Abstract
Bioisosteric deaza analogues of 6-methyl-9-β-D-ribofuranosylpurine, a hydrophobic analogue of adenosine, were synthesized and evaluated for antiviral activity. Whereas the 1-deaza and 3-deaza analogues were essentially inactive in plaque assays of infectivity, a novel 7-deaza-6-methyl-9-β-D-ribofuranosylpurine analogue, structurally related to the natural product tubercidin, potently inhibited replication of poliovirus (PV) in HeLa cells (IC50 = 11 nM) and dengue virus (DENV) in Vero cells (IC50 = 62 nM). Selectivity against PV over cytotoxic effects to HeLa cells was >100-fold after incubation for 7 h. Mechanistic studies of the 5'-triphosphate of 7-deaza-6-methyl-9-β-D-ribofuranosylpurine revealed that this compound is an efficient substrate of PV RNA-dependent RNA polymerase (RdRP) and is incorporated into RNA mimicking both ATP and GTP.
Introduction
Synthetic mimics of adenosine (1) and adenosine triphosphate (ATP) constitute an important class of therapeutics. A wide variety of kinase inhibitors such as the anticancer drug imatinib mimic recognition properties of ATP in binding to these enzymes.1 Similarly, phosphorylated derivatives of antiviral agents such as Adefovir, a sugar-modified analogue of adenosine, inhibit actions of viral polymerases. Many nucleoside-based mimics of adenosine are substrates of adenosine kinase and other kinases, and these compounds are often converted intracellularly into phosphorylated metabolites that exhibit potent biological activities.2, 3
Although sugar-modified mimics of adenosine have been extensively studied,2, 4, 5 less is known about how subtle structural modifications to the nucleobase of adenosine collectively impact both cellular proliferation and antiviral activity. However, minor changes to the adenosine nucleobase can confer substantial antiproliferative and antiviral effects. For example, bioisosteric replacement of the exocyclic amino group of 1 with a methyl group yields 6-methyl-9-β-D-ribofuranosylpurine (2), a potent but non-selective and highly cytotoxic inhibitor of replication of a number of viruses, including herpes simplex virus-1 (HSV-1) and vaccinia virus. However, the mechanisms of antiviral and cytotoxic action of 2 are poorly understood.6, 7
Numerous other mimics of adenosine bearing deazapurine modifications are known.8 The natural product tubercidin (3), representing a 7-deazapurine analogue, is a potent antineoplastic9 and antiviral agent.10 Tubercidin is phosphorylated intracellularly to the corresponding nucleoside triphosphate,11 is incorporated into DNA and RNA, and exhibits some polymerase inhibition activity.12, 13 However, this compound is non-selective and too toxic for clinical use as an antiviral agent.10 Interest in the broad-spectrum antiviral activity and cytotoxicity of tubercidin has led to a number of studies of nucleobase and sugar-modified tubercidin analogues.14–17 Olsen and coworkers identified the tubercidin analogue 7-deaza-2'-C-methyladenosine as an inhibitor of replication of hepatitis C virus (HCV, IC50 = 0.108 µM) without appreciable cytotoxicity.13 The structurally similar 7-deaza-2'-C-ethynyladenosine (NITD008) was recently identified as a potent inhibitor of dengue virus type 2 (DENV2, IC50 = 0.64 µM).18 A series of 7-deazapurine ribonucleosides with alkyl, aryl, or heteroaryl groups in the 6-position and H, F, or Cl substitution at the 7-position have also been evaluated in a HCV replicon system.19 However, the decrease in the replicon reporter signal caused by some of these compounds was attributed to cytostatic effects. Other deaza adenosine analogues such as 1-deaza and 3-deaza variants exhibit some antiproliferative activity against certain mammalian cell lines,8 and 3-deaza adenosine is known to inhibit HIV replication in peripheral blood mononuclear cells.20, 21
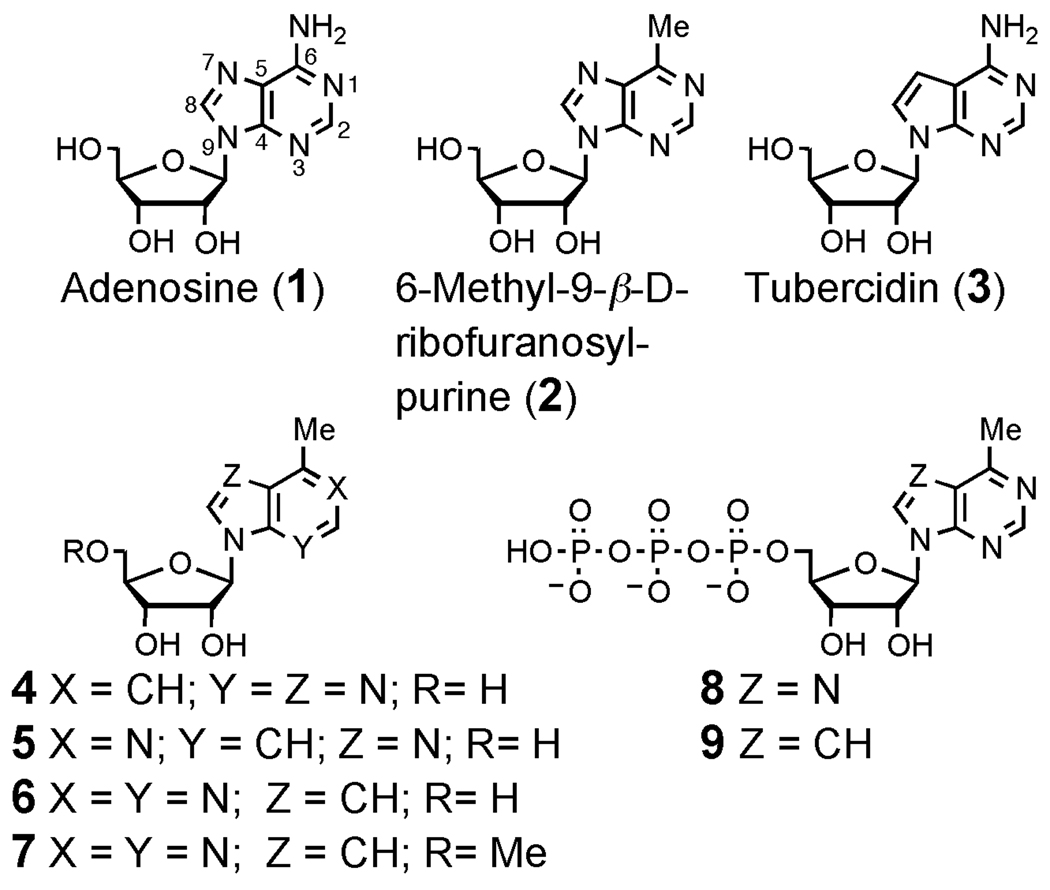
We report here the synthesis and biological evaluation of three deazapurine analogues of 2 (compounds 4–6) and related chemical probes (7–9). These hydrophobic mimics of adenosine were designed to examine the relationship between antiproliferative activity and antiviral activity against the RNA viruses poliovirus (PV) and dengue virus (DENV) in vitro. To further investigate the antiviral activity of the novel nucleoside 6, a compound structurally related to tubercidin (3), we additionally synthesized the 5'-O-methyl analogue 7 as well as the structurally related triphosphate derivatives 8 and 9.
Chemical synthesis
We employed a fusion glycosylation method to synthesize compounds 4 (Scheme 1) and 5. The known22, 23 nucleobase 7-methyl-3H-imidazo[4,5-b]pyridine (15) was prepared in 5 steps from 2-amino-4-picoline (10) analogous to a previously reported route (Scheme 1).24 Another approach for the preparation of 4 has been published,25 but this method generated a mixture of isomers at the 7- and 9-positions. To avoid this limitation, we used the fusion coupling method to exclusively generate the 9- substituted isomer of 16.26 Deprotection of 16 under basic conditions afforded 4 (Scheme 1). The readily prepared nucleobase 4-chloro-1H-imidazo[4,5-c]pyridine27 was used as an intermediate to yield 3-deaza-6-methyl-9-β-D-ribofuranosylpurine (5), which was synthesized by known methods.28
Scheme 1a.
a(a) 1,3-dibromo-5,5-dimethylhydantoin, CH2Cl2; (b) i. HNO3, H2SO4; ii. H2SO4; (c) SnCl2, HCl; (d) HCOOH; (e) PtO2, Pd/C, NaOH; (f) p-TsOH, β-D-ribofuranose 1,2,3,5-tetraacetate; (g) NH3, MeOH; (h) Ac2O, pyridine; (i) Pd(PPh3)4, AlMe3, THF; (j) NH3, MeOH; (k) acetone, p-TsOH, 2,2-dimethoxypropane; (l) NaH, MeCN, MeI; (m) TFA, H2O; (n) i. PCl3, CH2Cl2; ii. TFA, H2O; (o) TMSCl, pyridine, I2, DMF, (NBu4)3HP2O7.
The synthesis of the novel nucleoside 7-deaza-6-methyl-9-β-D-ribofuranosylpurine (6) employed the commercially available nucleoside 17 (Scheme 1).19 Protection as the peracetate, cross coupling with trimethyl aluminum, and deprotection yielded 6. The 5'-O-methyl derivative 7 was synthesized by protection of 6 as the acetonide, deprotonation with sodium hydride, and methylation at the 5'-position to produce intermediate 21. Treatment with aqueous trifluoroacetic acid afforded 7. The 5'-triphosphate derivative 8, prepared as previously described,29 and triphosphate 9 were synthesized via the H-phosphonate monoesters (e.g. 22) using our previously reported one-pot method.29
Biological evaluation of compounds
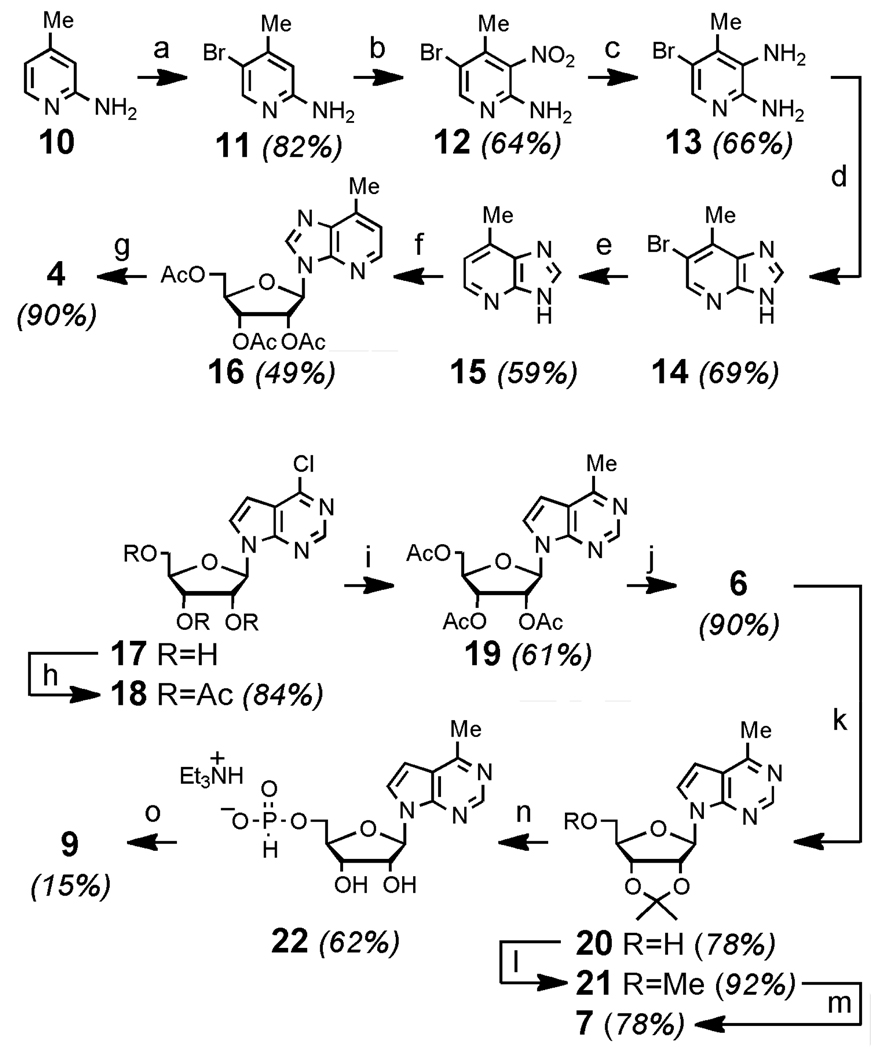
As shown in Figure 1 (panel A), compounds 2–7 were evaluated against infectious PV in a whole-cell assay. HeLa S3 cells were pretreated with 2–7 for 1 h before administration of a high multiplicity of infection (MOI) of PV. After treatment of cells with PV for 15 min, fresh media containing 2–7 was added, the infection was allowed to progress for another 6 h, and the cell-associated viral titer was determined as previously reported.30 Under these conditions, as listed in Table 1, compounds 2 and 3 inhibited replication of PV (IC50 (2)=1.01 µM; IC50 (3)=0.030 µM). The known compounds 4 and 5 were inactive against PV at concentrations below 10 µM. However, the novel nucleoside 6 proved to be the most potent inhibitor of PV replication in this series of compounds (IC50 = 0.011 µM). Methylation of the 5'-hydroxyl group of 6 to afford 7 eliminated the antiviral effect, suggesting that like other structurally related compounds, intracellular phosphorylation of 6 is required to generate a biologically active metabolite.
Figure 1.
Panel A: Antiviral effects of compounds against poliovirus. HeLa S3 cells were incubated with 2–7 for 1 h at the concentrations shown, and subsequently infected with at least 106 PFU of PV. After infection for 15 min, fresh media containing 2–7 was added and the infection continued for 6 h. Cell-associated virus was titered by plaque assay. Panels B–D: Cytotoxicity to HeLa S3 cells after treatment with 2–7 for 7 h (B), 24 h (C) or 48 h (D).
Table 1.
| Compound | HeLa IC50 (7 h, µM) |
HeLa IC50 (24 h, µM) |
HeLa IC50 (48 h, µM) |
PV IC50 (µM) |
DENV2 IC50 (µM) |
|---|---|---|---|---|---|
| 2 | 17.66 | 6.68 | 2.20 | 1.01b | 5.46 (replicon) |
| 3 | 1.44 | 0.819 | 0.400 | 0.030c | ND |
| 4 | >100 | >100 | >10 | >10 | >10 (replicon) |
| 5 | >100 | >100 | >100 | >10 | >10 (replicon) |
| 6 | >100 | 0.986a | 0.224 | 0.011d | 0.877 (replicon) 0.062 (plaque) 0.039 (PCR) |
| 7 | >100 | >100 | >100 | >10 | >10 (replicon) |
Viability of cells was 33% after treatment with 6 at 100 µM.
SEM (standard error of the mean) = 16%
SEM = 36%
SEM = 6%
ND: not determined
Compounds 2 and 3 are known to be highly cytotoxic or cytostatic to mammalian cells.7, 10 To compare these compounds with analogues 4–7, HeLa S3 cells, the cell line used in the PV assays, were treated with 2–7 for 7 h, 24 h, and 48 h. Cellular viability was evaluated with a luciferase assay that quantifies the abundance of ATP present in living cells (Figure 1, panels B–D). These experiments confirmed previous reports7, 10 that although both 2 and 3 are highly cytotoxic, 2 is ~5-fold less toxic than 3 after treatment of cells for 48 h. In contrast, compounds 4, 5 and 7 showed essentially no cytotoxicity below 10 µM under these conditions. Interestingly, compound 6 showed a unique bioactivity profile. Unlike compounds 2 and 3, essentially no cytotoxic effects of 6 to HeLa cells were observed after treatment for 7 h, the conditions used for the assay against PV. Only after treatment for longer periods of 24 h or 48 h was the toxicity of 6 evident, and 6 was comparable to 3 only after 48 h in culture.
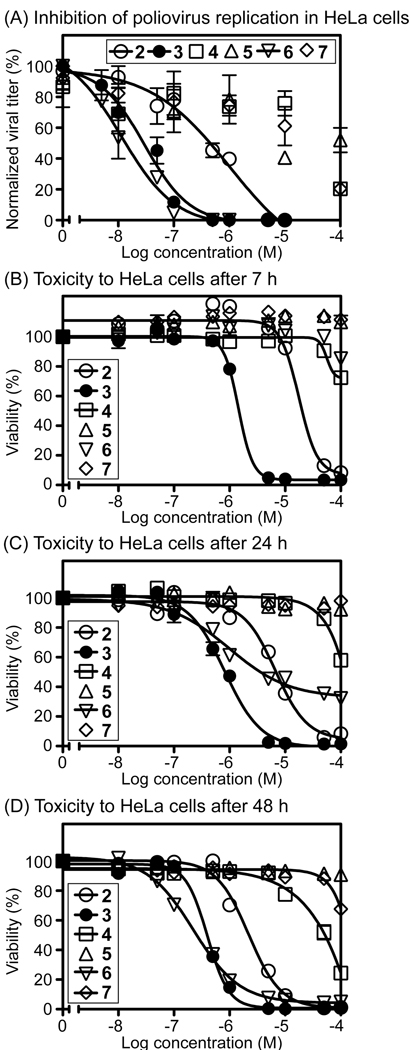
The antiviral activities of compounds 2 and 4–7 were further evaluated in a dengue replicon assay (Figure 2). In this assay, BHK-21 cells stably expressing a luciferase-driven DENV2 replicon were treated with these compounds for 24 h. The cells were subsequently lysed, and the activity of luciferase was quantified. As listed in Table 1, compound 6 was the most potent inhibitor (IC50 = 0.88 µM), and compound 2 showed some inhibition of replicon activity (IC50 = 5.46 µM), but compounds 4, 5, and 7 showed essentially no inhibition below 10 µM.
Figure 2.
Panel A: Dengue replicon assay. BHK-21 cells expressing DENV2 replicon were treated with nucleosides for 24 h. After cell lysis, the luciferase signal was measured. Panel B: Infectivity assay of compound 6 with infectious dengue virus.
To further probe the antiviral activity of the novel nucleoside 6, this compound was evaluated in an infectivity assay with live dengue virus (Figure 2B). Vero cells infected with DENV2 were treated with 6 for 24 h, and the viral titer was determined by counting viral plaques present after cell lysis31 and through quantification of DENV2 RNA by real time PCR.32 In these assays, compound 6 was a highly potent inhibitor of DENV2 replication (plaque assay IC50 (6) = 0.062 µM; real time PCR IC50 (6) = 0.039 µM). To examine whether the antiviral mechanism of action of 6 or 2 might involve lethal viral mutagenesis, these compounds were compared with ribavirin in a guanidine resistance assay.30 Ribavirin, a clinically employed antiviral drug, becomes phosphorylated intracellularly, mimics purine ribonucleotides, and becomes misincorporated into the genome of RNA viruses, resulting in loss of function due to error catastrophe.30, 33 However, compared to ribavirin, neither 6 nor 2 increased the frequency of viral mutagenesis (Supporting information, Table S1), indicating that the antiviral activity of these compounds against PV is mechanistically distinct from ribavirin.
Intracellular adenosine kinase is known to phosphorylate a wide variety of base-modified mimics of adenosine, including tubercidin (3).34, 35 Following this rate-limiting phosphorylation step, monophosphorylated mimics of adenosine are generally converted to triphosphates by other nucleotide kinases. Because 2, 3, 6 and adenosine are highly structurally similar, these compounds are very likely to be converted to triphosphate metabolites that function as the biologically active species in cells. Consistent with this idea, methylation of the 5'-alcohol of 6 to yield 7 was found to reduce biological activity by over 1000-fold (Table 1).
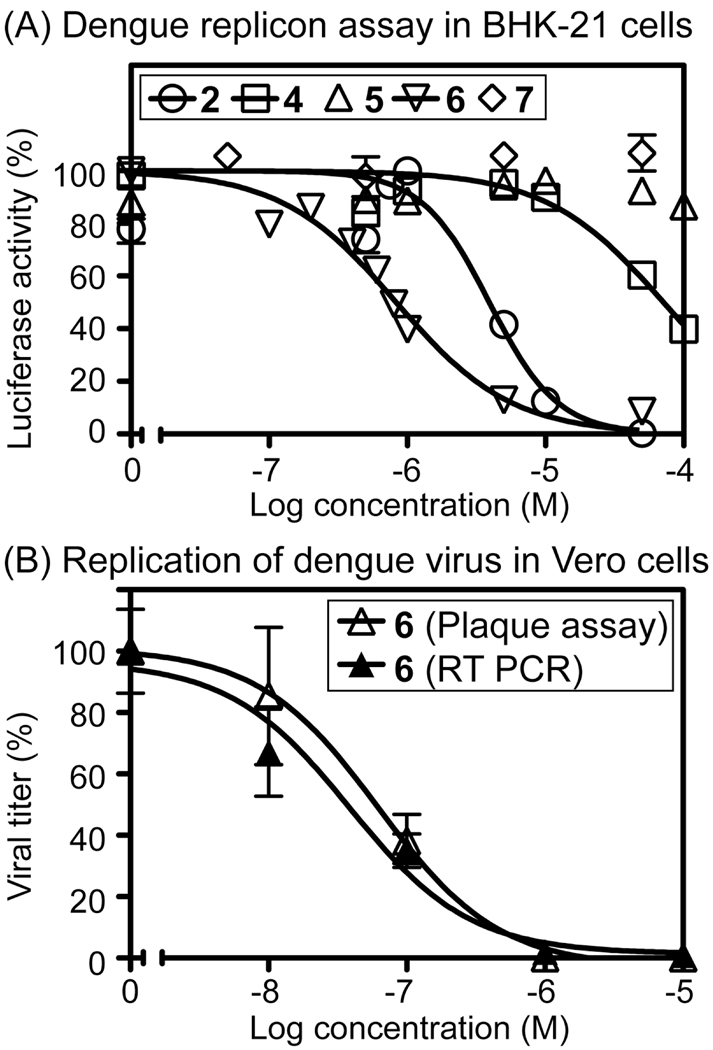
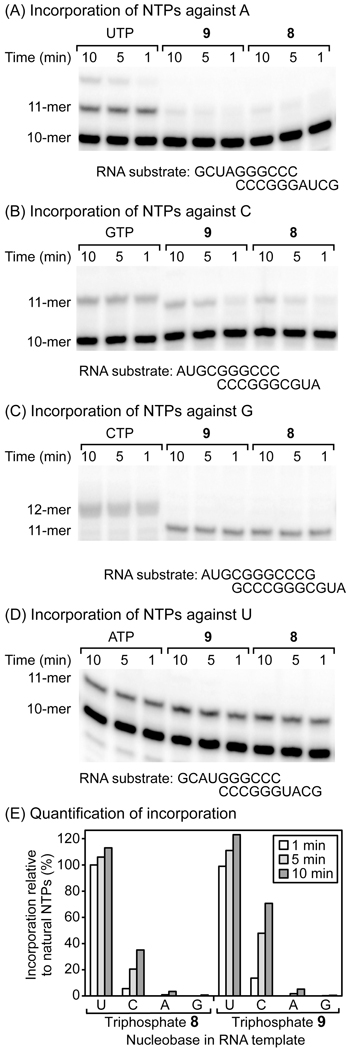
To further examine the biological properties of triphosphate derivatives of 2 and 6, we synthesized 5'-triphosphates 8 and 9. Using a previously reported primer-extension assay,36 we studied the in vitro incorporation of these compounds into RNA by poliovirus RNA-dependent RNA polymerase (PV RdRP, 3Dpol). In this assay, both 8 and 9 became incorporated opposite uracil and cytosine, but exhibit almost no incorporation opposite adenine and guanine (Figure 3). As expected for compounds structurally related to ATP, the incorporation efficiency of both 8 and 9 across from uracil was comparable to natural ATP on this time scale. Compared with GTP, incorporation opposite cytosine after reaction for 10 min was ~40% for 8 and ~60% for 9, illustrating the importance of the exocyclic amino group of ATP for discrimination of the complementary nucleobase in RNA by PV RdRP.
Figure 3.
Panels A–E: Incorporation of 8 and 9 into double-stranded RNA by PV RdRP. Complexes of RNA template and PV RdRP were incubated for 3 min, 8, 9 or the natural NTP (0.5 mM) was added respectively, and the reaction was allowed to proceed at 30 °C for 1, 5 or 10 min prior to separation of oligonucleotides by PAGE. In panel E, the percent incorporation relative to natural NTPs measured from the gel data in A–D is compared.
To assess the efficiency of 8 and 9 as substrates for RNA extension by PV RdRP when templated by U, pre-steady state kinetics of incorporation were measured. Values for kpol, the maximal rate constant for single nucleotide addition, Kd,app, the kinetically-measured binding constant for 8 or 9 to the PV RdRP-RNA primer/template complex, and kpol/Kd,app, a measure of substrate efficiency, are summarized in Table 2 (data shown in Figures S8 and S9). Also shown are previously published kinetic values for complementary natural ATP and the purine mimic ribavirin triphosphate (RTP). Efficiencies of 8 and 9 are only ten-fold lower than natural ATP, indicating that these analogues are robust substrates for PV RdRP when templated by U. For comparison, the efficiency of RTP is approximately three orders magnitude lower than 8 or 9. In contrast to the robust incorporation of 8 and 9 by PV RdRP opposite U, incorporation opposite C is extremely slow (Figure S10).
Table 2.
Kinetic parameters for compounds 8 and 9 as substrates by PV RdRP
Additionally, although other antiviral nucleosides are known to function by causing termination of the growing RNA chain,37 both 8 and 9 were capable of extending RNA oligomers to the +4 position (Supporting information, Figure S11), indicating that neither of these compounds functions as a RNA chain terminator.
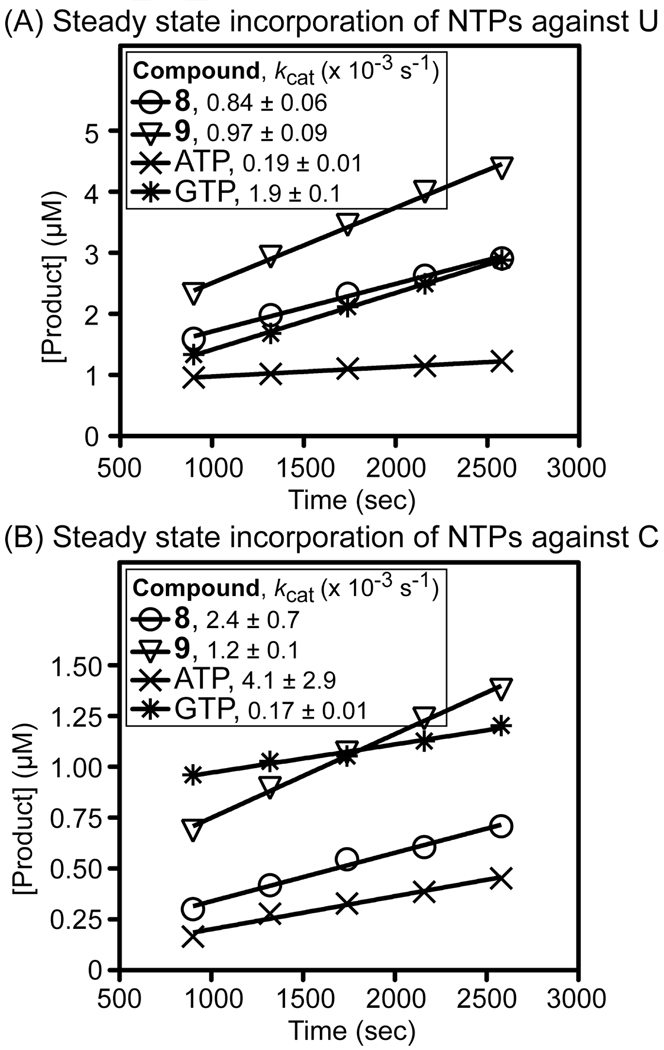
To investigate the effects of triphosphates 8 and 9 on PV RdRP-RNA primer/template complexes and inhibition of RdRP, these compounds were assessed in steady state incorporation assays (Figure 4). The concentration of products of incorporation of 8, 9, ATP, and GTP templated by uracil or cytosine was plotted versus time, and the calculated kcat (quotient of slope/y-intercept from linear fits to data points), which is related to the dissociation rate constant of RdRP from RNA primer/template, was compared (Figure 4). These experiments revealed that both 8 and 9 destabilize PV RdRP-RNA primer/template complexes, as evidenced by increases in kcat compared with natural correct nucleotides (ATP opposite U; GTP opposite C). It should be noted that the concentrations of 8 and 9 with template U approached saturation, yielding true estimates of kcat. Due to the extremely slow incorporation opposite C, whether or not saturation of 8 and 9 with template C was approached was not determined. Nevertheless, the slopes of the time courses of steady state incorporation of both 8 and 9 were constant over time, indicating that neither 8 nor 9 inhibit PV RdRP.
Figure 4.
Steady state incorporation of 8, 9, ATP or GTP against uracil (panel A) and cytosine (panel B). 3Dpol (1 µM) was added to the NTP (1 mM) and sym/sub duplex RNA (10 µM) to initiate the reaction. EDTA was added at 50 mM (final conc.) to stop the reaction at the time points shown.
Discussion
Examined collectively, the kinetics of incorporation of triphosphates 8 and 9 into RNA, the known structure-activity relationships governing phosphorylation of ATP, tubercidin, and related compounds, and the lack of biological activity of control compound 7 suggest that triphosphate 9 represents the active antiviral metabolite of compound 6. This compound (9) functions as a robust substrate of PV RdRP, and it likely engenders antiviral effects not by inhibiting the viral polymerase or by chain termination of viral RNA but through a downstream mechanism that may involve RNA-RNA or protein-RNA interactions, or possible effects on host enzymes essential for viral replication. Once incorporated into RNA, the product of 9 may alternatively function as a defective templating base in duplex synthesis because of the missing C6 amino group involved in Watson-Crick base pairing and the missing nitrogen atom at the 7-position. This RNA modification may lead to slow or defective synthesis of complementary RNA, or stalling or dissociation of PV RdRP when an analogue-templated site is encountered, or possibly result in other effects that block successful replication of the viral genome.
Interestingly, the cytotoxicity profiles of compounds 2–7 differ substantially. The cytotoxicity of tubercidin (3) is thought to derive from suppression of a number of cellular processes including mitochondrial respiration, de novo purine synthesis, processing of rRNA, tRNA methylation and protein and RNA synthesis.10 Whereas 2 and 3 show pronounced effects on proliferation of HeLa cells after treatment for only 7 h, compounds 4–7 show little cytotoxicity during that time period. Moreover, significant cytotoxic effects of 6 are evident only after 24 to 48 h. These results suggest that subtle modifications to the adenine heterocycle show promise for separation of antiviral from cytotoxic effects on host cells. In addition, robust use of 8 and 9 as substrates by PV RdRP suggests the possibility that cytotoxicity may result from off-target effects on human RNA polymerases, including human mitochondrial RNA polymerase.
Conclusion
A novel 7-deaza derivative of 6-methyl-9-β-D-ribofuranosylpurine (6) was synthesized. This compound (6) exhibits highly potent activity against PV and DENV2 in cell culture assays and possesses an improved cytotoxicity profile compared to 6-methyl-9-β-D-ribofuranosylpurine (2) and tubercidin (3). Studies of the putative triphosphate metabolites of 2 and 6 revealed that these compounds (8, 9) do not inhibit the viral polymerase, do not serve as RNA chain terminators, and are not lethal mutagens, but rather are robust substrates for PV RdRP. These results suggest antiviral mechanisms of action that involve downstream effects on RNA-RNA interactions, RNA-protein interactions, or perturbation of endogenous targets in host cells.
Experimental section
General
Chemical reagents and solvents were obtained from Acros, Aldrich, TCI America or Fisher Scientific without further purification unless otherwise noted. Anhydrous solvents were obtained after passage through a drying column of a solvent purification system from GlassContour (Laguna Beach, CA). Reactions were performed under an atmosphere of dry nitrogen. Reactions were monitored by analytical thin-layer chromatography on plates coated with 0.25 mm silica gel 60 F254 (EMD Chemicals). TLC plates were visualized by UV irradiation (254 nm) or stained with 20% sulfuric acid in ethanol. Nuclear magnetic resonance (NMR) spectroscopy employed a Bruker DRX 400 spectrometer. NMR peaks are reported as parts per million (ppm) referenced to internal CHCl3, MeOH or H2O peaks. Mass spectral data was obtained from The University of Kansas Mass Spectrometry Facility. Melting points were determined on a Thomas Hoover apparatus and are uncorrected. Column chromatography employed silica gel (ICN SiliTech, 32–63 µm) or Sephadex LH-20 (GE Healthcare, column dimensions: 2.5 cm × 40 cm). Purification by preparative HPLC employed an Agilent 1100 series instrument equipped with an Atlantis C18 preparative column (19 × 150 mm, 5 µm; Waters Corporation). The HPLC flow rate was maintained at 25 mL/min for the duration of the run. Determination of purity of compounds 2 and 4–7 by analytical HPLC used a PRP-1 (polystyrene-divinylbenzene) reverse-phase column (4.1 × 250 mm, 7 µm; Hamilton) and a gradient of 1% to 99% CH3CN (containing 0.1% TFA) in nanopure water (containing 0.1% TFA, 0 to 30 min) followed by 99% CH3CN (containing 0.1% TFA) from 30–40 min at a flow rate of 0.8 mL/min. Analysis of purity of compounds 8–9 by HPLC used an Aquasil column (4.6 × 250 mm, 5 µm; Thermoelectron Corporation) at a flow rate of 1.0 mL/min with a gradient of 1% to 50% CH3CN in aqueous KH2PO4 (100 mM, pH = 6.0) over 30 min followed by 50% CH3CN in aqueous KH2PO4 from 30–40 min. 6-Methyl-9-β-D-ribofuranosylpurine (2)7 and 6-methyl-9-β-D-ribofuranosylpurine 5'-triphosphate (8)29 were prepared as previously reported. All compounds studied in biological assays were of •95% purity as evidenced by analytical HPLC.
5-Bromo-2-amino-4-methylpyridine (11)
This known compound was prepared by modification of a previously reported38 route. 2-Amino-4-methylpyridine (Aldrich, 10, 2.49 g, 23.0 mmol) in CH2Cl2 (30 mL) was cooled to −45 °C with dry ice in a bath of CH3CN. 1,3-Dibromo-5,5-dimethylhydantoin (6.58 g, 23.0 mmol) was added and the mixture was stirred at −45 °C for 30 min. Saturated aqueous NaHCO3 (40 mL) was added, the mixture was allowed to warm to room temperature (22 °C), and CH2Cl2 (100 mL) was added. The organic layer was separated, dried over anhydrous sodium sulfate and evaporated. The resulting residue was recrystallized from EtOAc to yield 11 (3.53 g, 82%) as a pale brown solid. 1H NMR (400 MHz, CDCl3) δ 8.10 (s, 1H), 6.42 (s, 1H), 4.37 (s, 2H), 2.29 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 149.2 (×3), 110.3 (×2), 22.3.
5-Bromo-4-methyl-3-nitropyridin-2-amine (12)
This known compound was prepared by modification of a previously reported39 route. 5-Bromo-2-amino-4-methylpyridine (11, 2.43 g, 13.0 mmol) was dissolved in concentrated H2SO4 (20 mL), and the solution was cooled to −10 °C with a bath of ice and NaCl. HNO3 (14 mL) was added dropwise, the reaction mixture was stirred for 30 min and allowed to warm to room temperature (22 °C). The mixture was poured into an ice / water mixture (50 mL), and the pH was adjusted to 3 by addition of concentrated aqueous ammonium hydroxide (22%). A light yellow precipitate was collected by filtration, washed with water and dried in vacuo. The resulting solid was dissolved in H2SO4 (20 mL), and the mixture was stirred at room temperature (22 °C) for 2 h. The reaction solution was poured onto ice and neutralized with aqueous ammonium hydroxide (22%). After extraction with CH2Cl2 (3 × 70 mL), the combined organic layers were dried over anhydrous sodium sulfate and evaporated. The crude product was recrystallized from CH2Cl2 to afford 12 (1.93 g, 64%) as a yellowish brown solid. 1H NMR (400 MHz, CDCl3) d 8.30 (s, 1H), 5.88 (s, 2H), 2.55 (s, 3H); 13C NMR (100 MHz, CDCl3) d 153.4 (×2), 151.5, 144.1, 111.8, 20.4.
5-Bromo-4-methylpyridine-2,3-diamine (13)
This known compound was prepared by modification of a previously reported40 route. SnCl2 (6.23 g, 27.6 mmol) was dissolved in concentrated aqueous hydrochloric acid, and the mixture was cooled to 0 °C. 5-Bromo-4-methyl-3-nitropyridin-2-amine (12, 1.60 g, 6.90 mmol) was added to this solution in four portions, the reaction mixture was warmed to room temperature (22 °C) and placed in an oil bath maintained at 100 °C for 1 h. After cooling, the solution was adjusted to pH = 11 with aqueous NaOH solution (40%). The precipitate was collected by filtration, washed with water, and the crude product was further purified by flash chromatography (eluent: hexane, EtOAc, 1:1) to yield 13 (0.989 g, 66%) as a light brown solid. 1H NMR (400 MHz, CD3OD) d 7.49 (s, 1H), 2.34 (s, 3H); 13C NMR (100 MHz, CD3OD) d 147.3, 135.4, 127.6, 120.1, 111.8, 15.2.
6-Bromo-7-methyl-3H-imidazo[4,5-b]pyridine (14)
This known compound was prepared by modification of a previously reported40 route. 5-Bromo-4-methylpyridine-2,3-diamine (13, 0.695 g, 3.20 mmol) was dissolved in formic acid (5 mL) and heated to reflux for 1 h. The formic acid was removed in vacuo, and the residue was recrystallized from methanol to give 14 (250 mg) as an orange solid. Chromatographic purification of the mother liquor yielded another 220 mg of the product (yield: 69%). 1H NMR (400 MHz, CD3OD) d 8.43 (s, 1H), 8.35 (s, 1H), 2.68 (s, 3H); 13C NMR (100 MHz, CD3OD) d 145.0 (×6), 17.1.
7-Methyl-3H-imidazo[4,5-b]pyridine (15)
6-Bromo-7-methyl-3H-imidazo[4,5-b]pyridine (14, 403 mg, 1.90 mmol) in aqueous NaOH (1%, 20 mL) containing PtO2 (29.5 mg, 0.130 mmol) and palladium on carbon (10%, 298 mg, 0.280 mmol) was hydrogenated in a Parr apparatus for 30 min at 50 psi. The catalyst was removed by filtration through celite, and the filtrate was neutralized by dropwise addition of aqueous hydrochloric acid (2 M). After extraction with CH2Cl2 (3 × 50 mL), the organic layers were combined, dried over anhydrous sodium sulfate and evaporated in vacuo. The crude product was recrystallized from CH2Cl2 to afford 15 (148 mg, 59%) as a white solid. 1H NMR (400 MHz, CD3OD) d 8.34 (s, 1H), 8.22 (d, 1H, J = 5.0 Hz), 7.12 (d, 1H, J = 5.0 Hz), 2.61 (s, 3H); 13C NMR (100 MHz, CD3OD) d 143.7 (×2), 142.8, 119.2 (×3), 15.1.
7-Methyl-3-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)-3H-imidazo[4,5-b]pyridine (1-deaza-6-methyl-9-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)purine, 16)
A mixture of 7-methyl-3H-imidazo[4,5-b]pyridine (15, 70.2 mg, 0.520 mmol), β-D-ribofuranose 1,2,3,5-tetraacetate (Aldrich, 318 mg, 1.00 mmol), and p-toluenesulfonic acid (19.0 mg, 0.10 mmol) was heated to 170 °C under reduced pressure (12–15 mm Hg) with stirring for 20 min. The resulting solid was neutralized with saturated aqueous NaHCO3 and extracted with CH2Cl2 (4 × 40 mL). The combined organic layers were dried over anhydrous sodium sulfate, and removed in vacuo to afford a residue that was purified by flash column chromatography (eluent: hexane, EtOAc, 4:1) to yield 16 (101 mg, 49%) as a white foam. mp 53–55 °C; 1H NMR (400 MHz, CDCl3) d 8.27 (d, 1H, J = 4.9 Hz), 8.17 (s, 1H), 7.09 (d, 1H, J = 4.9 Hz), 6.30 (d, 1H, J1', 2' = 5.3 Hz), 6.04 (dd, 1H, J2', 1' = 5.3 Hz, J2', 3' = 5.4 Hz), 5.74 (dd, 1H, J3', 2' = 5.4 Hz, J3', 4' = 2.8 Hz), 4.49-4.37 (m, 3H), 2.68 (s, 3H), 2.15 (s, 3H), 2.13 (s, 3H), 2.08 (s, 3H); 13C NMR (100 MHz, CDCl3) d 170.4, 169.6, 169.4, 145.9, 144.6, 141.2, 140.1, 135.8, 120.1, 86.3, 80.0, 73.0, 70.8, 63.2, 20.8, 20.6, 20.4, 16.2; IR (film) l max 3412, 2912, 2846, 1748, 1608, 1498, 1432, 1372, 1228, 1092, 1045 cm−1; HRMS (ESI+) m/z 392.1444 (M+, C18H22N3O7 requires 392.1458).
4-Chloro-7-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (7-deaza-6-chloro-9-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)purine, 18)
A solution of 17 (prepared as previously reported19 or from TRC Biomedical Research Chemicals, 300 mg, 1.00 mmol) in dry pyridine (5 mL) was cooled to 4 °C, and Ac2O (0.501 mL, 5.30 mmol) was added dropwise. The mixture was allowed to warm to room temperature (22 °C) and continuously stirred for 4 h. The reaction solution was cooled to 4 °C, and H2O (10 mL) was added to quench the reaction. The reaction was extracted with CH2Cl2 (4 × 40 mL), and the combined organic layers were washed with H2O (2 × 30 mL), dried over anhydrous sodium sulfate, evaporated to dryness, and the product was purified by flash chromatography (eluent: hexane, EtOAc, 3:1) to yield a white foam (18, 344 mg, 84%). 1H NMR (400 MHz, CDCl3) d 8.67 (s, 1H), 7.43 (d, 1H, J = 3.8 Hz), 6.70 (d, 1H, J = 3.8 Hz), 6.46 (d, 1H, J1', 2' = 6.0 Hz), 5.76 (dd, 1H, J2', 1' = 6.0 Hz, J2', 3' = 5.8 Hz), 5.57 (dd, 1H, J3', 2' = 5.8 Hz, J3', 4' = 4.4 Hz), 4.44-4.34 (m, 3H), 2.16 (s, 3H), 2.15 (s, 3H), 2.05 (s, 3H); 13C NMR (100 MHz, CDCl3) d 170.3, 169.7, 169.4, 152.6, 151.5, 151.2, 126.2, 118.5, 101.5, 85.9, 79.9, 73.1, 70.7, 63.3, 20.8, 20.6, 20.4.
4-Methyl-7-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (7-deaza-6-methyl-9-(2',3',5'-tri-O-acetyl-β-D-ribofuranosyl)purine, 19)
A mixture of 18 (285 mg, 0.690 mmol), Pd(PPh3)4 (160 mg, 0.140 mmol) and trimethyl aluminum (2 M solution in hexanes, 0.690 mL, 1.380 mmol) in THF (10 mL) was heated to reflux for 7 h. Saturated aqueous NH4Cl was added slowly to quench the reaction, and the volatile fraction was removed in vacuo. After extraction with EtOAc (3 × 50 mL), the organic layers were combined, washed with saturated aqueous sodium chloride solution, dried over anhydrous sodium sulfate and evaporated to a colorless oil, which was purified by flash chromatography (eluent: hexane, EtOAc, 1:3) to give 19 (165 mg, 61%) as a white foam. mp 48–49 °C; 1H NMR (400 MHz, CDCl3) d 8.80 (s, 1H), 8.34 (d, 1H, J = 3.8 Hz), 6.66 (d, 1H, J = 3.8 Hz), 6.52 (d, 1H, J1', 2' = 6.0 Hz), 5.79 (dd, 1H, J2', 1' = 6.0 Hz, J2', 3' = 5.8 Hz), 5.60 (dd, 1H, J3', 2' = 5.8 Hz, J3', 4' = 3.9 Hz), 4.44-4.36 (m, 3H), 2.75 (s, 3H), 2.18 (s, 3H), 2.17 (s, 3H), 2.06 (s, 3H); 13C NMR (100 MHz, CDCl3) d 170.4, 169.8, 169.5, 159.9, 151.7, 150.9, 124.5, 118.7, 101.5, 85.3, 79.6, 73.0, 70.8, 63.4, 21.6, 20.8, 20.6, 20.4; IR (film) l max 3434, 3132, 2928, 2846, 1748, 1684, 1586, 1514, 1429, 1374, 1229, 1095, 1042 cm−1; HRMS (ESI+) m/z 392.1443 (M+, C18H22N3O7 requires 392.1458).
General procedure for deprotection of compounds 16 and 19 (0.220 mmol scale) to yield 4 and 6
These compounds were treated with ammonia in methanol (7 N, 5 mL), and stirred at room temperature (22 °C) for 4 h. The solvent was removed in vacuo and the residue was purified by flash column chromatography (eluent: CH2Cl2, MeOH, 9:1). The resulting solid was further purified by recrystallization from methanol.
7-Methyl-3-β-D-ribofuranosyl-3H-imidazo[4,5-b]pyridine (1-deaza-6-methyl-9-β-D-ribofuranosylpurine, 4)
After deprotection of 16, compound 4 was obtained as a white solid (52.5 mg, 90%). mp 198–200 °C. 1H NMR (400 MHz, CD3OD) d 8.55 (s, 1H), 8.22 (d, 1H, J = 5.0 Hz), 7.21 (d, 1H, J = 5.0 Hz), 6.06 (d, 1H, J1', 2' = 6.4 Hz), 4.82 (dd, 1H, J2', 1' = 6.4 Hz, J2', 3' = 5.2 Hz), 4.34 (dd, 1H, J3', 2' = 5.2 Hz, J3', 4' = 2.6 Hz), 4.19 (ddd, 1H, J4', 3' = 2.6 Hz, J4', 5' = 2.4, 2.6 Hz), 3.89 (dd, 1H, Jgem = 12.5 Hz, J5'a, 4' = 2.4 Hz), 3.76 (dd, 1H, Jgem = 12.5 Hz, J5'b, 4' = 2.6 Hz), 2.66 (s, 3H); 13C NMR (100 MHz, CD3OD) d 145.1, 143.5, 143.4, 140.4, 135.6, 119.7, 90.0, 73.7, 71.3, 62.1, 20.6, 14.9; IR (film) l max 3271, 2912, 2852, 1610, 1501, 1206, 1081 cm−1; HRMS (ESI+) m/z 266.1138 (M+, C12H16N3O4 requires 266.1141).
4-Methyl-7-β-D-ribofuranosylpyrrolo[2,3-d]pyrimidine (7-deaza-6-methyl-9-β-D-ribofuranosylpurine, 6)
After deprotection of 19, compound 6 was obtained as a white solid (52.7 mg, 90%). mp 175–176 °C; 1H NMR (400 MHz, CD3OD) d 8.61 (s, 1H), 7.70 (d, 1H, J = 3.8 Hz), 6.74 (d, 1H, J = 3.8 Hz), 6.20 (d, 1H, J1', 2' = 6.2 Hz), 4.61 (dd, 1H, J2', 1' = 6.2 Hz, J2', 3' = 5.5 Hz), 4.29 (dd, 1H, J3', 2' = 5.5 Hz, J3', 4' = 3.3 Hz), 4.10 (ddd, 1H, J4', 3' = 3.3 Hz, J4', 5' = 3.0, 3.3 Hz), 3.84 (dd, 1H, Jgem = 12.2 Hz, J5'a, 4' = 3.0 Hz), 3.74 (dd, 1H, Jgem = 12.2 Hz, J5'b, 4' = 3.3 Hz), 2.72 (s, 3H); 13C NMR (100 MHz, DMSO-d6) d 175.0, 159.4, 149.8, 127.3, 119.0, 100.0, 89.0, 85.6, 74.3, 71.0, 62.0, 20.7; IR (film) l max 3293, 2923, 1594, 1569, 1514, 1432, 1361, 1232, 1125, 1084, 1042 cm−1; HRMS (ESI+) m/z 266.1133 (M+, C12H16N3O4 requires 266.1141).
4-Methyl-7-(2',3'-O-isopropylidene-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (7-deaza-6-methyl-9-(2',3'-O-isopropylidene-β-D-ribofuranosyl)purine, 20)
Compound 6 (450 mg, 1.70 mmol) was dissolved in acetone (5 mL). 1,2-Dimethoxy propane (5 mL) and p-toluenesulfonic acid (442 mg, 4.20 mmol) were added. The mixture was stirred at room temperature (22 °C) for 20 min. MeOH (20 mL) was added to dilute the solution, and Amberlite IRA-400 (OH) resin (from Supelco) was used to modify the pH value to 7. The resin was removed by filtration and washed with MeOH (30 mL), and the filtrate was evaporated in vacuo. The product was purified by flash chromatography (eluent: CH2Cl2, MeOH, 25:1) to afford 404 mg of 20 as a white foam (78%). mp 67–68 °C; 1H NMR (400 MHz, CD3OD) d 8.64 (s, 1H), 7.69 (d, 1H, J = 3.8 Hz), 6.74 (d, 1H, J = 3.8 Hz), 6.34 (d, 1H, J1', 2' = 3.6 Hz), 5.21 (dd, 1H, J2', 1' = 3.6 Hz, J2', 3' = 6.3 Hz), 5.03 (dd, 1H, J3', 2' = 6.3 Hz, J3', 4' = 2.9 Hz), 4.30 (dd, 1H, J4', 3' = 2.9 Hz, J4', 5' = 3.9 Hz), 3.78 (dd, 1H, Jgem = 12.0 Hz, J5'a, 4' = 3.9 Hz), 3.72 (dd, 1H, Jgem = 12.0 Hz, J5'b, 4' = 3.9 Hz), 2.72 (s, 3H), 1.63 (s, 3H), 1.39 (s, 3H); 13C NMR (100 MHz, CD3OD) d 159.4, 150.1, 149.9, 127.3, 118.7, 114.0, 100.2, 90.6, 85.8, 84.0, 81.3, 62.0, 26.2, 24.2, 19.5; IR (film) l max 3275, 2984, 2928, 1676, 1588, 1566, 1427, 1374, 1210, 1073 cm−1; HRMS (ESI+) m/z 306.1443 (M+, C15H20N3O4 requires 306.1454).
4-Methyl-7-β-D-ribofuranosylpyrrolo[2,3-d]pyrimidine 5'-H-phosphonate (7-deaza-6-methyl-9-β-D-ribofuranosylpurine 5'-H-phosphonate, 22)
To a solution of phosphorus trichloride (0.500 mL, 5.73 mmol) in anhydrous CH2Cl2 at −20 °C, 4-methyl-7-(2',3'-O-isopropylidene-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (20, 174 mg, 0.570 mmol) was added and the solution was stirred for 1 h at −20 °C. The reaction was warmed to room temperature (22 °C) and stirred for 4 h. The solvent was evaporated in vacuo and the residue was treated with aqueous TFA (2 mL, 30%) for 20 min. The reaction mixture was concentrated in vacuo, the crude product was dissolved in aqueous triethylammonium bicarbonate (TEAB) buffer (1 mL, 10 mM, pH = 8), and the product was purified by Sephadex LH-20 column (2.5 cm × 40 cm). Elution with 10 mM aqueous TEAB buffer and lyophilization yielded 22 as the triethylammonium salt, a colorless glassy solid (139 mg, 62%). 1H NMR (400 MHz, D2O) d 8.37 (s, 1H), 7.55 (d, 1H, J = 3.8 Hz), 6.62 (d, 1H, J = 3.8 Hz), 6.61(d, 1H, JP, H = 638 Hz), 6.19 (d, 1H, J1', 2' = 6.4 Hz), 4.57 (dd, 1H, J2', 1' = 6.4 Hz, J2', 3' = 5.6 Hz), 4.36 (dd, 1H, J3', 2' = 5.6 Hz, J3', 4' = 3.2 Hz), 4.22 (ddd, 1H, J4', 3' = 3.2 Hz, J4', 5' = 1.5, 1.2 Hz), 3.98 (dd, 1H, Jgem = 2.8 Hz, J5'a, 4' = 1.5 Hz), 3.97 (dd, 1H, Jgem = 2.8 Hz, J5'b, 4' = 1.2 Hz), 3.05 (q, 6H, J = 7.4 Hz, N(CH2CH3)3), 2.46 (s, 3H), 1.13 (t, 9H, J = 7.4 Hz, N(CH2CH3)3); 13C NMR (100 MHz, D2O) d 154.7, 150.3, 143.7, 130.3, 118.4, 104.1, 87.1, 84.0 (Jc, p = 8 Hz), 74.3, 70.6, 63.1 (Jc, p = 4 Hz), 46.6, 16.7, 8.2; 31P NMR (162 MHz, D2O), 6.72 (s); HRMS (ESI-) m/z 328.0706 (M−, C12H15N3O6P requires 328.0689).
4-Methyl-7-β-D-ribofuranosylpyrrolo[2,3-d]pyrimidine 5'-triphosphate (7-deaza-6-methyl-9-β-D-ribofuranosylpurine 5'-triphosphate, 9)
Prior to the reaction, tris(tetra-n-butylammonium) hydrogen pyrophosphate (prepared as previously described,41 dried over P2O5, and stored at −20 °C, 116 mg, 0.13 mmol) and compound 25 (55.0 mg, 0.130 mmol) were dried overnight under vacuum at room temperature (22 °C) in two separate round bottom flasks. Anhydrous DMF (1.5 mL) and pyridine (250 µL, 3.00 mmol) were added to the round bottom flask containing 22. After the solid was dissolved, TMSCl (132 µL, 1.04 mmol) was added by gas tight microsyringe. After 5 min, a solution of I2 in DMF (0.2 M, 910 µL) was added dropwise. The mixture was stirred for 5 min at 22 °C before a solution of tris(tetra-n-butylammonium) hydrogen pyrophosphate in DMF (0.5 mL) was added. The reaction mixture was stirred at room temperature (22 °C) for 30 min and concentrated in vacuo. The residue was dissolved in cold deionized water (1 mL) by sonication for 1 min. The precipitated iodine was removed by filtration through a small plug of cotton inserted in the bottom of a 12 cm glass Pasteur pipet. The product was purified by Sephadex LH-20 column with ice-cold aqueous TEAB buffer (10 mM, pH = 8) as the eluent. The fractions containing the triphosphate were identified by mass spectrometry, combined and lyophilized to yield the crude product, which was further purified by preparative reverse-phase HPLC with a linear gradient of 0% to 10% CH3CN in aqueous TEAB buffer (10 mM, pH = 7.5, adjusted with acetic acid) over 30 min (Retention time = 10.8 to 12.0 min). Repeated lyophilization and resuspension in deionized water (3 × 3 mL) yielded 9 as the triethylammonium salt, a colorless glassy solid. 1H NMR (400 MHz, D2O) d 8.56 (s, 1H), 7.76 (d, 1H, J = 3.8 Hz), 6.82 (d, 1H, J = 3.8 Hz), 6.30 (d, 1H, J1', 2' = 7.0 Hz), 4.65 (dd, 1H, J2', 1' = 7.0 Hz, J2', 3' = 4.7 Hz), 4.48 (dd, 1H, J3', 2' = 4.7 Hz, J3', 4' = 2.8 Hz), 4.27-4.26 (m, 1H), 4.18-4.04 (m, 2H), 3.08 (q, 18H, J = 7.3 Hz), 2.65 (s, 3H), 1.15 (t, 27H, J = 7.3 Hz); 31P NMR (162 MHz, D2O) d −10.2 (d, Jp, p = 19.9 Hz), −11.3 (d, Jp, p = 19.9 Hz), −23.1 (t, Jp, p = 19.9 Hz); HRMS (ESI-) m/z 503.9962 (M−, C12H17N3O13P3 requires 503.9974).
4-Methyl-7-(2',3'-O-isopropylidene-5'-O-methyl-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (7-deaza-6-methyl-9-(2',3'-O-isopropylidene-5'-O-methyl-β-D-ribofuranosyl)purine, 21)
4-Methyl-7-(2',3'-O-isopropylidene-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (20, 44.0 mg, 0.144 mmol) was dissolved in CH3CN (1 mL), and cooled to 4 °C. NaH (11.6 mg, 0.288 mmol) was added, and the resulting suspension was stirred at 4 °C for 0.5 h before adding MeI (0.180 mL, 0.288 mmol). The mixture was stirred at room temperature (22 °C) for 2 h. MeOH (3 mL) was added to quench excess NaH. After removal of solvent in vacuo, the crude product was purified by flash chromatography (eluent: hexane, EtOAc, 1:1) to afford 40.0 mg of a colorless oil (86%). 1H NMR (400 MHz, CDCl3) δ 8.78 (s, 1H), 7.38 (d, 1H, J = 3.7 Hz), 6.58 (d, 1H, J = 3.7 Hz), 6.38 (d, 1H, J1', 2' = 2.9 Hz), 5.21 (dd, 1H, J2', 1' = 2.9 Hz, J2', 3', = 6.4 Hz), 5.00 (dd, 1H, J3', 2' = 6.4 Hz, J3', 4' = 3.4 Hz), 4.39 (ddd, 1H, J4', 3' = 3.4 Hz, J4', 5' = 3.9, 4.8 Hz), 3.63 (dd, 1H, Jgem = 10.3 Hz, J5'a, 4' = 3.9 Hz), 3.59 (dd, 1H, Jgem = 10.3 Hz, J5'b, 4' = 4.8 Hz), 3.39 (s, 3H), 2.73 (s, 3H), 1.66 (s, 3H), 1.39 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 159.5, 151.5, 150.3, 125.9, 118.6, 114.4, 100.4, 90.4, 84.7, 84.5, 81.5, 72.9, 59.3, 27.3, 25.4, 21.5; IR (film) λ max 2984, 2928, 1586, 1512, 1460, 1374, 1347, 1215, 1092 cm−1; HRMS (ESI+) m/z 320.1595 (M+, C16H22N3O4 requires 320.1610).
4-Methyl-7-(5'-O-methyl-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (7-deaza-6-methyl-9-(5'-O-methyl-β-D-ribofuranosyl)purine, 7)
Compound 21 (40.0 mg, 0.125 mmol) was treated with aqueous TFA (70%, 1 mL), and stirred at room temperature (22 °C) for 15 min. The solvent was removed in vacuo, and the residue was purified by flash chromatography (eluent: CH2Cl2, MeOH, 20:1) to yield 30.4 mg of 7 as a white foam (87%). mp 63–65 °C; 1H NMR (400 MHz, CD3OD) δ 8.89 (s, 1H), 8.01 (d, 1H, J = 3.9 Hz), 7.06 (d, 1H, J = 3.9 Hz), 6.42 (d, 1H, J1', 2' = 5.6 Hz), 4.48 (dd, 1H, J2', 1' = 5.6 Hz, J2', 3' = 5.3 Hz), 4.29 (dd, 1H, J3', 2' = 5.3 Hz, J3', 4' = 3.6 Hz), 4.19 (ddd, 1H, J4', 3' = 3.6 Hz, J4', 5' = 3.0, 3.4 Hz), 3.71 (dd, 1H, Jgem = 10.8 Hz, J5'a, 4' = 3.0 Hz), 3.63 (dd, 1H, Jgem = 10.8 Hz, J5'b, 4' = 3.4 Hz), 3.45 (s, 3H), 2.91 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 155.3, 150.5, 145.3, 129.5, 118.1, 102.3, 88.0, 84.3, 75.3, 72.2, 71.2, 58.2, 16.8; IR (film) λ max 3354, 3110, 2923, 2890, 2066, 1682, 1643, 1602, 1515, 1463, 1201, 1131 cm−1; HRMS (ESI+) m/z 280.1285 (M+, C13H18N3O4 requires 280.1297).
Cytotoxicity assays
HeLa S3 cells (ATCC# CCL-2.2) were maintained in DMEM/F-12 media supplemented with 2% dialyzed fetal bovine serum and 1% penicillin/streptomycin (1 ×, Invitrogen). HeLa S3 cells (4 × 103) in 100 µL media were loaded on a 96-well plates 24 h before use. Cells were incubated with ribonucleosides at various concentrations for 7 h, 24 h and 48 h respectively at 37 °C. All wells were adjusted to a final concentration of 0.1% DMSO. Media was removed and cells were washed with PBS (100 µL). 100 µL media, 75 µL PBS and 25 µL CellTiter-Glo reagent (Promega) were added to each well, and the 96-well plate was shaken on a titer plate shaker for 2 min to detach the cells. The cell suspension (200 µL) was transferred to an opaque 96-well plate according to the Promega protocol provided with the reagent. After incubation at room temperature (22 °C) for 10 min, the luminescence of the samples was measured with a Packard Fusion microtiterplate reader. The IC50 values were calculated by with a nonlinear four-parameter curve fit (GraphPad Prism 5.0) of luciferase activity vs. concentration of nucleoside.
Antiviral assay against poliovirus (PV)
HeLa S3 cells (1 × 105) were plated 1 day prior to treatment in a 24-well plate. Cells were pretreated with different concentrations of nucleosides in fresh media containing 0.1% DMSO. After 1 h incubation at 37 °C, media was removed and cells were infected with PV (1 × 106 PFU) in PBS (100 µL). Plates were incubated for 15 min at room temperature (22 °C), PBS was removed by aspiration, and fresh media containing the specified amount of nucleoside was added. The infection was allowed to proceed at 37 °C for 6 h. Cells were washed with PBS and collected by treatment with trypsin. Cells were pelleted by centrifugation, resuspended in PBS (500 µL), and subjected to 3 freeze-thaw cycles. Cell debris was removed by centrifugation and the supernatant containing the cell-associated virus was saved. Viral titer was determined by applying serial dilutions of supernatant to HeLa S3 monolayers (plated in 6-well plates 1 day before at 5 × 105 cells/well) and overlaying with growth media containing low melting point agarose (1%). Plates were incubated for 2 days at 37 °C. The agarose was removed and plaques were visualized by staining with crystal violet (1%) in aqueous ethanol (20%). The IC50 values were calculated by with a nonlinear four-parameter curve fit (GraphPad Prism 5.0) of viral titer vs. concentration of nucleoside.
Antiviral assays against dengue (DENV2)
DENV2 replicon assay
BHK-21 cells stably expressing the DENV2 replicon were thawed and cultured in DMEM/F-12 media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (1×, Invitrogen). Cells (6 × 104) were seeded in a 48-well plate 24 h before use. Nucleosides at different concentrations in media containing 1% DMSO were used to treat cells for 24 h. Cells were washed with PBS once and lysed by adding lysis buffer (Promega, 60 µL/well) and passing the lysate through 27-gauge needle fitted with a 1 mL syringe. Lysate (20 µL) was added to a opaque 96-well reader plate, and the luciferase activity was measured by Luminometer (Centro LB 960, Berthold Technologies).
DENV2 infectivity plaque assay31
BHK-21 cells (5 × 105) were plated one day prior to treatment in a 12-well plate. Cells were infected with DENV2, and treated with nucleosides at different concentrations in media containing 1% DMSO for 24 h at 37 °C. Cell culture supernatant was covered with a first overlayer of media containing 0.9% low-melting point agarose, 1 × EMEM and 0.5% FBS, and incubated at 37 °C for 6 days. A second layer containing 0.9% low melting point agarose, 1% NaCl, 0.015% neutral red dye was added, and the plate was incubated at 37 °C overnight. The number of plaques were counted and calculated as PFU/ml. The IC50 values were calculated with a nonlinear four-parameter curve fit (GraphPad Prism 5.0) of viral titer vs. concentration of nucleoside.
DENV2 infectivity real time PCR assay
The DENV2 RNA copy number was measured using primers and probe specific to DENV2 NS1 region. The primers and probe were designed using the Beacon designer 4.0 program. PCR amplification and real-time data collection and analysis were performed according to iQ5 multicolor real-time PCR detection system (Bio-Rad). Briefly, a reverse-transcriptase (RT) reaction was carried out using a RT kit from Bio-Rad (iScript cDNA synthesis). RNA samples (5 µL) were mixed with the iScript cDNA synthesis mixture and incubated at 42 °C for 30 min. The cDNA product (2 µL) was subjected to PCR using conditions as previously reported.32 Real-time detection of the DENV2 PCR product was correlated with the input cDNA copy number and the results were plotted.
Guanidine resistance assay30
Resistance to 2 mM guanidine is conferred by a single C to U transition mutation in the 2C protein in poliovirus genome. HeLa S3 cells were plated one day before the experiment at 25% confluence in 10-cm dishes. Cells were infected with 1 × 106 PFU poliovirus from the appropriate viral stock previously obtained in the presence or absence of the nucleosides at the concentrations shown in Table S1 (Supporting information). Cells were covered with 20-mL DMEM/F12 media supplemented with 10% FBS, 1% agarose, 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin and 2 mM guanidine hydrochloride. The guanidine resistant viral titer (Guar plaques) was determined.
Nucleotide incorporation by PV RdRP (3Dpol) in vitro
Incorporation of NTPs (8, 9) opposite each of the four templating bases (A, C, G and U) was examined using pre-assembled 3Dpol-primer/template complexes.36 As previously described, symmetrical substrate (sym/sub) served as the primer and template RNA. Annealing of 32P-end-labeled and unlabeled sym/sub oligos to form the primer/template duplex was preformed as previously described. 3Dpol was allowed to preassemble with sym/sub duplex for 3 min at 30 °C. Incorporation assays with A, C, G and U as templating bases were performed at 30 °C in HEPES buffer (pH 7.5, 50 mM) containing 2-mercaptoethanol (10 mM), MgCl2 (5 mM), 3Dpol (1 µM), sym/sub duplex RNA (0.5 µM) and nucleotide (0.5 mM). All incorporation assays were initiated by addition of nucleotide, reaction products were separated by denaturing PAGE, gels were visualized using a phosphorimager, and radioactivity was quantified using ImageQuant software (Molecular Dynamics). Measurement of pre-steady state kinetic parameters for 8 and 9 are described in Figure S8 and S9 and references cited therein.
Steady state incorporation assay
Steady-state incorporation of 8 and 9 by PV RdRP (3Dpol) opposite templating bases U and C was examined. Incorporation of the correct nucleotide (ATP templated by U and GTP templated by C) and an incorrect nucleotide (GTP templated by U and ATP templated by C) served as comparative controls. Nucleotide incorporation reactions contained HEPES buffer (pH 7.5, 50 mM), 2-mercaptoethanol (10 mM), MgCl2 (5 mM), ZnCl2 (60 µM), NTP (1 mM), sym/sub duplex RNA (10 µM) and 3Dpol (1 µM). All incorporation assays were initiated by addition of 3Dpol, and stopped with 50 mM EDTA (final concentration). For steady state assays involving addition of more than one nucleotide, and where the total nucleotide concentration was greater than 1 mM, the concentration of MgCl2 was increased to maintain the concentration of free Mg2+ at 4 mM above the total NTP concentration. The reaction products were separated by denaturing PAGE, gels were visualized using a phosphorimager, and radioactivity was quantified using ImageQuant software (Molecular Dynamics). Data were fit to a linear model using GraphPad Prism 5.0.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge the NIH (AI054776, AI070791, and AI082068) for financial support.
Abbreviations
- PV
poliovirus
- DENV
dengue virus
- HSV
herpes simplex virus
- HCV
hepatitis C virus
- p-TsOH
p-toluenesulfonic acid
- HMPT
hexamethylphosphorous triamide
- TDA-1
tris[2-(2-methoxyethoxy)ethyl]amine
- TFA
trifluoroacetic acid
- MeCN
acetonitrile
- TMSCl
trimethylsilyl chloride
- MOI
multiplicity of infection
- RdRP
RNA-dependent RNA polymerase
- S/S
symmetrical RNA substrate
- NTP
nucleoside triphosphate
- RT
reverse transcriptase
- PFU
plaque-forming unit
- PAGE
polyacrylamide gel electrophoresis
Footnotes
Supporting Information Available: Purity data for synthetic compounds, guanidine resistance assay data, and supporting figures for NTP incorporation assays. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zuccotto F, Ardini E, Casale E, Angiolini M. Through the "Gatekeeper Door": Exploiting the Active Kinase Conformation. J. Med. Chem. 2010;53:2681–2694. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 2.De Clercq E, Bergstrom DE, Holy A, Montgomery JA. Broad-Spectrum Antiviral Activity of Adenosine Analogues. Antiviral Res. 1984;4:119–133. doi: 10.1016/0166-3542(84)90012-3. [DOI] [PubMed] [Google Scholar]
- 3.Serafinowski P, Dorland E, Harrap KR, Balzarini J, De Clercq E. Synthesis and Antiviral Activity of some New S-Adenosyl-L-Homocysteine Derivatives. J. Med. Chem. 1992;35:4576–4583. doi: 10.1021/jm00102a010. [DOI] [PubMed] [Google Scholar]
- 4.De Clercq E. John Montgomery's Legacy: Carbocyclic Adenosine Analogues as SAH Hydrolase Inhibitors with Broad-Spectrum Antiviral Activity. Nucleos. Nucleot. Nucl. 2005;24:1395–1415. doi: 10.1080/15257770500265638. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SS, La Femina RL. Nucleoside Analog Inhibitors of Hepatitis C Viral Replication. Antiviral Res. 2009:153–166. [Google Scholar]
- 6.Montgomery J, Hewson K. Analogs of 6-Methyl-9-beta-D-Ribofuranosylpurine. J. Med. Chem. 1968;11:48–52. doi: 10.1021/jm00307a010. [DOI] [PubMed] [Google Scholar]
- 7.Van Aerschot A, Mamos P, Weyns N, Ikeda S, De Clercq E, Herdewijn P. Antiviral Activity of C-Alkylated Purine Nucleosides Obtained by Cross-coupling with Tetraalkyltin Reagents. J. Med. Chem. 1993;36:2938–2942. doi: 10.1021/jm00072a013. [DOI] [PubMed] [Google Scholar]
- 8.Vittori S, Dal Ben D, Lambertucci C, Marucci G, Volpini R, Cristalli G. Antiviral Properties of Deazaadenine Nucleoside Derivatives. Curr. Med. Chem. 2006;13:3529–3552. doi: 10.2174/092986706779026228. [DOI] [PubMed] [Google Scholar]
- 9.Turk SR, Shipman C, Jr, Nassiri R, Genzlinger G, Krawczyk SH, Townsend LB, Drach JC. Pyrrolo[2,3-d]pyrimidine Nucleosides as Inhibitors of Human Cytomegalovirus. Antimicrob. Agents Chemother. 1987;31:544–550. doi: 10.1128/aac.31.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergstrom DE, Brattesani AJ, Ogawa MK, Reddy PA, Schweickert MJ, Balzarini J, De Clercq E. Antiviral activity of C-5 substituted tubercidin analogues. J. Med. Chem. 1984;27:285–292. doi: 10.1021/jm00369a010. [DOI] [PubMed] [Google Scholar]
- 11.Smith CG, Reineke LM, Burch MR, Shefner AM, Muirhead EE. Studies on the Uptake of Tubercidin (7-Deazaadenosine) by Blood Cells and its Distribution in Whole Animals. Cancer Res. 1970;30:69–75. [PubMed] [Google Scholar]
- 12.Seibert G, Maidhof A, Zahn RK, Muller WE. Tubercidin Metabolism in Mouse L5178y Cells in vivo and in vitro. Gann. 1978;69:739–747. [PubMed] [Google Scholar]
- 13.Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL. A 7-Deaza-Adenosine Analog is a Potent and Selective Inhibitor of Hepatitis C Virus Replication with Excellent Pharmacokinetic Properties. Antimicrob. Agents Chemother. 2004;48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Clercq E, Balzarini J, Madej D, Hansske F, Robins M. Nucleic Acid Related Compounds. 51. Synthesis and Biological Properties of Sugar-Modified Analogs of the Nucleoside Antibiotics Tubercidin, Toyocamycin, Sangivamycin, and Formycin. J. Med. Chem. 1987;30:481–486. doi: 10.1021/jm00386a007. [DOI] [PubMed] [Google Scholar]
- 15.Butora G, Olsen D, Carroll S, McMasters D, Schmitt C, Leone J, Stahlhut M, Burlein C, MacCoss M. Synthesis and HCV Inhibitory Properties of 9-Deaza-and 7, 9-Dideaza-7-oxa-2'-C-methyladenosine. Bioorg. Med. Chem. 2007;15:5219–5229. doi: 10.1016/j.bmc.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Seela F, Peng X, Budow S. Advances in the Synthesis of 7-Deazapurine-pyrrolo [2,3-d] pyrimidine 2-Deoxyribonucleosides Including D-and L-Enantiomers, Fluoro Derivatives and 2,3-Dideoxyribonucleosides. Curr. Org. Chem. 2007;11:427–462. [Google Scholar]
- 17.Leroy F, Chaves D, Dukhan D, Storer R, Sommadossi JP, Loi A, Cadeddu A, Fanti M, Boscu N, Bassetti F. Synthesis and Antiviral Evaluation of 7-Fluoro-7-deaza-2-aminopurine Nucleoside Derivatives. Vol. 2008. Oxford Univ Press; 2008. pp. 595–596. [DOI] [PubMed] [Google Scholar]
- 18.Yin Z, Chen Y, Schul W, Wang Q, Gu F, Duraiswamy J, Kondreddi RR, Niyomrattanakit P, Lakshminarayana SB, Goh A. An Adenosine Nucleoside Inhibitor of Dengue Virus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naus P, Pohl R, Votruba I, Dzubak P, Hajduch M, Ameral R, Birkus G, Wang T, Ray AS, Mackman R, Cihlar T, Hocek M. 6-(Het)aryl-7-Deazapurine Ribonucleosides as Novel Potent Cytostatic Agents. J. Med. Chem. 2010;53:460–470. doi: 10.1021/jm901428k. [DOI] [PubMed] [Google Scholar]
- 20.Mayers DL, Mikovits JA, Joshi B, Hewlett IK, Estrada JS, Wolfe AD, Garcia GE, Doctor BP, Burke DS, Gordon RK. Anti-Human Immunodeficiency Virus 1 (HIV-1) Activities of 3-Deazaadenosine Analogs: Increased Potency against 3'-Azido-3'-deoxythymidine-resistant HIV-1 Strains. Proc. Natl. Acad. Sci. U.S.A. 1995;92:215–219. doi: 10.1073/pnas.92.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon RK, Ginalski K, Rudnicki WR, Rychlewski L, Pankaskie MC, Bujnicki JM, Chiang PK. Anti-HIV-1 Activity of 3-Deaza-adenosine Analogs. Eur. J. Biochem. 2003;270:3507–3517. doi: 10.1046/j.1432-1033.2003.03726.x. [DOI] [PubMed] [Google Scholar]
- 22.Brekiesz-Lewandowska B, Talik Z. Pyridotriazoles and pyridoimidazoles. III. 2-Methylpyridine, 3-methylpyridine, and 4-methylpyridine derivatives. Roczniki Chemii. 1970;44:69–75. [Google Scholar]
- 23.Deady L, Korytsky O, Rowe J. Substituent Effects on the Isomer Ratios in the Rearrangement of some 2-and 4-Nitraminopyridines. Aust. J. Chem. 1982;35:2025–2034. [Google Scholar]
- 24.Graboyes H, Day AR. Metabolite Analogs .8. Syntheses of Some Imidazopyridines and Pyridotriazoles. J. Am. Chem. Soc. 1957;79:6421–6426. [Google Scholar]
- 25.Urban M, Joubert N, Hocek M, Alexander RE, Kuchta RD. Herpes Simplex Virus-1 DNA Primase: A Remarkably Inaccurate yet Selective Polymerase. Biochemistry. 2009;48:10866–10881. doi: 10.1021/bi901476k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cristalli G, Franchetti P, Grifantini M, Vittori S, Bordoni T, Geroni C. Improved Synthesis and Antitumor Activity of 1-Deazaadenosine. J. Med. Chem. 1987;30:1686–1688. doi: 10.1021/jm00392a029. [DOI] [PubMed] [Google Scholar]
- 27.Crey-Desbiolles C, Kotera M. Synthesis of 3-deaza-3-nitro-2'-deoxyadenosine. Bioorg. Med. Chem. 2006;14:1935–1941. doi: 10.1016/j.bmc.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 28.Naus P, Kuchar M, Hocek M. Cytostatic and Antiviral 6-Arylpurine Ribonucleosides IX. Synthesis and Evaluation of 6-Substituted 3-Deazapurine Ribonucleosides. Coll. Czech. Chem. Comm. 2008;73:665–678. [Google Scholar]
- 29.Sun Q, Edathil JP, Wu R, Smidansky ED, Cameron CE, Peterson BR. One-pot synthesis of nucleoside 5'-triphosphates from nucleoside 5'-H-phosphonates. Org. Lett. 2008;10:1703–1706. doi: 10.1021/ol8003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crotty S, Maag D, Arnold JJ, Zhong W, Lau J, Hong Z, Andino R, Cameron CE. The Broad-Spectrum Antiviral Ribonucleoside Ribavirin is an RNA Virus Mutagen. Nature Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 31.Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. Inhibition of Dengue Virus Replication by Mycophenolic Acid and Ribavirin. J. Gen. Virol. 2006;87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- 32.Houng H, Hritz D, Kanesa-thasan N. Quantitative Detection of Dengue 2 Virus Using Fluorogenic RT-PCR Based on 3'-Noncoding Sequence. J. Virol. Meth. 2000;86:1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 33.Graci JD, Cameron CE. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir. Chem. Chemoth. 2004;15:1–13. doi: 10.1177/095632020401500101. [DOI] [PubMed] [Google Scholar]
- 34.Lindberg B, Klenow H, Hansen K. Some Properties of Partially Purified Mammalian Adenosine Kinase. J. Biol. Chem. 1967;242:350–356. [PubMed] [Google Scholar]
- 35.Miller RL, Adamczyk DL, Miller WH, Koszalka GW, Rideout JL, Beacham LM, 3rd, Chao EY, Haggerty JJ, Krenitsky TA, Elion GB. Adenosine Kinase from Rabbit Liver. II. Substrate and Inhibitor Specificity. J. Biol. Chem. 1979;254:2346–2352. [PubMed] [Google Scholar]
- 36.Arnold JJ, Cameron CE. Poliovirus RNA-Dependent RNA Polymerase (3D(pol)). Assembly of Stable, Elongation-Competent Complexes by Using a Symmetrical Primer-Template Substrate (sym/sub) J. Biol. Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 37.De Clercq E, Neyts J. Antiviral Agents Acting as DNA or RNA Chain Terminators. Handb. Exp. Pharmacol. 2009;189:53–84. doi: 10.1007/978-3-540-79086-0_3. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J, Xu L, Stevens ED, Trudell ML, Izenwasser S, Wade D. Stereoselective Synthesis of Conformationally Constrained Tropane Analogues: 6-Chloro-2,5-diazatetracyclo[8.5.0.02,13.04,9]pentadeca-4,6,8-triene-11-one and 6-Chloro-2,7-diazatetracyclo[8.5.0.02,13.04,9]pentadeca-4,6,8-triene-11-one. J. Heterocycl. Chem. 2004;41:569–574. [Google Scholar]
- 39.Bhattacharya A, Purohit VC, Deshpande P, Pullockaran A, Grosso JA, DiMarco JD, Gougoutas JZ. An Alternate Route to 2-Amino-3-nitro-5-bromo-4-picoline: Regioselective Pyridine Synthesis via 2-Nitramino-picoline Intermediate. Org. Process Res. Dev. 2007;11:885–888. [Google Scholar]
- 40.Israel M, Day AR. Preparation of Pyrido-(2,3)-pyrazines, Pyrido-(3,4)-pyrazines and Imidazo-(b)-pyridines. J. Org. Chem. 1959;24:1455–1460. [Google Scholar]
- 41.Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, Poulter CD. Phosphorylation of Isoprenoid Alcohols. J. Org. Chem. 1986;51:4768–4779. [Google Scholar]
- 42.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.