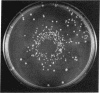
Abstract
Three recombination events, reciprocal recombination, sister-chromatid recombination, and gene conversion, were studied using substrates designed in vitro. Each type of recombination event can be monitored at any chromosomal location. We have shown that sister-chromatid recombination is induced mitotically by DNA damaging agents, such as methyl methanesulfonate and gamma-rays, but is decreased mitotically in strains defective in rad52. Reciprocal recombination by which circular plasmids integrate into the genome is unaffected by rad52 defective alleles and occurs by a different recombination pathway. Mechanisms are suggested by which gene conversion between sister chromatids can generate chromosome rearrangements.
Full text
PDF

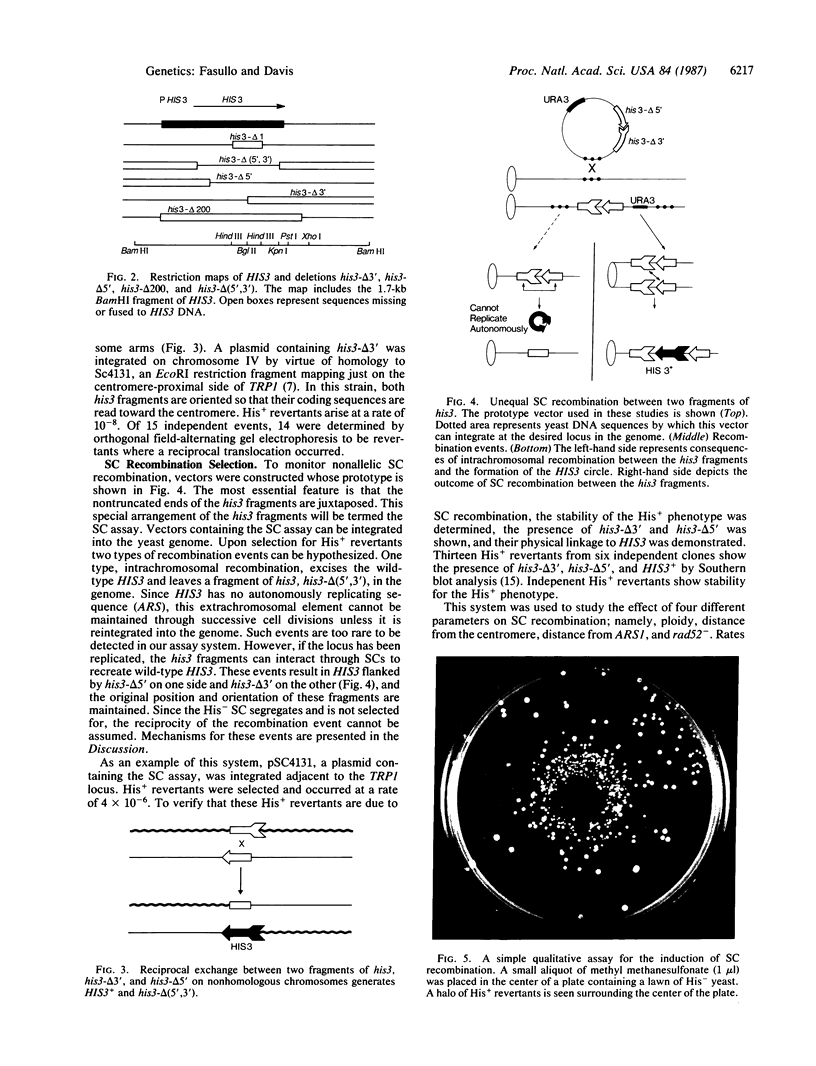

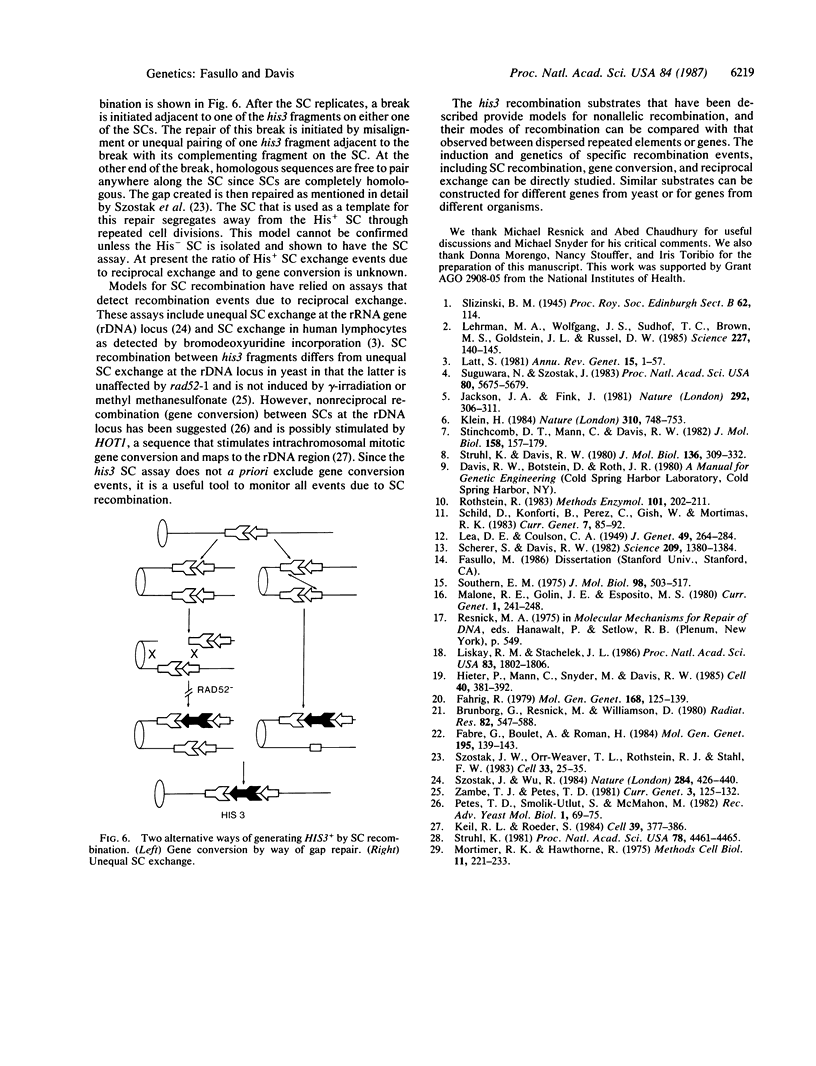
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunborg G., Resnick M. A., Williamson D. H. Cell-cycle-specific repair of DNA double strand breaks in Saccharomyces cerevisiae. Radiat Res. 1980 Jun;82(3):547–558. [PubMed] [Google Scholar]
- Fabre F., Boulet A., Roman H. Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(1-2):139–143. doi: 10.1007/BF00332736. [DOI] [PubMed] [Google Scholar]
- Fahrig R. Evidence that induction and suppression of mutations and recombinations by chemical mutagens in S. cerevisiae during mitosis are jointly correlated. Mol Gen Genet. 1979 Jan 10;168(2):125–139. doi: 10.1007/BF00431439. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Klein H. L. Lack of association between intrachromosomal gene conversion and reciprocal exchange. 1984 Aug 30-Sep 5Nature. 310(5980):748–753. doi: 10.1038/310748a0. [DOI] [PubMed] [Google Scholar]
- Lehrman M. A., Schneider W. J., Südhof T. C., Brown M. S., Goldstein J. L., Russell D. W. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985 Jan 11;227(4683):140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L. Information transfer between duplicated chromosomal sequences in mammalian cells involves contiguous regions of DNA. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1802–1806. doi: 10.1073/pnas.83.6.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic mapping in yeast. Methods Cell Biol. 1975;11:221–233. doi: 10.1016/s0091-679x(08)60325-8. [DOI] [PubMed] [Google Scholar]
- Nanney D. L. T. M. Sonneborn: an interpretation. Annu Rev Genet. 1981;15:1–9. doi: 10.1146/annurev.ge.15.120181.000245. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Recombination of dispersed repeated DNA sequences in yeast. Science. 1980 Sep 19;209(4463):1380–1384. doi: 10.1126/science.6251545. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. A physical, genetic and transcriptional map of the cloned his3 gene region of Saccharomyces cerevisiae. J Mol Biol. 1980 Jan 25;136(3):309–332. doi: 10.1016/0022-2836(80)90376-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Szostak J. W. Recombination between sequences in nonhomologous positions. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5675–5679. doi: 10.1073/pnas.80.18.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]