Abstract
The impact of a bulky DNA lesion on the structure and dynamics of a nucleosome core particle (NCP) containing a lesion derived from the unusually potent tumorigen dibenzo[a,l]pyrene that resists nucleotide excision repair (NER) in free DNA, was investigated using 65-ns molecular dynamics simulations. Our results reveal that, relative to unmodified NCP, the lesion stabilizes the nucleosome via stacking interactions, improved Watson-Crick base pairing, hydrogen bonding between DNA and histones, as well as damped dynamics. These findings suggest that such lesions should be as resistant to NER in the nucleosome environment as they are in free DNA.
The origins of the acute tumorigenic potencies of certain polycyclic aromatic hydrocarbon (PAH) environmental pollutants remain elusive. It has been hypothesized that the resistance to nucleotide excision repair (NER) (1), a prominent mechanism for excising the bulky mutagenic DNA lesions that are produced by these substances, is key to their cancer-initiating properties (2). While resistance to NER has been reported for some PAH-derived DNA lesions embedded in free DNA (3, 4), the repair of PAH lesions in the protein environment of nucleosomes, the basic building blocks of chromatin. The relationship between structural features of protein-free PAH-DNA lesions in aqueous environments and their resistance to NER have been discussed (5). However, nothing is known about the impact of the adduct on the structure and dynamics of the nucleosome remains to be expolored. Here, we have addressed these issues for the first time by investigating the properties of such an adduct utilizing molecular dynamics (MD) simulations.
It has been hypothesized that the relative susceptibility to NER of a bulky lesion is determined by the extent of local thermodynamic destabilization induced by the lesion (4–7) and that a repair-resistant lesion scarcely destabilizes the DNA duplex (4). It has been shown that NER activity is observable when the damaged DNA is packaged in nucleosomes, although at a slower rate than in the case of free DNA (8). Our objectives were to determine how this higher order DNA organization might affect the conformational characteristics of the PAH-DNA lesion, local stability, and the dynamic properties of the nucleosomes (9).
As a model system, we have focused on dibenzo[a,l]pyrene (DB[a,l]P), the most potent tumorigen among all carcinogenic PAHs tested to date (10, 11). PAHs are present in cigarette smoke, coal particulates and in soil or sediment samples (2, 11). This non-planar fjord region PAH is metabolically activated, through the predominant diol epoxide (DE) pathway ( 11, 12), primarily to (−)-anti-DB[a,l]PDE. Reactions with DNA produces both adenine and guanine adducts, with adenine adducts frequently predominant. The diol epoxides are highly mutagenic (12) and tumorigenic, and cancer can be initiated by the bulky adducts if they are not repaired (Reviewed in (11)).
Here we investigated the prominent 14S (−)-trans-anti-DB[a,l]P-N6-dA adduct (Figure 1a) with 65-ns MD simulations in the nucleosome core particle (NCP). This adduct shows poor susceptibility to NER in the human HeLa cell extract system in protein-free DNA (3, 13). Furthermore, thermal melting data shows that repair-resistant lesions cause little destabilization of DNA duplexes (14). Molecular modeling with MD simulations suggests that the DB[a,l]P aromatic ring system interacts strongly with DNA bases by assuming a classically intercalated conformation (15) and (Cai Y, Broyde S. et al, manuscript in preparation): the DB[a,l]P ring system is inserted into the helix from the major groove on the 3′-side of the damaged base without disrupting Watson-Crick hydrogen bonding, and there are strong van der Waals interactions with the bases that form the intercalation pocket (Figure 1b). This structure was modeled based on the NMR solution structure of the analogous fjord region benzo[c]phenanthrene (B[c]Ph)-derived adduct, 1S (−)-trans-anti-B[c]Ph-N6-dA (16) by adding two aromatic rings (Figure S1 and Methods, Supporting Information). Spectroscopic observations reveal that the UV absorption maximum for this adduct is red-shifted (Figure S2, Supporting Information), which is fully consistent with an intercalative conformation.
Figure 1.
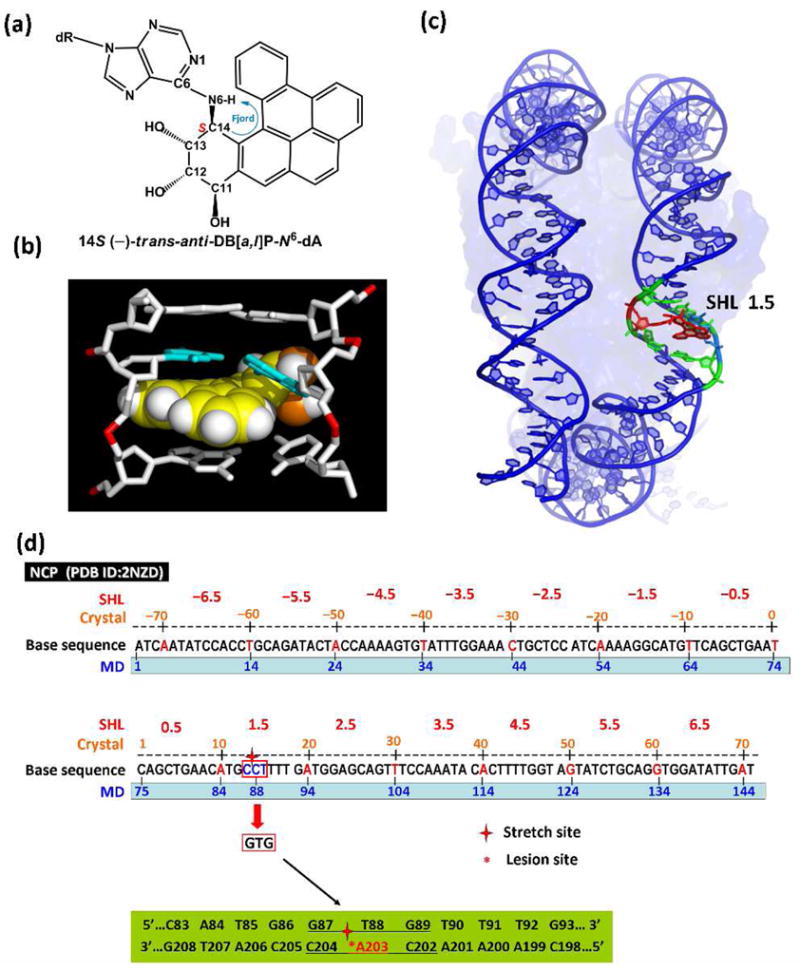
(a) chemical structure of the 14S (−)-trans-anti-DB[a,l]P-N6-dA adduct, and intercalation models for (b) the uncomplexed duplex (see Supporting Information) and (c) the nucleosome following 65-ns of MD simulations. See also Movie S1 Supporting Information. (d) Base sequence contexts of the NCP with a 145-mer DNA duplex (PDB ID : 2NZD) (17) with numbering schemes in the crystal structure and MD simulations. The sequences surrounding the stretch site at SHL 1.5, circled in red, were remodeled to the …GTG… : …CAC… duplex (…G87-T88-G89… : …C204-A203-C202…). The 11-mer centered at the lesion-modified base pair A203* : T88 is highlighted in green.
As the starting structure for the MD simulation, we used the classical intercalation structure of the 14S (−)-trans-anti-DB[a,l]P-N6-dA adduct shown in Figure 1b, which was modeled into a NCP crystal structure that contained a base-stretched segment of nucleosomal DNA (Figures 1d and S3 and Methods, Supporting Information) (PDB (16) ID: 2NZD) (18). This segment, which can serve as an intercalation pocket upon alkylation by an aromatic compound (19), is at the 1.5 superhelix location (SHL) (Figures 1c and 1d) (18). The local duplex sequence context was changed to …GTG…:…CA*C… (Figure 1d), which has been studied by us extensively in the past (reviewed in (20)). This sequence-modified crystal structure also served for the unmodified simulation that was studied as a control and for comparison.
MD simulations for 65-ns at 300 K and 1 atm with explicit solvent and counterions utilizing AMBER 9.0 (21) were carried out as detailed in full in Supporting Information. Figure S3 and Movie S1 show how the DB[a,l]P aromatic rings adapt to the curvature of the DNA in the nucleosome following 65-ns of MD. Details concerning force field parametrization and all analyses are given in Supporting Information.
Our simulation results show that the lesion stabilizes the NCP in comparison with the unmodified NCP (Figure 2) in a number of ways. The overall stacking interaction energy in the intercalation pocket of the lesion-containing NCP is −20.3 ± 2.2 kcal/mol, to which the DB[a,l]P aromatic rings contribute −18.0 kcal/mol (Table S1, Supporting Information). However, for the unmodified NCP, the stacking interaction energy involving the same base pairs is only −13.0 ± 1.7 kcal/mol (Figure 2b and Table S1, Supporting Information). Notably, over half (−9.4 ± 0.7 kcal/mol) of the stabilization provided by the aromatic DB[a,l]P ring system in the modified NCP comes from cross-strand interactions with the thymine partner (T88) to the modified adenine; these interactions act as a “bridge” between the two strands (Figure 2a, Movie S2 and Table S1, Supporting Information). As a result, the Watson-Crick hydrogen bonding at the lesion site in the lesion-containing NCP is more ideal than in the unmodified NCP (Figure 2c). Further details are given in Figure S4 and Table S2, Supporting Information.
Figure 2.
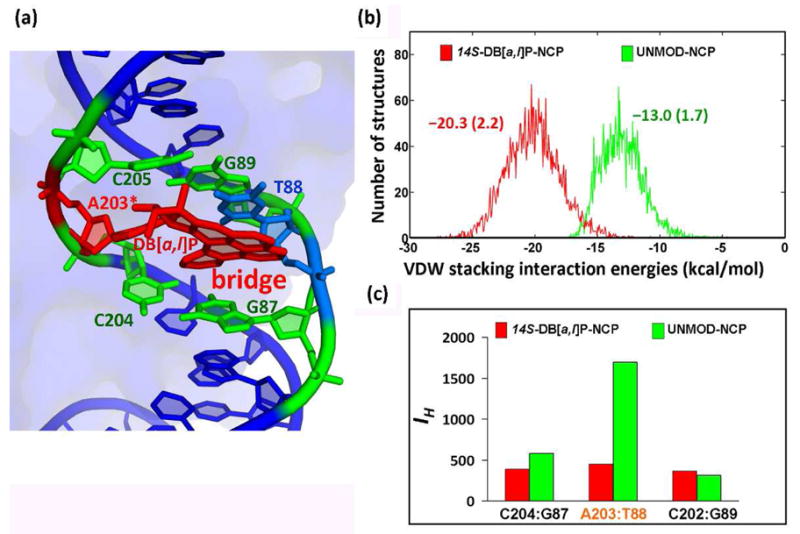
(a) the DB[a,l]P aromatic ring system acts as a “bridge” between the two DNA strands through stacking interactions with base T88 of the partner strand (See also Movie S2, Supporting Information). (b) van der Waals stacking interactions involving the DB[a,l]P aromatic ring system and all bases (A203, T88, A204 and G87) comprising the intercalation pocket, and interactions of the same bases in the unmodified NCP (see also Table S1, Supporting Information). Ensemble average values and standard deviations are given. (c) Hydrogen bond quality index (IH) (See Supporting Information) at designated base pairs in the modified and unmodified NCP showing the better hydrogen bond quality at the lesion site in the modified NCP. The larger IH corresponds to more disturbed hydrogen bonding.
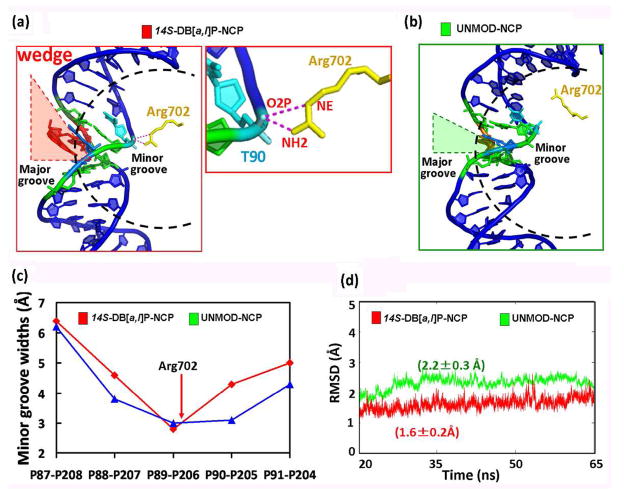
In addition to its stabilizing “bridge” effect (Figure 2), the DB[a,l]P, intercalated from the major groove, acts as a “wedge” (Figure 3a and Movie S4, Supporting Information) which bends the DNA toward the minor groove; this further stabilizes the NCP via newly formed hydrogen bonding interactions between the histone H3 Arg702 and the DNA backbone (Figure 3a). The wedge causes the local minor groove to narrow sharply compared to the unmodified NCP (Figure 3c), producing a more negative electrostatic potential in the groove through electrostatic focusing; this has been shown with many examples involving protein and peptide-DNA interactions (22, 23) including nucleosomes (23). In turn, the more negative potential draws the positively charged Arg702 side chain closer to the backbone to form an additional bifurcated hydrogen bond that stabilizes the lesion-containing NCP (Figure 3a and Table S3, Supporting Information). As a consequence of these stabilizing “bridge” and “wedge” effects induced by the lesion, the dynamics are damped in the lesion-containing NCP as compared to the unmodified NCP. This is shown in the time-dependence of the RMSD which shows both a smaller RMSD as well as a smaller standard deviation in the RMSD for the lesion-containing NCP (Figure 3d and Figure S5, Supporting Information)).
Figure 3.
(a) Wedge induced by the DB[a,l]P aromatic ring system on the major groove side in the modified NCP with a close-up view to highlight the new bifurcated hydrogen bonds between Arg702 and DNA backbone (See also Movies S3 and S4, Supporting Information). (b) the unmodified NCP in similar view. (c) Sharp narrowing of the minor groove at the lesion site in the modified NCP. (d) RMSDs for the local region (within 15 Å) of the damaged residue and its undamaged counterpart (See also Figure S5, Supporting Information).
We have placed the adduct in a stretched position of the nucleosome; this is a hotspot for intercalation and alkylation (19), and this particularly plausible for the intercalative DB[a,l]P adduct. However, the impact of the adduct at other positions in the nucleosome is an area for future investigation.
The stabilizing impact of the lesion on the NCP structure and dynamics manifested in our MD simulations suggests that the lesion may remain repair-resistant in the nucleosome. The nucleosome may be prevented from spontaneously unwrapping (24) for lesion recognition by the NER XPC/hHR23B factor, as suggested from the fluorescence resonance energy transfer (FRET)-observed increased dynamics of UV-damaged nucleosomes; this dynamic effect enhances spontaneous unwrapping of the DNA from the histones (9). Furthermore, the crystal structure of the Rad4/Rad23 (25), yeast homolog of the human XPC/hHR23B NER recognition factor, shows that a β-hairpin inserts between the two strands at the lesion site, and two partner bases in the complementary strand flip out and bind to the protein. Our results suggest that the thymine base partner to the damaged adenine is well-stacked with the DB[a,l]P aromatic ring system; these stabilizing interactions would impede the flipping of the complementary strand bases opposite the lesion out of the helix and thus inhibit their interactions with amino acid residues of the XPC/hHR23B NER recognition factor (25). Consequently, the formation of a stable DNA NER-recognition complex would be inhibited. The stabilizing properties of the lesion in the NCP fit well with the hypothesis that resistance to NER is conferred by lesion-induced local thermodynamic stabilization (4, 7, 25), and that the lack of thermodynamic destabilization is a hallmark of repair resistance (4). In this connection, future experimental NER studies with nucleosomes containing lesions that are repair-resistant in uncomplexed DNA will be of great interest.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants CA-28038 to S.B. and CA-099194 to N.E.G. Computational infrastructure and systems management was partially supported by CA-75449 to S.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supplementary methods, Figures S1–S5, Tables S1–S6 and Movies S1–S4 are provided. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gillet LC, Scharer OD. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 2.Luch A. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 3.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 4.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 5.Geacintov NE, Naegeli H, Patel DJ, Broyde S. In: DNA Damage and Recognition. Siede W, Kow YW, Doetsch PW, editors. Taylor & Francis; London: 2006. [Google Scholar]
- 6.Gunz D, Hess MT, Naegeli H. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 7.Scharer OD. Mol Cell. 2007;28:184–186. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Hara R, Mo J, Sancar A. Mol Cell Biol. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan MR, Smerdon MJ. J Biol Chem. 2010;285:26295–26303. doi: 10.1074/jbc.M110.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin S, Krzeminski J, Rivenson A, Kurtzke C, Hecht SS, el-Bayoumy K. Carcinogenesis. 1995;16:1971–1974. doi: 10.1093/carcin/16.8.1971. [DOI] [PubMed] [Google Scholar]
- 11.Luch A. Exs. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 12.Yoon JH, Besaratinia A, Feng Z, Tang MS, Amin S, Luch A, Pfeifer GP. Cancer Res. 2004;64:7321–7328. doi: 10.1158/0008-5472.CAN-04-1094. [DOI] [PubMed] [Google Scholar]
- 13.Kropachev K, Kolbanovskiy M, Rodriguez FA, Cai Y, Ding S, Zhang L, Amin S, Broyde S, Geacintov NE. American Chemical Society 238th National Meeting & Exposition. 2009;23:282. [Google Scholar]
- 14.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Chem Res Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 15.Cai Y, Ding S, Kropachev K, Schwaid A, Kolbanovksiy M, Rodriguez FA, Amin S, Geacintov NE, Broyde S. American Chemical Society 238th National Meeting & Exposition; 2009. pp. 285–286. [Google Scholar]
- 16.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong MS, Richmond TJ, Davey CA. J Mol Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 19.Davey GE, Wu B, Dong Y, Surana U, Davey CA. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Geacintov NE, Broyde S. In: The Chemical Biology of DNA Damage. NEG, SB, editors. Wiley-VCH; 2010. pp. 261–298. [Google Scholar]
- 21.Case DA, Darden TA, Cheatham III TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco, CA: 2006. [Google Scholar]
- 22.Huang H, Kozekov ID, Kozekova A, Rizzo CJ, McCullough AK, Lloyd RS, Stone MP. Biochemistry. 2010;49:6155–6164. doi: 10.1021/bi100364f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buning R, van Noort J. Biochimie. 2010 doi: 10.1016/j.biochi.2010.08.010. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 25.Min JH, Pavletich NP. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.