature 389, 963-966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supp D. M., Brueckner M., Kuehn M. R., Witte D. P., Lowe L. A., McGrath J., Corrales J., Potter S. S. (1999). Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development 126, 5495-5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. (2000). Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005-1009 [DOI] [PubMed] [Google Scholar]
- Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. (1999). Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A–/– mice analysis. J. Cell Biol. 145, 825-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar J. W., Lim L., Costa R. H., Whitsett J. A. (1999). HNF-3/forkhead homologue-4 influences lung morphogenesis and respiratory epithelial cell differentiation in vivo. Dev. Biol. 213, 405-417 [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454-2472 [DOI] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J., Davidson E. H., Wong W. H., McMahon A. P. (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977-1989 [DOI] [PubMed] [Google Scholar]
- Weinstein D. C., Ruiz i Altaba A., Chen W. S., Hoodless P., Prezioso V. R., Jessell T. M., Darnell J. E., Jr (1994). The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78, 575-588 [DOI] [PubMed] [Google Scholar]
- Xie J., Murone M., Luoh S. M., Ryan A., Gu Q., Zhang C., Bonifas J. M., Lam C. W., Hynes M., Goddard A., et al. (1998). Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90-92 [DOI] [PubMed] [Google Scholar]
- Yin Y., Bangs F., Paton I. R., Prescott A., James J., Davey M. G., Whitley P., Genikhovich G., Technau U., Burt D. W., et al. (2009). The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development 136, 655-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Ng C. P., Habacher H., Roy S. (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445-1453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.