Abstract
Cap’n’collar (Cnc) transcription factors are conserved in metazoans and have important developmental and homeostatic functions. The vertebrate Nrf1, Nrf2, and Nrf3, the Caenorhabditis elegans SKN-1, and the Drosophila CncC comprise a subgroup of Cnc factors that mediate adaptive responses to cellular stress. The most studied stress-activated Cnc factor is Nrf2, which orchestrates the transcriptional response of cells to oxidative stressors and electrophilic xenobiotics. In rodent models, signaling by Nrf2 defends against oxidative stress and aging-associated disorders, such as neurodegeneration, respiratory diseases, and cancer. In humans, polymorphisms that decrease Nrf2 abundance have been associated with various pathologies of the skin, respiratory system, and digestive tract. In addition to preventing disease in rodents and humans, Cnc factors have lifespan-extending and anti-aging functions in invertebrates. However, despite the pro-longevity and antioxidant roles of stress-activated Cnc factors, their activity paradoxically declines in aging model organisms and in humans suffering from progressing respiratory disease or neurodegeneration. We review the roles and regulation of stress-activated Cnc factors across species, present all reported instances in which their activity is paradoxically decreased in aging and disease, and discuss the possibility that the pharmacological restoration of Nrf2 signaling may be useful in the prevention and treatment of age-related diseases.
The Cnc Family of Transcription Factors
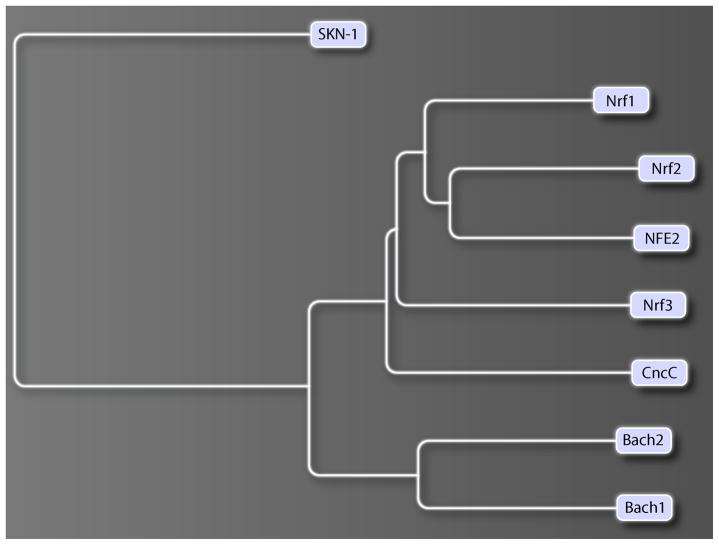
Cap’n’collar (Cnc) proteins are a family of basic leucine zipper transcription factors that are conserved in worms, insects, fish, birds, and mammals, including humans, but are not present in plants or fungi. They are defined by the presence of a conserved 43-amino acid Cnc domain located N-terminally to the DNA binding domain. Cnc transcription factors comprise the Caenorhabditis elegans SKN-1 (Skinhead family member 1) (1), the Drosophila Cnc (2), and four vertebrate counterparts-- the p45 NFE2 (nuclear factor erythroid-derived 2) and the NFE2-related factors Nrf1, Nrf2 and Nrf3 (hereafter referred to as “the Nrfs”) (Fig. 1) (3–10). The related vertebrate transcription factors Bach1 (BTB and CNC homology 1) and Bach2 are characterized by the additional presence of a BTB (Broad complex, Tramtrack, Bric-a-brac) protein interaction domain (11). Most Cnc factors are transcriptional activators; however, Bach 1, Bach2, a naturally occurring truncated isoform of Nrf1, and a caspase-cleaved form of Nrf2 function as transcriptional repressors (11–16). One study has suggested that Bach1 can activate transcription in erythroid cells (11).
Fig. 1. A phylogenetic tree of the Cnc and Bach transcription factors.
A multiple species alignment was constructed using ClustalW (www.ebi.ac.uk/clustalw) and a phylogenetic tree was generated using the Jalview applet. The tree was based on the largest gap-free block of aligned sequences, which contained the DNA binding domains of the Cnc and Bach factors. Accession numbers were: mouse p45 NFE2, NP_032711.2; mouse Nrf1, NP_032712.2; mouse Nrf2, NP_035032.1; mouse Nrf3, NP_035033.1; mouse Bach1, NP_031546.1; mouse Bach2, NP_001103131.1; Drosophila CncC, NP_732833.1; C.elegans SKN-1, NP_741404.1.
Cnc proteins function during development, or contribute to the maintenance of homeostasis in response to some types of environmental stress, or both. Worms and flies have a single gene encoding their respective Cnc factors, which function both in development and in stress responses. In contrast, each Cnc protein is encoded by a separate gene in vertebrates. Though Bach1 is expressed ubiquitously and is abundant in hematopoietic organs, such as bone marrow and fetal liver (11, 17), it is dispensable for development and reproduction in mice (18). Bach2 is most abundant in the brain and in the B-cell lineage where it is required for antibody class switching (15, 19). The p45 NFE2 functions in development; it is present only in hematopoietic progenitor, erythroid, megakaryocytic, and mast cells, and it is required for proper development of platelets (4, 20, 21). The three Nrfs have broad and partly overlapping expression patterns and function as stress-activated transcription factors (5, 7, 9, 22). We focus on the vertebrate stress-activated Nrfs and their invertebrate homologs, and discuss their conserved roles in the cellular response to oxidative stress, with particular emphasis on their paradoxical dysfunction in aging and human disease.
Cnc Transcription Factors as Mediators of the Antioxidant Response
Cells experience oxidative stress when pro-oxidant and electrophilic reactive species overwhelm the cell’s antioxidant and detoxification proteins (23). In addition to causing protein and lipid damage, oxidative stress can cause mutations and epigenetic perturbation by damaging DNA and proteins that modify chromatin. Therefore, oxidative stress can be a causative or exacerbating factor in a range of diseases, including respiratory and metabolic disorders, neurodegenerative diseases, and cancer (24, 25). Cells possess signaling pathways that can sense oxidative stress and launch adaptive responses that bolster the antioxidant defense networks (26). Nrf2 is the central mediator of a prominent antioxidant response system (27, 28). In response to oxidative stress, Nrf2 accumulates in the nucleus, where it binds to Antioxidant Response Element (ARE) sequences in the regulatory sequences of its target genes. Nrf2 activates transcription primarily as a dimer with a member of the small Maf (musculo-aponeurotic fibrosarcoma oncogene) family of proteins (29–31). Binding of the small Maf-Nrf2 dimers to ARE sequences results in the coordinated transcriptional activation of a battery of antioxidant enzymes and detoxifying proteins. This regulated adaptive response has been termed “the electrophile counterattack”, and is also known as the “Phase 2 detoxification response” (32). Activation of Nrf2 increases the abundance of thioredoxins and glutathione-synthesizing enzymes (which maintain the redox balance), glutathione S-transferases (which detoxify xenobiotics), molecular chaperones (which facilitate optimal protein folding), proteasome subunits (which remove damaged macromolecules) and various other cytoprotective proteins (33–36). In the absence of oxidative stress, the basal activity of Nrf2 maintains the housekeeping expression of many of the same antioxidant and detoxification genes (27, 28, 37, 38). Thus, Nrf2 controls the basal and inducible activity of the cell’s antioxidant defenses.
Regulation of Nrf2 activity and ARE-mediated transcription
The understanding of the molecular mechanisms by which ARE activity is regulated continues to evolve (39–42). In the absence of oxidative stress, Nrf2 binds to its cytoplasmic inhibitor Keap1 (Kelch-like ECH-associating protein 1), a protein tethered to the actin cytoskeleton (27, 43–45). Keap1 suppresses the activity of Nrf2 both passively by sequestering it in the cytoplasm, as well as actively by targeting it for polyubiquitination by a Cullin3-based E3 ligase complex and thus facilitating its proteasomal degradation (46–49) (Fig. 2). In addition to serving as an inhibitor of Nrf2, Keap1 also functions as a sensor of oxidants and electrophiles, which react with its redox-sensitive cysteine residues (50–52). Oxidative stresses or electrophilic xenobiotics abolish the inhibition of Nrf2 by Keap1. Although these inducers do not disassociate Nrf2 from Keap1, they impair the ability of Keap1 to target Nrf2 for ubiquitination and degradation, presumably by triggering a conformational change in Keap1 structure (53–55). The “hinge and latch” structural model for the Keap1-Nrf2 association proposes that a single Nrf2 molecule makes contacts of different strength with the two molecules of a Keap1 dimer (Fig. 3A) (54, 56). The “hinge” is a high affinity interaction that is not affected by inducers; in contrast, inducers abolish the low-affinity interaction mediated by the “latch,” thereby disrupting the presentation of Nrf2 to the ubiquitination machinery (57). The “hinge and latch” model was originally based on in vitro biochemical and structural evidence using purified peptides (53, 56) and this model is supported by the finding that cancer-associated somatic mutations specifically altering amino acids in either the DLG or the ETGE motif result in aberrant cellular accumulation of Nrf2 (58). The genetic data suggest that the interaction mediated by the DLG is relevant in vivo, even though in vitro it is two orders of magnitude weaker than the ETGE interaction (59). Further supporting the “hinge and latch” model, it was shown that the p21 protein directly interacts with the DLG and ETGE motifs and thus competes with Keap1 for Nrf2 binding (60). On the other hand, cell culture studies using forced expression of differentially tagged or mutated forms of Keap1 and Nrf2 have concluded that the stoichiometry of the Keap1-Nrf2 association in cells is not 2:1 but is rather 2:2 (61). In addition, Keap1 binds simultaneously one Nrf2 molecule and one molecule of phosphoglycerate mutase 5 (PGAM5) (62, 63) (Fig. 3B), which also conflicts with the “hinge and latch model”. Thus, it remains to be resolved which of the two models best represents the interaction between Keap1 and Nrf2 in vivo. However, it should be noted that the two models are not necessarily mutually exclusive, because each might be valid for different Keap1 dimers or higher order multimers that Keap1 might potentially form. The transcriptional activation of genes through the ARE by oxidative stress is regulated at multiple levels, from transcription to protein localization to protein degradation and posttranslational modifications. The shuttling of Nrf2 into and out of the nucleus has been linked to the redox status of the cell. Nrf2 possesses both nuclear localization signals (NLSs) and nuclear export signals (NESs), which are balanced under basal conditions (64–67). Oxidative conditions inactivate one of the Nrf2 NESs through modification of a reactive cysteine, which facilitates the nuclear accumulation of Nrf2 (67). Thus, the oxidative stress-sensing function of Keap1 may have a permissive role for Nrf2 activation, whereas the direct effects of oxidative stress on Nrf2 itself may fine tune the speed, magnitude, and duration of the antioxidant response (40). Keap1 also exhibits nucleocytoplasmic shuttling (68, 69), although the physiological relevance of this observation has been challenged (70). Nuclear localization of Keap1 might serve to target Nrf2 to specific chromatin areas through the Keap1 BTB protein interaction domain (40, 71). Alternatively, it may promote the degradation of Nrf2 inside the nucleus or facilitate the nuclear export of Nrf2, or both, thereby terminating the antioxidant response after the redox balance has been restored (40, 69, 72).
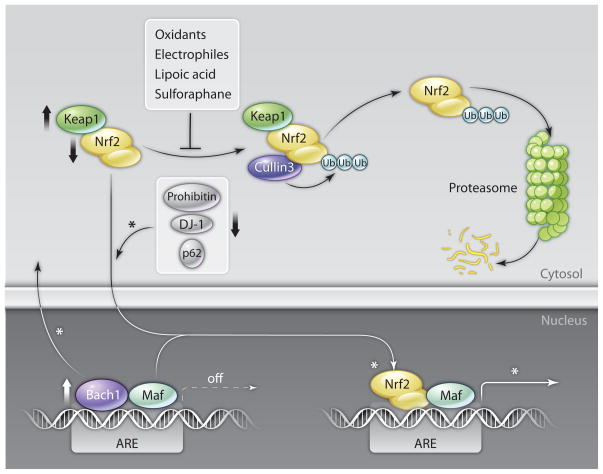
Fig. 2. Regulation of the Keap1-Nrf2 antioxidant response pathway.
Signaling through the Keap1-Nrf2 antioxidant response pathway is shown in a simplified manner. In basal conditions, Nrf2 is targeted by Keap1 for Cullin3-mediated polyubiquitination and proteasomal degradation. Phase 2 genes are mostly inactive and Antioxidant Response Elements (AREs) in their enhancers are largely bound by small Maf dimers or other repressive factors like Bach1. Oxidants, electrophiles, cancer chemopreventive agents, and other “inducers” impair Nrf2 degradation and facilitate the nuclear accumulation of Nrf2. The activation of Nrf2 is further promoted by positive pathway modulators like DJ-1, p62, and prohibitin. As a result, antioxidant response genes are transcriptionally induced in a coordinated manner through Nrf2-small Maf dimers bound to their AREs, the redox balance is restored, and oxidative damage is minimized. Chronic oxidative stress associated with aging or degenerative diseases handicaps the expression or function (↓) of critical Nrf2 pathway modulators, or increases the abundance (↑) of negative regulators like Keap1 and Bach1. Nrf2 proteolysis is increased, while several other steps (*) of the pathway are reduced. As a result, Nrf2 activity is compromised despite the presence of oxidative conditions. Nrf2-stimulating compounds, like sulforaphane and lipoic acid, induce Nrf2 activity and restore the Cnc antioxidant response.
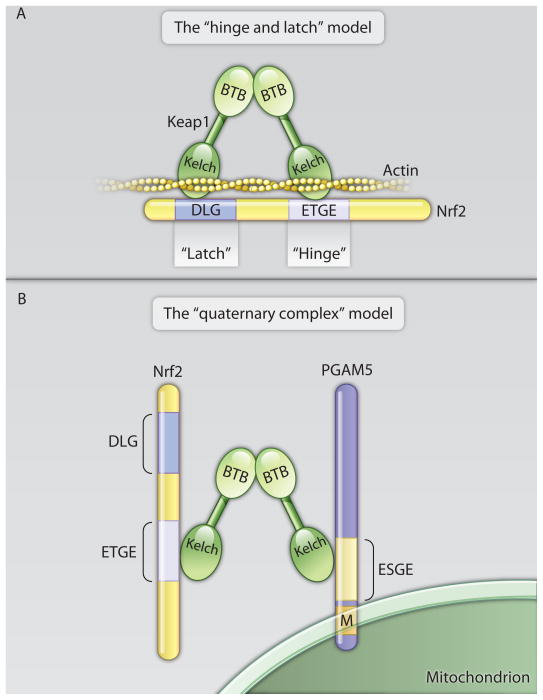
Fig. 3. Two models for the Keap1-Nrf2 interaction.
The molecules are not drawn to scale. (A) Keap1 molecules form dimers through their BTB domains. The “hinge and latch” model proposes that a Keap1 dimer binds a single Nrf2 molecule through high and low affinity interactions with the Neh2 domain’s ETGE (“hinge”) and DLG (“latch”) motifs, respectively. In addition to Nrf2, the Keap1 Kelch domain binds actin, thus tethering the Keap1-Nrf2 complex to the cytoskeleton. Oxidative stress impairs Nrf2 ubiquitination and proteasomal degradation by abolishing only the low affinity “latch” interaction. (B) The “quaternary complex” model proposes that a Keap1 dimer binds two molecules of substrate(s) through high affinity interactions with E(S/T)GE motifs. A Keap1 dimer can bind one Nrf2 molecule and one PGAM5 molecule as shown here, or two Nrf2 molecules (not shown). PGAM5 possesses an N-terminal membrane targeting signal (M), through which the Nrf2-Keap1-PGAM complex is tethered to the cytosolic surface of the outer mitochondrial membrane. Oxidative stress impairs Nrf2 degradation without abolishing the high affinity interaction.
Phosphorylation also contributes to the cellular response to oxidative stress, by targeting Keap1, Nrf2, other leucine zipper proteins, or their transcriptional cofactors, and thus directly or indirectly activating or repressing ARE activity. The mitogen-activated protein kinases (MAPKs) p38, ERK, and JNK, as well as protein kinase C (PKC), glycogen synthase kinase 3β (GSK-3β), the tyrosine kinase Fyn, casein kinase 2 (CK-2), phosphoinositide 3-kinase (PI3K), and protein kinase-like endoplasmic reticulum kinase (PERK) have all been implicated in ARE regulation (73–94). Mostly, the phosphorylation targets of these kinases that are relevant for the response to oxidative stress remain poorly characterized, with the exceptions of PKC, Fyn, and the MAPKs, which phosphorylate specific Nrf2 residues (81–83, 88, 92). Different kinase pathways are involved in ARE regulation in a manner that is both inducer- and tissue-type specific. For example, p38 has been found to either positively or negatively regulate the Nrf2-mediated transcriptional induction of the gene encoding the antioxidant enzyme heme oxygenase 1 (HO-1) (76, 95–97).
The abundance of Keap1, Nrf2, small Mafs, Bach1, Bach2, and factors with which these proteins interact is also regulated and may influence the cellular sensitivity to oxidative stress. The genes encoding Keap1, Nrf2, and MafG (a small Maf protein) contain the ARE and thus their expression is stimulated by Nrf2 (98–100); the nrf2 gene is also transcriptionally induced by the aryl hydrocarbon receptor (101). Moreover, oxidative stress stimulates the translation of Nrf2, increasing its abundance (102). In response to ARE inducers, Bach1 undergoes rapid nuclear export and proteasomal degradation (103, 104), and bach1 gene transcription is also stimulated, such that Bach1 abundance can be ultimately restored (104). Conversely, oxidative stress induces nuclear accumulation of Bach2, which suppresses ARE activity and promotes apoptosis (105, 106). The chromatin remodeling factor BRG1 and the transcriptional repressor SMRT can interact with Nrf2 and enhance or silence ARE activity, respectively (107, 108). Various other transcription factors interact with Nrf2, the ARE, or both, including the activator protein 1 (AP-1) transcription factors Fos and Jun, the activating transcription factors 1, 2, 3 and 4 (ATF-1, ATF-2, ATF-3 and ATF-4), the estrogen receptors α and β (ERα and ERβ), the peroxisome proliferator-activated receptor γ (PPAR), and the retinoic acid receptor α (RARα) (109–120). The relative cellular abundance and activity of these ARE-regulating proteins may fine tune transcriptional responses mediated by Cnc factors (13, 121–124). In addition to phosphorylation, acetylation and regulated cleavage influence Nrf2 activity. The posttranslational acetylation of Nrf2 by its transcriptional co-activator p300/CBP promotes its DNA binding to specific ARE promoters (125). The caspase-mediated removal of its N-terminal transactivation domains converts Nrf2 to an apoptosis-promoting ARE repressor (16). Thus, molecular processes regulating ARE activity can be broadly classified into ubiquitous mechanisms that involve the oxidative stress-sensing functions of Keap1 and Nrf2, and tissue-specific mechanisms that involve (i) the abundance of factors that interact with Nrfs or the ARE and (ii) the posttranslational modification of proteins involved in mediating the antioxidant response.
Functions and Regulation of Other Vertebrate Cnc Proteins
In addition to Nrf2, Nrf1 and Nrf3 also stimulate transcription of ARE-containing genes (126, 127). In contrast, the Bach factors repress ARE activity (11, 12, 128), as do dimers of small Maf proteins (121, 122) (Fig. 2). Whereas full-length Nrf1 activates the ARE, a naturally occurring truncated Nrf1 isoform functions as a transcriptional repressor (14). Because the genes encoding Keap1, Nrf2, and MafG can be induced through AREs in their regulatory sequences (98–100), transcriptional cross-regulation among the Cnc proteins might also occur. Thus, for example, it is possible that Nrf1 might contribute to Nrf2 abundance by inducing ARE-mediated nrf2 gene transcription, or Nrf1 might reduce Nrf2 abundance by inducing ARE-mediated keap1 gene transcription. Although all Nrfs can regulate the ARE, the distinct phenotypes of the corresponding mouse knockouts indicate that they are not functionally redundant (Table 1). Whereas nrf1−/− mice die during mid- to late embryonic development due to a non-cell autonomous defect in liver erythropoiesis (129), Nrf2 is not required for development (130). Instead, aged nrf2−/− mice develop a neurodegenerative disorder characterized by destruction of the myelin sheath in the nervous system, which is accompanied by the widespread presence of reactive astroglia and is presumably caused by oxidative stress (131). In addition, aged female nrf2−/− mice develop a multi-organ autoimmune disease similar to human Systemic Lupus Erythematosus (132–134). Mice lacking Nrf2 are also highly sensitive to environmental stressors. nrf3−/− mice are also viable and have no obvious phenotypes under nonstressful conditions (22).
Table 1.
Summary of the mouse Cnc knockout phenotypes.
| Genotype | Phenotype | Reference |
|---|---|---|
| nrf1−/− | Mid- to late embryonic lethal; defective erythropoiesis; oxidative stress | (129) |
| nrf2−/− | Viable and fertile; age-related lupus-like syndrome (♀) and neurodegeneration (♀ and ♂); sensitivity to oxidative and ER stress | (130–134) |
| nrf1−/−, nrf2−/− | Enhancement of nrf1−/− phenotypes: early embryonic lethal; increased oxidative stress | (136) |
| p45−/− | Viable; mild anemia; thrombocytopenia (lack of platelets); death from hemorrhage | (20, 137) |
| p45−/−, nrf2−/− | No synthetic lethality; mild enhancement of p45−/− phenotype (thrombocytopenia) | (138–140) |
| nrf3−/− | No obvious phenotype | (22) |
| nrf3−/−, nrf2−/− | Viable and fertile | (22) |
| nrf3−/−, p45−/− | No synthetic lethality | (22) |
| nrf3−/−, nrf2−/−, p45−/− | No synthetic lethality | (22) |
Examination of double and triple knockout mice further supports that members of the Cnc family in mammals have mostly nonredundant functions (Table 1). Whereas nrf2−/− mice are born normally (130) and nrf1−/− mice suffer from oxidative stress (135) and die in late embryonic development (129), nrf1−/− nrf2−/− mice die in early development and their cells show elevated amounts of intracellular oxygen species and an increased rate of cell death compared to cells from either nrf2−/− or nrf1−/− mice (136). Thus, although Nrf2 does not rescue the lethality of Nrf1 deficiency, Nrf2 does partially ameliorate the nrf1−/− phenotype, indicating that Nrf2 can partially compensate for the absence of Nrf1. (136). Thus far, there is no genetic evidence in support of a similar functional overlap between Nrf3 and the other Cnc factors. The lack of Nrf3 did not cause lethality in nrf2−/−, p45−/−, or nrf2−/− p45−/− mice, and no changes in the transcript levels of p45, nrf1, and nrf2 were detected in nrf3−/− mice (22).
The phenotypic differences in the mouse knockouts, which are consistent with functional specificity in the Nrf family, are unlikely to relate to tissue-specific expression, because their expression patters are largely overlapping. Rather, Nrfs differ in their modes of regulation, subcellular localizations, target gene complements, and transactivation potencies (Table 2). The Neh2 (Nrf2-ECH homology 2) domain, which mediates the interaction with the Kelch domain of Keap1 (10), is not conserved in Nrf3 (127). Although Nrf1 has a Neh2-like domain, Nrf1 is not regulated by Keap1 (141, 142). Nrf2 is soluble and localized mostly to the cytoplasm and partly to the mitochondria under basal conditions, whereas Nrf1 and Nrf3 are integral membrane proteins targeted to the endoplasmic reticulum (ER) through a conserved NHB1 (N-terminal homology box 1) domain (141, 143–145). The precise mechanisms by which Nrf1 and Nrf3 shuttle from the ER to the nucleus to regulate transcription are not yet well understood. What is known is that ER stress (caused by the accumulation of misfolded proteins in the ER lumen) activates Nrf3 but suppresses Nrf1 activity (145, 146), and that Nrf2 is also induced by ER stress through activation of the ER transmembrane kinase PERK (86, 147). Although Nrf1 and Nrf2 regulate many of the same genes, each of these factors also has unique transcriptional targets (148), as does Nrf3 (149). Nrf2 is a more potent transcriptional activator than either Nrf1 or Nrf3 (9), which may relate to the membrane association of Nrf1 and Nrf3 or the presence of two transcription activation domains in Nrf2, or both (150).
Table 2.
Characteristics of Cnc and Bach transcription factors.
| Family member | Expression regulated through ARE | Inhibition by Keap1 | Dimerization with small Maf | Soluble or TM | Localization under basal conditions | Effect of ER stress | Effect of oxidative stress | Transcriptional activity | Function in development |
|---|---|---|---|---|---|---|---|---|---|
| p45 NFE2 | Not known | No | yes | Soluble | Nucleus of hematopoietic cells | Not known | Not known | Activator | Megakar-yopoiesis in mouse |
| Nrf1 | Not known | No | yes | TM | ER | Inhibition | Activation | Activator | Erythropoiesis in mouse |
| Nrf2 | Yes | Yes | Yes | Soluble | Cytosol and mitochondria | Activation | Activation | Activator | Not required |
| Nrf3 | Not known | No | Yes | TM | ER | Activation | Activation | Activator | Not required |
| Nrf1 truncated isoform | Derived from same transcript as full-length Nrf1 | No | Yes | Soluble | Nucleus | Not known | None | Repressor; lacks transcripttion activation domain | None known |
| Caspase-cleaved Nrf2 | Cleavage fragment of full length Nrf2 | No | Not known; retains bZip domain | Soluble | Nucleus; formed during apoptosis | Not known | Not known | Repressor; lacks transcripttion activation domain | None known |
| Bach1 | Yes | No | Yes | Soluble | Nucleus | Not known | Nuclear export | Repressor (Activator?) | Not required |
| Bach 2 | Not known | No | Yes | Soluble | Cytoplasm | Not known | Nuclear import | Repressor | Not required |
| SKN-1 | Not known | No | No | Soluble | Cytoplasm | Not known | Activation | Activator | Digestive system development of worm |
| CncA | Not known | No | Not known | Soluble | Not known | Not known | Not known | None known; lacks transcripttion activation domain | None known |
| CncB | Not known | No | Yes | Soluble | Nucleus | Not known | Not known | Activator | Labral and mandibular segments of fly head |
| CncC | Not known | Yes | Not known | Not known | Not known | Not known | Activation | Activator | Required for viability; function unknown |
ARE, antioxidant response element; TM, transmembrane; ER, endoplasmic reticulum
Invertebrate Cnc Factors
Oxidative stress poses a threat to all aerobic species, thus it is not surprising that Cnc transcription factors regulate antioxidant responses in invertebrate organisms (Fig. 1). The C. elegans Cnc homolog is SKN-1, a protein initially characterized as critical to the formation of the digestive system during worm embryogenesis (1). In addition to this early developmental function, SKN-1 regulates a Phase 2 detoxification response in the digestive tract of larvae and adults, thereby conferring resistance against various pro-oxidants, such as hyperbaric oxygen, hydrogen peroxide, paraquat, arsenic, and juglone (151, 152).
The regulation of SKN-1 and its mechanism of action are divergent from that of vertebrate Nrf2. The worm does not possess Keap1 and small Maf homologs, and SKN-1 binds ARE half-sites as a monomer (153). Phosphorylation by diverse protein kinases is a central mechanism of SKN-1 regulation. In the absence of stress, GSK-3, the serum/glucocorticoid regulated kinase 1 (SGK-1), and the serine-threonine kinases AKT-1 and AKT-2 are all needed to repress SKN-1 activity by preventing its nuclear accumulation (154, 155). In contrast, the kinases p38, MAP kinase kinase 4 (MKK-4), inhibitor of nuclear factor κB kinase ε-1 (IKKε-1), never in mitosis kinase like 2 (NEKL-2), and pyruvate dehydrogenase kinase 2 (PDHK-2) all contribute to the activation of SKN-1 in response to stress (156, 157). The abundance SKN-1 is also controlled by the cell’s protein degradation machinery: Proteasome core and regulatory subunits, ubiquitin-specific hydrolases, and chaperonin components repress SKN-1 activity under normal conditions (152). In the nuclei of intestinal cells, SKN-1 specifically interacts with the WD40 repeat protein WDR-23, which in turn binds to a Cullin4-based ubiquitin ligase complex and presumably targets SKN-1 for ubiquitination and proteasomal degradation (158). Under conditions of oxidative stress, SKN-1 escapes from WDR-23-mediated inhibition, accumulates in the nucleus, and transactivates its target genes (158). Although the Cnc family of transcription factors was named after the Drosophila Cnc protein nearly two decades ago (2), the functional homology of Drosophila Cnc to the C. elegans SKN-1 and to the vertebrate Nrfs was only recently demonstrated experimentally (159). In flies, the cnc locus encodes three protein isoforms: CncA, CncB, and CncC, with CncC being the longest protein and CncB and CncA representing progressively truncated forms derived from different transcripts (160). CncB is active during embryogenesis in the labral and mandibular segments that develop into regions of the fly’s head. Thus, the embryonic expression pattern of CncB resembles a cap (labral segment) and a collar (mandibular segment), hence the name of the protein family (2, 160, 161). Like the truncated p65 isoform of Nrf1 and the caspase-cleaved form of Nrf2, Drosophila CncA lacks transcription activation domains (160, 162); it is yet unknown whether CncA functions as a transcriptional repressor like the short forms of Nrf1 and Nrf2.
CncC, the longest of the Drosophila Cnc proteins, is present from late embryogenesis through adult life. Unlike CncA or CncB, CncC contains a unique N-terminus with homology to the Keap1-binding domain of Nrf2 (10, 160). Furthermore, a Keap1 ortholog is present in Drosophila and functions as an inhibitor of CncC and ARE activity (159). Like SKN-1 and Nrf2, CncC is activated by various oxidants, such as paraquat, diethyl-maleate, and hydrogen peroxide. CncC is also induced by compounds, like oltipraz, that have cancer chemopreventive properties in rodents, and its activation augments the fly’s antioxidant defense systems (159). The N-terminus of CncC contains a stretch of amino acids that is homologous to the NHB1 domains of Nrf1 and Nrf3, through which these vertebrate factors are targeted to the ER (141, 142, 145). Whether CncC also localizes to the ER compartment and responds to ER stress or to oxidative stress in the ER has not been investigated, but the structural homology suggests that CncC may perform the functions of not only Nrf2, but also those of Nrf1 and/or Nrf3. Thus, Drosophila appears to encode in a single locus all of the diversified functions of Cnc proteins: CncB would represent the developmental effector that is not subjected to Keap1-mediated inhibition (reminiscent of p45 NFE2); CncC represents a stress-responsive homeostatic mediator (like the Nrfs); and CncA is a putative competitive repressor of CncB or CncC binding to the ARE (functionally analogous to p65 Nrf1, caspase-cleaved Nrf2, or the Bach factors). More work is needed to further characterize the functional correspondence between the invertebrate Cnc and SKN-1 isoforms and the vertebrate Cnc factors.
Activation and Paradoxical Dysregulation of Nrf2 in Disease
It has been appreciated for several years that the electrophile counterattack rallied by Nrf2 might be exploited as a strategy for the chemoprevention of cancer (163, 164). The rationale is that the administration of drugs that augment the cellular defense against electrophilic carcinogens could potentially prevent, ameliorate, or even reverse the tissue damage these chemicals cause (165). Various compounds that activate Nrf2 are regarded as promising chemopreventive agents, and some of these substances are being studied in human trials of cancer chemoprevention. Such Nrf2-activating drugs include synthetic compounds like oltipraz and related dithiolthiones (166) and novel triterpenoids (167), as well as natural products like curcumin (168) and sulforaphane (169). These substances are effective in mouse models of experimental carcinogenesis, and their chemopreventive actions are markedly reduced or abolished in nrf2 knockouts, indicating that their activities are mediated by the induction of Nrf2 (168, 170–172). Consistent with the role of Nrf2 as a central regulator of the adaptive response to oxidative stress, nrf2−/− mice are sensitive to diverse oxidative insults. When exposed to electrophilic xenobiotics or chemicals that generate intracellular oxidative stress, nrf2−/− mice display increased tissue damage and prolonged inflammation, high amounts of DNA, lipid, and protein oxidation, and increased incidence of cancers (171, 173–175).
Oxidative stress is linked not only to cancer but also to various nonmalignant diseases. Primarily on the basis of experiments with nrf2−/− mice and cells derived from them, there is additional interest in pharmacological manipulation of the Nrf2 system beyond cancer chemoprevention. For example, nrf2−/− mice are sensitive to experimental models of pulmonary diseases, such as asthma (176), emphysema (33), pulmonary fibrosis (177), hyperoxia (178), and acute lung injury (179). The protective role of Nrf2 has also been established in models of neurodegenerative disorders, such as Parkinson’s disease (180), Alzheimer’s disease (181), and Amyotrophic Lateral Sclerosis (182); inflammatory disorders, such as inflammatory bowel disease (183); models of liver toxicity including toxin- and alcohol-induced liver damage (184, 185); atherosclerosis (186); and insulin resistance (187), among others (28, 188). Thus, Nrf2 appears to have a broad protective function that could potentially be exploited therapeutically for the prevention or treatment of many diseases (28, 189, 190).
Analysis of disease-associated DNA polymorphisms in humans also supports a protective role of Nrf2 against multiple human diseases. Inherited DNA polymorphisms that reduce the abundance of Nrf2 (191, 192) have been associated with skin vitiligo (193), chronic gastritis (194), peptic ulcer (195), ulcerative colitis (196), and adult respiratory distress syndrome (192). Notably, the affected organs are ones through which the body comes into contact with environmental stressors. Strikingly, the allelic frequencies of the disease-associated polymorphisms are as high as 20% in Europeans, 40% in Asians, and 55% in Native Americans (192). Thus, there are many individuals in the general population who may have a hereditary predisposition to a reduced antioxidant response and can be at increased risk for diseases related to oxidative stress. Those subjects might benefit the most from pharmacological stimulation of Nrf2.
Extending the focus on Nrf2 signaling from a protective mechanism for cancer chemoprevention to a candidate drug target for numerous other clinical indications raises the question whether the pharmacological induction of Nrf2 will prove to be a practicable and efficacious strategy for the treatment of diseases related to oxidative stress even subsequent to their clinical manifestation. It could be argued that Nrf2 activation should not be expected to be therapeutically useful: In a disease caused or exacerbated by oxidative stress, the stress-responsive Nrf2 may be already activated fully during disease pathogenesis and progression. Thus, if the disease can progress despite the endogenous induction of the antioxidant response, then the exogenous administration of compounds that stimulate Nrf2 may have little added value. In stark contract to the large literature on the protective role of Nrf2 in experimental mouse models, few human studies have yet directly examined its involvement in disease. To date, the activity of the Nrf2 pathway in human disease has been investigated only in cancer, respiratory disease, neurodegeneration, and preeclampsia. Thus far, the data support the notion that the progression of nonmalignant human diseases related to oxidative stress can be associated with, and likely promoted by, paradoxically deficient Nrf2 function. Thus, the pharmacological restoration of Nrf2 activity might have not only preventive, but indeed also therapeutic applications in human diseases.
Nrf2 suppression in Chronic Obstructive Pulmonary Disease
Chronic Obstructive Pulmonary Disease (COPD) is a disorder of the lungs in which the airways become narrowed, leading to reduced air flow that causes shortness of breath (197). In contrast to asthma, the limitation of air flow is not fully reversible, and in most cases it worsens gradually over time. COPD is associated with an abnormal inflammatory response of the lung to noxious particles or gases, the most common risk factor being chronic cigarette smoking (198). The clinical presentation of the disease ranges from chronic bronchitis, which results from the inflammatory response in the larger airways, to emphysema, the destruction of lung tissue caused by the inflammatory response in the alveoli. The precise mechanism of the pathogenesis of emphysema is not clear, but it involves the recruitment and potent activation of alveolar macrophages, and it is also associated with an imbalance in the relative abundance of proteases versus antiproteases and oxidants versus antioxidants in the lung; namely, oxidative stress (199, 200).
Consistent with the involvement of oxidative stress in COPD, Nrf2 is protective in two different mouse models of the disease, in which emphysema is experimentally induced by exposure of the lung to neutrophil elastase or cigarette smoke. nrf2−/− mice were much more susceptible to elastase- or smoke-induced emphysema than their wild-type counterparts (33, 201). The cellular mechanism by which Nrf2 activity protects against COPD is not fully known, but macrophages have an important role. Transplantation of wild-type bone marrow cells with an intact nrf2 locus into nrf2−/− mice instilled with elastase retarded the development of the initial lung inflammation and subsequent emphysema (201). This improvement was associated with the appearance of macrophages expressing Nrf2-regulated genes encoding antiproteases and antioxidant proteins in the lung, which suggests that Nrf2 signaling in pulmonary macrophages has a crucial role for the prevention of emphysema.
Another study with the cigarette smoke-induced mouse model of COPD demonstrated that the induction of Nrf2 activity during the course of emphysema development is not only pharmacologically feasible but also beneficial (202). This work confirmed that the alveolar destruction, declining lung function, and severe emphysema caused by chronic exposure to cigarette smoke are significantly more pronounced in nrf2−/− mice than in wild-type animals. At the cellular level, tobacco smoke exposure causes protein oxidation and leads to cell death, and these effects were more dramatic in the lungs of nrf2−/− mice than in wild-type tissue (202). Despite the clearly protective function of Nrf2 and ARE-regulated antioxidant genes, the endogenous activity of the transcriptional targets of Nrf2 in lungs chronically or acutely exposed to tobacco smoke was surprisingly modest. Importantly, the administration of CDDO-imidazolide, a synthetic triterpenoid, significantly activated Nrf2 and boosted the expression of its target genes. Treatment of mice with this small molecule compound over the course of tobacco smoke exposure showed significant therapeutic benefit (202). These findings suggest that the endogenous induction of Nrf2 during the development and progression of experimental emphysema is suboptimal, and that further pharmacological stimulation of the antioxidant response could improve lung function.
The connection between decreased Nrf2 activity and susceptibility to COPD has also been demonstrated in humans. When the abundance of Nrf2, Keap1, and Bach1 in patients with severe emphysema was compared to those in smokers and non-smokers without the disease (203), Nrf2 was decreased in whole lung tissue of emphysema patients, whereas Keap1 and Bach1 were increased. Similarly, the abundance of Nrf2 was decreased in the cytoplasm and nucleus of alveolar macrophages in subjects with emphysema, whereas that of Keap1 and Bach1 was increased. The combined decrease in both nuclear and cytoplasmic Nrf2 in the macrophages of emphysema patients suggests a global reduction in the abundance of Nrf2 rather than a defect in nuclear localization, which is consistent with the increased Keap1 that would reduce Nrf2 protein abundance by promoting proteasomal degradation. The reduced abundance of Nrf2 was accompanied by reduced expression of its target genes, such as those encoding the antioxidant proteins HO-1, NAD(P)H dehydrogenase quinone 1 (NQO1), and glutathione peroxidase 2 (GPX2), in alveolar macrophages from emphysema patients, and this correlated with clinical indices of COPD severity. In addition, macrophages from emphysema patients displayed higher amounts of the lipid peroxidation product 4-hydroxynonenal (203) which is an indication of oxidative stress. Thus, patients with severe emphysema have a pulmonary phenotype of excessive oxidative stress and suppressed antioxidant response. This may be due to compromised signaling through the Nrf2 pathway caused by increased abundance of Keap1 and Bach1, which negatively regulate Nrf2 and the ARE, respectively. Therefore, therapies that restore Nrf2 activity might prove valuable in the treatment of this disease.
A related study provided additional evidence supporting a reduced antioxidant response in COPD patients and suggested another potential mechanism underlying Nrf2 dysfunction in the disease (204). This work compared whole lung tissue samples from patients with mild and advanced COPD to tissue from smokers and former smokers without the disease. Compared to the control subjects’ lungs, the COPD patients’ lungs showed a marked reduction in the expression of the antioxidant genes encoding HO-1, NQO1, and the glutamate cysteine ligase regulatory subunit (GCLM), which is critical for the synthesis of the abundant antioxidant glutathione. In tissues from patients with advanced COPD, the concentration of glutathione was significantly reduced and lipid peroxidation was significantly increased (204), confirming that COPD is associated with a phenotype of excessive oxidative stress and deficient antioxidant response in the lung (203). This study also found that the abundance of Nrf2 was decreased in the COPD patients’ lungs, although the amount of Nrf2 mRNA was similar in COPD and control tissue (204). Whereas the abundance of Keap1 mRNA and protein was also similar in the COPD and control samples (204), the mRNA and protein abundance of DJ-1, a protein that stabilizes Nrf2 by interfering with its Keap1-mediated proteasomal degradation (Fig. 2), was significantly reduced (205). Oxidative modification of DJ-1 decreases its functionality and promotes its degradation (206, 207), and prolonged exposure of a human lung epithelial cell line to cigarette smoke extract caused oxidation, carbonylation, and degradation of DJ-1 (204). Thus, the loss of the Nrf2-dependent antioxidant response in COPD could be due to a disease-associated reduction in DJ-1 abundance. In support of this model, in mouse lungs, mouse embryonic fibroblasts, and human lung cells, the genetic disruption of DJ-1 decreased Nrf2 protein stability and impaired the antioxidant response to cigarette smoke. In the DJ-1-disrupted cells, it was still possible to relieve Nrf2 inhibition and restore the expression of Nrf2-dependent antioxidants by targeting Keap1, either genetically with small interfering RNAs to knock down its expression or pharmacologically with sulforaphane (204). This confirms that DJ-1 is epistatic to Keap1 and Nrf2, and suggests that the suppressed antioxidant response phenotype in COPD might be reversible by pharmacological intervention at the level of Keap1 or at other points downstream of DJ-1.
Further evidence for a dysfunction of the Nrf2 response in COPD was provided by an independent study that found reduced Nrf2 mRNA and protein in alveolar macrophages from COPD patients compared to control subjects (208). These three reports on Nrf2 in human COPD have some discrepancies; for example, the abundance of Nrf2 mRNA was reportedly normal (204), reduced (208), or not tested (203), and the abundance of Keap1 protein was normal (204), increased (203), or not tested (208). Nevertheless, a common theme of the studies and an important concept in Cnc biology is that Nrf2 function and the antioxidant response can be unexpectedly compromised during the progression of an oxidative stress-related disease like COPD.
Nrf2 suppression in neurodegenerative diseases
Studies on neurodegeneration in humans suggest that the paradoxical dysfunction of Nrf2 signaling in disease is not specific to COPD, but rather a more general phenomenon. Oxidative stress in the nervous system is associated with neuronal cell death during the pathogenesis of multiple neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS) (189, 209). These are all age-related degenerative disorders leading to progressive functional impairment and ultimate mortality, for which effective treatments are lacking. Oxidative stress activates Nrf2 in both neuronal and glial cells, and the Nrf2 pathway is critical for the protection of neurons from degeneration-related oxidative insults in cell culture systems (210–215). Nrf2 contributes to neuronal homeostasis through the adaptive response mounted by glia, which exert prosurvival actions on stressed neurons (213). Genetic and pharmacological approaches have also demonstrated the importance of Nrf2 as a protective factor against neurodegeneration with in vivo models of AD, PD, HD, and ALS (180–182, 215–219).
Even though compounds that are approved treatments of PD, such as bromocryptine (220) and selegiline (221), activate Nrf2, little is known about the activity of the Nrf2 pathway in human neurodegenerative diseases. Only three human studies have so far investigated Nrf2 in neurodegeneration (222–224). One of these studies found that cultured fibroblasts from patients with Friedreich’s ataxia, a genetic neurodegenerative disease associated with decreased abundance of the mitochondrial protein ataxin, showed impaired Nrf2 antioxidant response (222). The normal cytoplasmic colocalization of Nrf2 and Keap1 with actin fibers was not observed in the patients’ cells, and the ability of Nrf2 to accumulate in the nucleus in response to oxidative challenges was significantly impaired. The suppressed Nrf2 pathway activity was restored pharmacologically by exposing the cells to Euk-134, a small molecule compound that mimics catalase and scavenges hydrogen peroxide (222).
Neuronal degeneration in AD initiates in the entorhinal cortex and hippocampus (225). In a comparative study of postmortem brains of AD patients and control subjects, differences in Nrf2 abundance and subcellular localization were detected (223). In control hippocampi, Nrf2 was detected in both the cytoplasm and nucleus of neurons and astrocytes. Few astrocytes were positive for Nrf2 in AD hippocampi, and this minimal Nrf2 abundance in glia may contribute to neuronal impairment. Moreover, Nrf2 was mostly cytoplasmic in hippocampal neurons from patients with AD or the Lewy Body Variant of Alzheimer’s Disease (LBVAD), and its abundance in the nucleus was dramatically reduced. Neurons in the AD or LBVAD samples that had cytoplasmically localized Nrf2 were under substantial oxidative stress, as indicated by the increased amount of the lipid peroxidation product 7-hydroxynonenol. In light of the protective role of Nrf2 against oxidative stress and the abundant oxidative damage of hippocampal neurons in AD, the reduced nuclear localization of Nrf2 indicates that the antioxidant response is paradoxically compromised in human AD (223). This conclusion is supported by the observation that in a transgenic mouse model of AD the abundance of Nrf2 and the expression of its target genes declines in an age-dependent manner (181).
In PD, severe damage occurs in the substantia nigra, resulting in loss of dopaminergic neurons (226). Nrf2 in the few surviving dopaminergic neurons of postmortem brains from PD patients was nuclear and increased compared to control subjects or patients with AD or LBVAD, in whom it was cytoplasmic and weak (223). Thus, in contrast to AD, Nrf2 accumulates in the nucleus in at least some of the neurons severely affected in PD. One explanation for these findings is that in PD the dopaminergic neurons that manage to accumulate Nrf2 in their nuclei are the ones that survive, whereas those who fail to do so are the ones that perish. Alternatively, the transcriptional activity of Nrf2 may be compromised in PD, such that its nuclear accumulation does not translate into increased target gene expression. Finally, it is possible that Nrf2 is transcriptionally competent but the antioxidant response is not sufficient to protect the neurons from the oxidative stress that underlies the pathology of PD. It is not straightforward to distinguish among these possibilities in a study of postmortem brain samples with substantial loss of dopaminergic neurons. Nevertheless, Nrf2 localization in the hippocampal neurons correlates with the degree of neuropathology. In control brains, Nrf2 was predominantly nuclear, presumably due to age-associated oxidative stress. In PD, where the hippocampal neurons are only secondarily affected, Nrf2 was both nuclear and cytoplasmic. In AD, however, where the hippocampal neurons are the primarily affected neurons, Nrf2 was predominantly cytoplasmic (223). These observations suggest that the subcellular localization of Nrf2 in neurons may indeed be compromised at early stages of PD pathogenesis. It is conceivable that individuals who develop PD may be those who have an inherently reduced ability to accumulate Nrf2 in the nucleus, either globally or in a critical subset of their neurons.
ALS is characterized by neuronal degeneration that affects motor neurons in the motor cortex, brain stem, and spinal cord (227, 228). ALS is usually a sporadic disease, although it is familial in 10% of cases. A subset of familial ALS cases have been associated with mutations in the antioxidant protein superoxide dismutase 1 (SOD1) (229). In postmortem samples from patients with ALS, the mRNA and protein abundance of Nrf2 in motor neurons of the motor cortex and spinal cord was decreased compared to age-matched controls (224). This finding did not simply reflect neuronal cell death, as Nrf2 was reduced at the individual cell level. For Keap1, a slight increase in mRNA was found in the motor cortex but not in the spinal cord, and no differences in protein abundance were detected. These findings indicate that Nrf2 signaling is suppressed in late stages of ALS in humans, and suggest that Nrf2 suppression may promote cell death due to unrestricted oxidative stress (224). Whether the five ALS patients examined in this study had familial disease or harbored SOD1 mutations was not determined (224). However, evidence for suppression of Nrf2 signaling in ALS has been found in some SOD1 transgenic and cellular models of familial ALS (230, 231), but not in others (217, 232). These discrepancies may be related to the inability of the rodent transgenic models of familial ALS to accurately reflect all aspects of the human disease. Interestingly, in a rodent model of ALS utilizing immune-mediated spinal cord motor neuron injury, the abundance of Nrf2 and the expression of its antioxidant target genes was significantly reduced (233, 234).
The paradoxical dysfunction of Nrf2 in neurodegeneration is a novel concept, and the comprehensive elucidation of its significance and underlying mechanisms deserves further investigation. Nevertheless, p62 and DJ-1 have already been identified as Nrf2 modulators whose expression or function is compromised in neurodegeneration leading to impaired antioxidant response. p62 is a cytoplasmic protein that mediates the formation of ubiquitinated aggregates that are removed by autophagy (235, 236), and it is localized to intracellular aggregates in various neurodegenerative diseases (237). Knock out of the p62 gene leads to increased oxidative DNA damage to the mouse brain and to biochemical and cognitive defects that resemble AD (238). Interestingly, p62 and Nrf2 are positively related: Nrf2 activates the transcription of p62 (239–241), and p62 promotes the nuclear localization of Nrf2 and the activation of ARE transcription (236, 242, 243). The abundance of p62 was decreased in brains from patients with AD relative to age-matched controls, and this decrease correlated with oxidative damage to the p62 promoter (243). A transgenic mouse model of AD also exhibited increased p62 promoter damage and reduced p62 abundance in the brain (243). These findings suggest that the decreased activation of Nrf2 in AD may be at least partially due to disease-associated oxidation of the p62 promoter and reduced p62 expression. Thus, a feed-forward cycle of decreased p62 expression and reduced Nrf2 activation could accelerate neuronal dysfunction and demise in AD (243). Because oxidation of the p62 promoter is a shared feature among neurodegenerative disorders, including AD, PD, and HD, this mechanism may account for Nrf2 dysfunction in multiple settings of neuronal degeneration (244). In PD, the dysfunction of Nrf2 is likely also related to DJ-1. Inherited loss-of-function mutations in PARK7, the gene encoding DJ-1, are associated with early-onset Parkinsonism (245, 246). DJ-1 protects neurons from oxidative stress, but its functionality is lost when the protein is oxidatively modified (206). Because DJ-1 stabilizes Nrf2 by preventing its association with Keap1, the oxidation of DJ-1 in PD could compromise Nrf2 signaling and precipitate the collapse of the antioxidant response system (205). Other studies have also implicated DJ-1 and p62 as defense mediators in ALS (247–249), which further supports the possibility that common mechanisms may underlie the suppression of Nrf2 signaling in neurodegenerative disorders. Thus, progressive disease processes related to oxidative stress, like COPD and neurodegeneration, are paradoxically associated with impaired Nrf2 signaling in humans. Similarly, advancing age also correlates with a suppression of the Nrf2 antioxidant response. This age-related suppression could in turn predispose to diseases linked to environmental exposures, including COPD, PD, cancer, and many others. Interestingly, the mouse p62 promoter exhibits elevated oxidative damage with increasing age, and the degree of p62 promoter oxidation also correlates with age in human brain samples (244). Likewise, increasing age correlates with oxidative modification and inactivation of the DJ-1 protein in flies, mice, and humans (206). Thus, the suppression of Nrf2 in aging and human disease may be mediated by mechanisms shared among pathophysiological contexts and conserved across species.
Stress-Activated Cnc Transcription Factors in Longevity
In addition to their roles in defending organisms against electrophilic insults and oxidation-related diseases, stress-activated Cnc transcription factors have anti-aging and pro-longevity functions. A popular hypothesis about the causes of aging is the “free radical theory” (250, 251), in which aging is the result of progressive damage to macromolecules and cellular structures that is caused by exposure to endogenous and exogenous pro-oxidant substances and free radicals. Oxidative damage accumulates with advancing age and progressively compromises various functions of the organism; aging can thus be viewed as the collective phenotypic manifestation of accumulated oxidative damage. The oxidative stress hypothesis predicts that bolstering the organism’s antioxidant defenses may retard the aging process and extend lifespan. This prediction has received considerable support in recent years, mainly from studies in model organisms showing that various genetic manipulations that extend lifespan also confer increased resistance to oxidative stress, and also by studies showing that the overexpression of antioxidant genes can augment oxidative stress tolerance and promote longevity (252–255). By extension, signaling pathways that regulate antioxidant responses, like stress-activated Cnc factors, are plausible regulators of the aging process (256).
Role for Nrf2 in longevity through calorie restriction
Despite the rational associations between aging, oxidative stress, and the Nrf2 pathway, the hypothesis that Nrf2 may have an impact on the aging of vertebrates has not yet been sufficiently investigated. One study examined the role of Nrf2 in the context of the anti-aging effects of caloric restriction in mice (257). Hypocaloric diets have impressive beneficial effects on experimental animals: They prevent the age-related decline in antioxidant capacity, increase the abundance of antioxidant enzymes, raise insulin sensitivity, reduce the incidence of various spontaneous and experimentally induced cancers, and significantly extend the lifespan (258–260). Consistent with its established role in cancer prevention, Nrf2 was required for the cancer-preventive effects of a reduction of ingested calories by 20%, 30%, or 40% (257). In contrast, the increase in insulin sensitivity was not linked to Nrf2 at any level of caloric restriction. nrf2−/− mice subjected to a 40% reduction in caloric intake lived significantly longer than ad libitum feeding nrf2−/− mice, which shows that the knockout mice still possess mechanisms for the lifespan-extending effects of caloric restriction (257). However, this finding cannot completely rule out a role of the antioxidant response in longevity. First, Nrf1 may have compensated for a pro-longevity function of Nrf2 in nrf2−/− mice. Second, the most convincing evidence for the role of Nrf2 in cancer prevention by caloric restriction is that the mildest intervention (20% reduction of calories) markedly reduced cancer incidence in wild-type but not in nrf2−/− mice. In contrast, a 40% reduction of calories significantly reduced cancer incidence in both wild-type and nrf2−/− mice. Because the lifespan experiment neither evaluated milder restrictions nor included wild-type mice as controls (257), an involvement of Nrf2 in the longevity of rodents caused by a hypocaloric diet or other interventions remains plausible.
Role of Cnc transcription factors in invertebrate longevity
Experiments in invertebrate model organisms, where genetic redundancies are less common, have provided compelling evidence for a function of Cnc factors in promoting longevity. C. elegans lives for a few weeks, and Drosophila lives for a few months, making the extensive lifespan studies required for an exhaustive test of the association between the antioxidant response and aging quite feasible. Indeed, studies in these organisms have linked Cnc homologs to longevity in various contexts: caloric restriction (261), reduced insulin- and insulin-like growth factor-1 (IGF-1)-like signaling (155), and normal growth conditions (159).
Caloric restriction significantly extends lifespan not only in mammals but in many organisms across the evolutionary spectrum. Longevity brought about by caloric restriction in these species has often been associated with increased oxidative stress resistance and with the activation of stress defense pathways, although the exact interplay between the antioxidant and lifespan-extending effects of caloric restriction is not thoroughly understood. Similarly to its effect on vertebrates, caloric restriction significantly extends lifespan in C. elegans (262). SKN-1, the worm’s Cnc protein, is present in epithelial cells of the intestine and also in the ASI neurons, a set of cells in the brain that are critical in translating information about food availability into endocrine signals that influence the worm’s developmental and reproductive behavior (263). Importantly, SKN-1 is required in the ASI neurons for the extension of the worm’s lifespan by caloric restriction (261). In contrast, intestinal SKN-1 does not contribute to the longevity caused by a hypocaloric diet. The mechanism by which SKN-1 function in the ASI neurons promotes longevity is not known. One hypothesis is that SKN-1 specifically facilitates the expression of an unknown hormone or other signal that is produced by the ASI neurons in response to caloric restriction and that exerts beneficial effects on peripheral tissues (261). Another explanation is that the ASI neurons are subjected to intense cellular stress during caloric restriction and require the antioxidant function of SKN-1 to cope with the stress and sustain their functions, including their effects on survival under these conditions. Even though the underlying mechanism was not elucidated, this study linked an extension of lifespan with a Cnc transcription factor (261).
In addition to caloric restriction, a related intervention that has positive effects on longevity in various species is the reduction of signaling through the insulin and IGF-1 (or IGF-1-like) signaling (IIS) pathway (264). In C. elegans, mutations that reduce IIS activity increase resistance to oxidative stress and extend lifespan even in the absence of caloric restriction. These effects require the expression and activity of the transcription factor DAF-16, a homolog of vertebrate FOXO, which is normally phosphorylated and inhibited by IIS (265). IIS causes the phosphorylation and inhibition not only of FOXO but also of SKN-1 (155). Conversely, SKN-1 is activated in conditions of reduced IIS and accumulates in the nuclei of the intestinal cells of the worm where it transcriptionally induces its target genes. In this manner, SKN-1 contributes to the increase of antioxidant resistance and extension of lifespan that are associated with reduced IIS. Under conditions of normal insulin signaling, the transgenic overexpression of SKN-1 in the intestine extended C. elegans lifespan (155). Conversely, the lack of SKN-1 shortened C. elegans lifespan (151). Consistent with these findings, genetically targeting the SKN-1 inhibitor WDR-23 also increased oxidative stress resistance and extended lifespan in worms (158). Finally, the activation of SKN-1 was suggested to contribute to the “soma-to-germline transformation” observed in long-lived C. elegans mutants, in which somatic cells modify their transcriptional program to resemble that of the more stress-resistant and “immortal” germline (266).
Work in Drosophila has provided independent evidence supporting the notion that the antioxidant program controlled by Cnc transcription factors contributes to the regulation of longevity (159). Heterozygous mutations in the fruit fly keap1 gene, whose protein product suppresses CncC and ARE activity, result in elevated mRNA levels of the CncC target gene gstD1 (encoding a glutathione S-transferase). This implies that loss of one keap1 copy is sufficient to activate CncC. The oxidative stress tolerance and the lifespan of adult keap1 heterozygotes were compared to those of their otherwise genetically identical wild-type siblings. Male keap1 heterozygous flies showed significantly increased survival rates after exposure to the free radical generator paraquat, and they also lived significantly longer than controls under normal growth conditions (159). For reasons that are not well understood, female keap1 heterozygotes did not show significant differences in either paraquat resistance or longevity. These findings demonstrate that partial loss of function of the Nrf2 pathway’s main negative regulator has beneficial effects on the oxidative stress tolerance and longevity of male Drosophila. Thus, this study showed that manipulation of the Cnc antioxidant response system can have lifespan-extending effects under normal laboratory growth conditions (without caloric restriction (261), reduced IIS (155), or other interventions) and demonstrated that targeting the Nrf2 pathway at the level of Keap1 can promote longevity (159).
Reduced stress-activated Cnc transcription factor activity in mammalian aging
Although Nrf2 protects the organism from disease, its activity paradoxically declines during the progression of COPD and neurodegeneration. It seems that the same paradox holds true in aging: Despite the role of Cnc factors in promoting longevity, their activity declines with advancing age. Evidence for an age-dependent dysfunction of the Cnc system was originally provided by studies in rats showing that the basal amount of Nrf2 protein declines with age and that this decline correlates with decreased expression of Nrf2-regulated genes, with lower concentrations of intracellular glutathione, and with increased abundance of various oxidation products (267, 268). The Nrf2 transcriptional response was restored pharmacologically by administration of the Nrf2 activator lipoic acid (267), which is consistent with the ability of other pharmacological agents to reactivate Nrf2 activity, such as sulforaphane, which restores Nrf2 activity after it has been compromised by prolonged exposure to cigarette smoke (204), and Euk-134, which has a similar effect in Friedreich’s ataxia (222). Thus, similar mechanisms may mediate the suppression of Nrf2 activity in the contexts of aging, neurodegeneration, and COPD, and the reversibility of this suppression by pharmaceuticals is likely to have general applicability and utility. This potential for beneficial pharmacological manipulation of Nrf2 activity is further supported by several studies. For example, one study administered to mice the Nrf2 inducer tert-butyl-hudroxyanisole (tBHA), an antioxidant widely used to preserve packaged foods, and showed that feeding mice with a diet supplemented with tBHA over 18 months stimulated the Nrf2 antioxidant response, decreased molecular indices of oxidative stress and inflammation, and led to healthier aging with reduced weight gain and improved locomotor function (269). Aging in mice is also associated with a decline in innate immunity, manifested as decreased hypersensitivity of the skin to contact with antigens (contact hypersensitivity, CHS). The age-associated decline of CHS was exacerbated in nrf2−/− mice; in contrast, the pharmacological activation of Nrf2 with sulforaphane restored the CHS response in aged animals (270). Furthermore, the abundance of Nrf2 and the expression of its target genes encoding HO-1 and NQO1 also decreases with age in mouse spinal cord astrocytes, but this decrease is reversible with the Nrf2 activator epigallocatechin gallate (271). Other studies have shown that the statins simvastatin and atorvastatin and the thiazolidinedione rosiglitazone, which are approved for use in humans with hypercholesterolemia and diabetes, respectively, also have the ability to increase Nrf2 activity in rodents (186, 272, 273). The antioxidant and anti-aging effects of these compounds in humans warrant further investigations. The loss of the Cnc antioxidant response with advancing age has been documented across the evolutionary spectrum. In worms, increasing age is associated with a reduced adaptive transcriptional response to the pro-oxidant xenobiotic juglone; this effect is mediated by suppression of the activity of both DAF-16 and SKN-1 (274). In humans, the dysfunction of the Nrf2 response in COPD was positively correlated with the age of the subjects and their years of exposure to cigarette smoke (208). Specifically, the abundance of Nrf2 and HO-1 mRNAs was reduced in alveolar macrophages from old, chronic smokers compared to young smokers; the reduction in Nrf2 mRNA levels correlated with an increase in the amount of oxidized glutathione and carbonylated albumin, which are markers of oxidative stress (208). Although these effects could in principle be attributable to either the age of the subjects or the duration of smoking, the Nrf2 system was differentially activated by cigarette smoke in alveolar macrophages from mice depending on their age. The induction of Nrf2 mRNA and protein by cigarette smoke was substantially reduced in macrophages from old mice compared to those from young animals (208). Reduced mRNA abundance of human Nrf2 has also been documented in liver transplants from older liver donors compared to those from younger ones (275). Consistent with the role of Nrf2 as a protective factor against ischemia-reperfusion injury, transplants from younger donors were less susceptible to this form of oxidative damage. However, whether the decrease in liver Nrf2 mRNA was directly related to the difference in the subjects’ age or was a result of the prolonged mechanical life support and more potent pharmacological stimulation of the younger donors could not be clarified (275).
Taken together, these discoveries support the notion that, in both aging and disease, the activity of stress-induced Cnc factors is paradoxically suppressed at the time when the organisms most need their function. Multiple mechanisms may contribute to the suppression of Nrf2 activity as a result of aging and oxidative stress, including, somatic loss-of-function mutations in the genes encoding Nrf2 or small Mafs, oxidation or epigenetic modification of the NRF2 promoter causing reduced expression, or changes in the abundance or activity of other factors (such as kinases, phosphatases, or DNA binding proteins) that regulate Nrf2 activity, ARE activity, or both, which leads to transcriptional silencing of the antioxidant response. Regardless of the precise mechanism, the impairment of Cnc activity during disease may accelerate the age-related deterioration of the affected tissue. Conversely, organismal aging may promote specific age-related diseases through the suppression of the antioxidant response in the relevant organs.
Homeostatic Regulation of the Cnc Antioxidant Response
The realization that stress-activated Cnc signaling can be paradoxically suppressed under conditions of oxidative stress, when it is most needed, poses puzzling questions. Why is the basal Cnc activity not set at a higher point? Why has the counterintuitive suppression of Cnc factors in aging and disease not been eliminated by evolution? Some insights into these intriguing enigmas are offered by observations in various organisms supporting the notion that, whereas the Cnc response is critical for cellular adaptation, organismal survival, and longevity, the prolonged and unopposed activation of Cnc signaling can be detrimental. In flies, the constitutive transgenic high overexpression of CncC can be lethal to both individual cells as well as the organism, whereas conditional moderate overexpression can confer oxidative stress resistance (159). Similarly, whereas the transgenic overexpression of SKN-1 in the intestine of worms promotes longevity, expression from high-copy transgenic arrays is toxic (155). Moreover, reducing WDR-23 with RNA interference increases the stress resistance and extends the lifespan of worms, but also retards growth in a SKN-1-dependent manner (158). These findings suggest that an optimal window of Cnc factor abundance is important for resistance to oxidative stress and for longevity; in contrast, excessive amounts of Cnc activity can have deleterious consequences. This concept is further supported by the detrimental phenotypes associated with loss of Keap1. In flies, whereas loss of one keap1 copy activates CncC and increases oxidative stress resistance and longevity, loss of both keap1 alleles leads to lethality during mid-larval development (159). Although it would be interesting to know whether this lethality depends on CncC, it is not straightforward to test this hypothesis, because cncC mutations also cause early developmental lethality (162). In mice, however, loss of keap1 causes Nrf2-dependent hyperplasia of the esophagus and occlusion of its lumen, which leads to early postnatal lethality from the inability to feed (276). Taken together, these observations indicate that the ability to limit the antioxidant response is as vital to homeostasis as the ability to mount it. This may partially rationalize the existence of mechanisms that suppress the activity of Cnc factors despite conditions of chronic oxidative stress.
A dramatic demonstration of the importance of accurate regulation of the antioxidant response pathway is provided by discoveries showing that Nrf2 is constitutively activated in diverse human cancers (42, 277). Although Nrf2 inherently prevents the malignant transformation of healthy cells, cancer cells hijack the Nrf2 pathway, presumably to protect themselves from the cellular stresses associated with their increased proliferation, hypoxic environment, and deregulated protein synthesis (278). This is accomplished through somatic genetic events, including KEAP1 or NRF2 mutations and KEAP1 loss of heterozygosity (57, 279–282), or through the epigenetic hypermethylation of the KEAP1 promoter (283). The loss of KEAP1 heterozygosity and KEAP1 promoter hypermethylation likely activate Nrf2 by decreasing Keap1 protein abundance. The KEAP1 and NRF2 mutations associated with human cancers impair the Keap1-mediated repression of Nrf2 (281). The clustering of NRF2 mutations in the two domains of high and low affinity for Keap1 supports the “hinge and latch” Keap1-Nrf2 protein association model (Fig. 3A) (58). Such paradoxical activation of the Nrf2 pathway has been documented in human cancers and cancer cell lines of the lung, breast, ovary, billiary tract, and head and neck. Furthermore, the constitutive activation of Nrf2 correlates with the inherent or acquired resistance of cancer cells to chemotherapy (284–289). Conversely, some cancer cell lines and human cancers with high sensitivity to chemotherapy exhibit reduced Nrf2 abundance (290). This chemosensitive phenotype depends on the increased abundance of Cullin3, which presumably increases degradation of Nrf2 (290). On the basis of these observations, the development of strategies to inhibit Nrf2 signaling in cancer has emerged as a logical strategy for the re-sensitization of tumors to chemotherapy (291). Furthermore, the association of Nrf2 activity with cancer and chemoresistance also warns against excessive pharmacological stimulation of Nrf2 in nonmalignant diseases, like COPD and neurodegeneration.
The Antioxidant Response as a Target for Drug Discovery
The fact that the activation of the Keap1-Nrf2 pathway confers cellular protection from oxidative stress and defends organisms against pathologies associated with stress and aging has not been lost on translational investigators interested in discovering novel treatments for various human disorders. Efforts are currently underway to discover compounds that activate Nrf2 and to develop them as treatments for oxidative stress-related diseases. These endeavors are largely guided by the elucidation of the mechanistic basis of Keap1-Nrf2 signaling at the biochemical and biophysical level (190).
The key role of Keap1 as the ubiquitous regulator of Nrf2 qualifies it as the central target in Nrf2-oriented drug discovery. One obvious strategy to activate the antioxidant response by targeting Keap1 is to knock down its expression using RNA interference, and proof-of-principle for this approach has been provided (292). In the more conventional arena of small molecule compounds, the detailed characterization of the mechanisms by which Keap1 integrates oxidative stress sensing with Nrf2 inhibition offers additional opportunities for the pharmacological activation of Nrf2. One such targeting strategy is highlighted by the discovery that the stability of Keap1 requires a specific phosphorylation event (293). Phosphorylation of Keap1 on Y141 has no direct effect on Keap1 dimerization or Nrf2 binding, but it stabilizes the Keap1 protein and thus prevents its rapid degradation. Conversely, oxidative stress triggers the dephosphorylation of Y141, through the presumed activation of a yet unidentified redox-sensitive phosphatase (293). These discoveries suggest that it should be possible to modulate the Keap1-Nrf2 system indirectly by targeting the Keap1 phosphorylation-dephosphorylation mechanism. Specifically, pharmacologically inhibiting the activity of the kinase that phosphorylates Keap1 would be expected to promote Keap1 degradation, thus preventing Keap1-mediated inhibition of Nrf2 and ultimately leading to ARE activation. Although the kinase that mediates Keap1 Y141 phosphorylation is currently unknown, a combination of chemical and genetic approaches employing kinase-specific inhibitors and RNA interference constructs should help uncover its identity.
Another approach to manipulation of ARE activation is to indirectly induce Nrf2 by targeting other proteins that serve as interaction partners of Keap1. For example, phosphoglycerate mutase 5 (PGAM5) is a Keap1-binding protein (62, 63). The N-terminal sequence of PGAM5 targets this protein to the outer membrane of mitochondria, where a Keap1 dimer simultaneously binds PGAM5 and Nrf2 in a ternary complex (Fig. 3B). Thus, PGAM5 serves to localize a subset of the total cellular Keap1 and Nrf2 pool to the outer membrane of the mitochondrion. Genetically reducing PGAM5 expression induced the antioxidant transcriptional program, presumably by selective activation of the mitochondrial Nrf2 fraction (63). Thus, even though only a small fraction of the total cellular Keap1 and Nrf2 pool localizes to mitochondria, its activation is sufficient to trigger the antioxidant response. This organelle-specific Keap1-Nrf2 system may, therefore, serve as a mechanism that elicits nuclear responses to leakage of reactive oxygen species from mitochondria into the cytoplasm. Pharmacologically targeting PGAM5 or its interaction with Keap1 would be expected to selectively activate this subcellular Nrf2 fraction. It would be interesting to test whether selective induction of mitochondrial Nrf2 can restore the suppressed antioxidant response in aging and disease. The most attractive strategy to activate Nrf2 entails the targeting of protein-protein interactions in its cytoplasmic degradation machinery. Logical targets for pharmacological disruption include the Keap1 dimerization interface, the Keap1-Cullin3 interaction, and the Keap1-Nrf2 association (190). Because Keap1 dimerization is a prerequisite for Nrf2 inhibition at basal conditions (294, 295), blocking this process with a small molecule compound should activate the antioxidant response. Similarly, because Keap1 targets Nrf2 for degradation through a physical association with Cullin3, disrupting the Keap1-Cullin3 interface should also lead to Nrf2 stabilization and target gene induction. However, because Keap1 has other binding partners and ubiquitination substrates in addition to Nrf2 (62, 72, 296, 297), inhibiting Keap1 dimerization or Cullin3 binding may also affect the protein stability or activity of those proteins, with unpredictable functional consequences. Thus, disrupting the Keap1-Nrf2 interaction is likely the most specific intervention. The in vitro structure of the Nrf2-binding domain of Keap1 alone and in complex with Keap1-associating fragments of Nrf2 has been established by crystallography, and the structural model has been validated by the mutagenesis of critical amino acid residues (56, 57, 61, 298). This is so far the only protein interaction in the Nrf2 pathway for which the crystal structure has been determined. Knowledge of this protein interaction interface can be exploited to discover small molecule compounds that can disrupt the association by occupying the Nrf2-binding pocket of Keap1 (190).
Notwithstanding the scientific promise of Nrf2 induction as a preventive and therapeutic strategy, it is prudent to consider caveats that could potentially apply to the pre-clinical development and clinical use of Nrf2-activating compounds. (i) It should be anticipated that the unchecked and long-term pharmacological stimulation of Nrf2 may be detrimental in vivo. The discovery of constitutively activated Nrf2 in cancer is especially disconcerting, and fuels an intimidating uncertainty as to whether Nrf2 activators will be safe or may accelerate the growth of pre-clinical malignant or pre-malignant lesions. Indeed, the administration of very high doses of cancer chemopreventive phenolic antioxidants promotes tumor development in the forestomach and bladder of rodents (299). (ii) The mechanism of action of electrophilic compounds is not likely to be selective for the Nrf2 system, because they are expected to react with many cellular proteins in addition to Keap1, and perhaps to irreversibly modify some of their targets. This expectation raises two issues: First, although previously unknown Nrf2 activators can be easily identified in cell-based screens by their ability to activate Nrf2 or its target genes (300, 301), it can prove very challenging to decipher the molecular target (or all targets) of each compound. Lack of knowledge of the direct target of a compound severely impairs the understanding of its mechanism of action. Second, the chronic and systemic exposure to electrophilic Nrf2 inducers may prove detrimental not only through sustained Nrf2 activation but also through the covalent modification of other cellular proteins (“off-target” effects). Both of these shortcomings can be critical liabilities for drug development. (iii) Drug-drug interactions may well turn out to be a hallmark of Nrf2-inducing compounds. Through the induction of Phase II enzymes, Nrf2 activators might promote the metabolism of other compounds. From a clinical standpoint this is a very important consideration, especially because it is likely that many of the patients with age-related, chronic, and degenerative diseases where Nrf2 stimulation may be indicated will be middle-aged or elderly and will also be taking additional medications. The impact of Nrf2 signaling on pharmacokinetics is evidenced by the altered disposition of acetaminophen and sulfobromophthalein in nrf2−/− and Keap1-knockdown mice (302, 303).
Although none of these considerations is sufficient reason to discredit the Keap1-Nrf2 signaling pathway as a drug target, it is important to take them all into account in the drug discovery and development process. Specifically, it seems logical to define the goal of treatment as the restoration of physiological Nrf2 activity or inducibility, rather than the hyperstimulation of the ARE. Pre-clinical disease models and trials of Nrf2 activators should also test whether the intermittent – rather than continuous – long-term induction of Nrf2 is safe and efficacious (304, 305). The discovery of small molecule disruptors of the Keap1-Nrf2 protein interaction should be vigorously pursued, because they are likely to be more specific and thus safer than electrophilic agents (190). In the clinical setting, the effects of Nrf2 induction on the pharmacokinetics of standard treatments for each contemplated indication should be thoroughly evaluated. Agents that activate Nrf2 only in specific tissues would be particularly attractive, as they could limit the antioxidant response to the diseased organ. However, the evidence for tissue-specific mechanisms activating Nrf2 in the human stems mainly from cultured cells derived from various tissues, and the in vivo relevance of such mechanisms needs further study. Moreover, tissue type-specific Nrf2 activation in these cellular systems is mediated by various protein kinases, which can be good targets for inhibition but not for activation. The GSK-3β and Fyn kinases are exceptions, because they promote suppression, rather than activation, of Nrf2, and are, therefore, reasonable targets for pharmacological inhibition (89, 306, 307).
Outlook
Multicellular organisms utilize Cnc transcription factors to defend themselves against oxidative stress. The antioxidant response orchestrated by Cnc proteins is essential for the prevention of diseases associated with oxidative and electrophilic insults. A collapse of the antioxidant response during the progression of such diseases may be a precipitating factor for their overt clinical presentation. In addition, converging evidence from invertebrate model organisms supports a role for Cnc factors in the regulation of longevity. The aging process itself correlates with a gradual breakdown of antioxidant responses, which may predispose to age- and oxidative stress-associated disorders. The pharmacological reactivation of the Nrf2 system may provide disease-preventive or therapeutic benefit in such settings, especially in individuals genetically predisposed to a suboptimal antioxidant response.
Since the cloning of the first mammalian Nrfs (Nrf1 and Nrf2) in the mid-1990s, research on stress-activated Cnc factors has been growing rapidly, and the field is now entering the important, yet challenging, arena of translational medicine. Whether the richness of basic research on Cnc proteins will ultimately lead to improved human health will depend critically on how much progress will be made in three key areas of clinical and translational investigation: human pathophysiology, the genetic basis of human disease, and drug discovery.
Human pathophysiology
The importance of the Nrfs as disease-preventing and longevity-promoting factors has been firmly established in model organisms. However, their roles in human disease have only recently begun to be probed, and whether they are also champions of human longevity is unknown. Much more research is needed in the domain of human pathophysiology: First, it will be important to identify additional human disease areas in which the Nrfs are important. Diseases most likely to be linked to the Nrfs are those affecting tissues that are exposed to the environment, such as the skin, lung, and digestive tract; generate high amounts of free radicals, such as muscle; have functions in detoxification, such as the liver, kidney, and placenta; or are particularly sensitive to oxidative stress, such as the nervous system. For example, the expression of human Nrf2 is highly up-regulated in the placenta in pregnancies complicated by preeclampsia, a disease known to be associated with oxidative stress (308). The degree to which the aging-associated physiological impairment of human tissues, manifested, for example, as sarcopenia in muscle or impaired wound healing in the skin, is linked to age-related decline in Nrf activity should also be investigated. Second, the relative contributions of the various Nrfs to human health must be characterized. The current disproportionate focus on Nrf2 is largely due to the serendipitous fact that nrf2−/− mice were viable whereas nrf1−/− mice were not. However, there is no scientific reason why Nrf1 should not be as important for human disease as Nrf2, and the same applies to Nrf3.
Finally, it will be very interesting to investigate whether the paradoxical suppression of the antioxidant response is a general occurrence in human aging and disease. Such studies could be informed by research on rodent models. For example, inflammatory bowel disease (IBD) is associated with oxidative stress, and Nrf2 protects mice from experimental IBD induced by dextran sulfate (183, 309, 310). Nevertheless, the expression of Nrf2 mRNA and protein paradoxically declines during the progression of dextran sulfate-induced colitis (311). The protein prohibitin was discovered as a positive regulator of Nrf2 in gut epithelial cells (311). Exposure of mice to dextran sulfate led to a decrease in the abundances of prohibitin and Nrf2 in the colonic mucosa. Conversely, the transgenic expression of prohibitin promoted the sustained presence of Nrf2 during experimental IBD and led to reduced oxidative stress and inflammation (311). These findings suggest that the suppression of Nrf2 during IBD is due to the loss of prohibitin (Fig. 2), and that the collapse of the antioxidant response can be prevented by maintaining prohibitin expression. It will be important to test whether the progression of human IBD is also associated with suppressed Nrf2 activity. If there are exceptions to or variations on the phenomenon of disease-associated Nrf2 suppression, then the comparative investigation of these settings could help elucidate the underlying mechanisms, and would thereby also highlight strategies for prevention or reversal. Ultimately, the comprehensive identification of the human diseases where the Nrfs are critical players will guide related research in both human genetics and drug discovery.
Human genetics
Equally important to deciphering the exact role of the Nrfs in human pathophysiology is investigating whether they also make genetic contributions to human disease. Common variants (single nucleotide polymorphisms, SNPs) in the NRF2 promoter have been associated with various diseases (192–196). These studies need to be replicated in independent and larger patient cohorts from different ethnic populations. One of the NRF2 promoter SNPs compromises a functional ARE, likely leading to impaired Nrf2 transcriptional autoregulation (192). Independent work has shown that numerous putative ARE sequences in the human genome are polymorphic, and that some of these sequence variations may contribute to variability in Nrf2-mediated transcription of antioxidant genes (312). Thus, the importance of SNPs in human AREs and their potential roles in gene-environment interactions in disease warrant further exploration. Moreover, it should be examined whether NRF1 and NRF3 also harbor SNPs that are associated with disease. SNPs in NRF2 and KEAP1 were associated with respiratory capacity in the general population; thus, genetic variation in the Cnc antioxidant system may be linked not only to disease predisposition but also to variability in physiological functions (313). Finally, whether rare variants (germline mutations) in KEAP1, NRF1, NRF2, or NRF3 underlie human disease has not been examined. It will be very interesting to test whether these genes contribute mutant alleles to human diseases other than cancer.
Drug discovery
Beyond cancer chemoprevention, the development of Nrf2-activating compounds seems to be focused mostly on chronic disorders, like COPD and neurodegeneration. These disease areas are fraught with inherent difficulties because of their chronic nature and need for long-term administration of compounds, their clinical manifestation in late stages of pathogenesis after substantial loss of functional tissue has occurred, and, for disorders of the nervous system, the requirement for compounds to pass through the blood-brain barrier. For cancer chemoprevention, a compound must also have an extremely safe profile, because it will be administered for months or years to individuals who may be at elevated risk for cancer but are not suffering from disease (165). These considerations can severely handicap preclinical and early clinical “proof-of-concept” trials of Nrf2 activators. In contrast, these shortcomings do not apply to acute settings where targeting Nrf2 over a short period of time may also be beneficial. Acute lung injury (also known as the adult respiratory distress syndrome) is an illustrative example of acute disease that has been linked to decreased Nrf2 in humans (192). Because this acute-onset disorder is associated with substantial mortality, it offers a unique opportunity for human proof-of-concept trials of Nrf2-inducing compounds. Evidence from mice shows that Nrf2 activation by the synthetic triterpenoid CDDO-imidazolide protects against acute lung injury caused by concomitant exposure to hyperoxia (314). In rodent models of brain injury (315, 316), cerebral ischemia (317), and intracerebral hemorrhage (318), the pharmacological activation of Nrf2 has been associated with improved outcomes even when initiated after the onset of the tissue-damaging event. These could be additional fruitful paradigms for trials of Nrf2 inducers in acute-onset human disease. Regardless of the disease indication, human trials also require the identification of biomarkers of target activity, which in the case of Nrf2 could be one or more of the ARE-regulated antioxidant enzymes. However, stress response genes are often regulated by multiple pathways in tissue-specific manners. Therefore, it may prove challenging to identify a biomarker that is both specific for Nrf2 and applicable to all pertinent human tissues. Because Keap1 is regulated by Nrf2 at the transcriptional level (98) and its known functions are quite specific to the Nrf2 pathway, the abundance of Keap1 mRNA may be useful as a general purpose biomarker for the pharmacological stimulation of Nrf2.
Finally, although the discovery of Nrf2 activators can be guided by information already available about obvious targets in the antioxidant response system, such as Keap1, the identification of previously unknown pathway modulators remains a priority, because they may also represent favorable targets for the ubiquitous or tissue-specific modulation of Nrf2 by compounds. The phylogenetic conservation of stress-activated Cnc factors in model organisms offers a unique opportunity for pathway expansion through whole-genome screens in C. elegans (using in vivo RNA interference), Drosophila (using traditional genetics or RNA interference in cell culture and in vivo), and zebrafish (using chemical mutagens). These approaches may facilitate the identification of targets for Nrf2 activation in stress- and age-related diseases, as well as for Nrf2 suppression in cancer. The development of Nrf2 inhibitors can also be informed by the successful method employed for the leucine zipper transcription factors Fos and Jun (319). It must be noted, however, that globally suppressing Nrf2 is likely to increase the toxicity of the chemotherapy to healthy tissues (320, 321); therefore, tumor- or tissue-specific rather than systemic delivery or activity of an Nrf2 inhibitor would be the desirable strategy. These are exciting times for research on stress-activated Cnc transcription factors. Translational investigations of the Nrfs promise to reveal insights useful for the prevention and treatment of diseases related to oxidative stress, and may also have implications for extension of the healthy lifespan.
GLOSS.
Cap’n’collar (Cnc) proteins are a family of basic leucine zipper transcription factors. Some Cnc factors have important functions in development, whereas others are critical for maintaining homeostasis in the face of environmental stresses. The “electrophile counterattack” is a conserved cellular response to oxidative stressors and electrophilic xenobiotics. In this adaptive process, Cnc factors transcriptionally activate protective genes through Antioxidant Response Elements (AREs) in their regulatory sequences. In vertebrates, the electrophile counterattack is largely mediated by Nrf2. By defending animals against oxidative stress, Nrf2 prevents DNA and protein damage, and protects from the development of cancer and many other oxidative stress-related disorders, including respiratory and neurodegenerative diseases. In animal models of disease, pharmacological activation of Nrf2 can prevent cancer and other pathologies linked to oxidative stress. In humans, inherited DNA sequence polymorphisms that decrease Nrf2 abundance have been linked to various diseases of the skin, lung, stomach, and intestine, which are all organs that are subjected to environmental stressors. Thus, individuals genetically predisposed to a suboptimal antioxidant response may be more susceptible to disorders caused or exacerbated by oxidative stress. Like many diseases, the aging process is also linked to oxidative stress, and studies in model organisms show that Cnc transcription factors promote longevity. Paradoxically, in aging and in advanced human respiratory and neurodegenerative disease, the Cnc antioxidant response is suppressed. Thus, Nrf2-activating compounds could be beneficial in the treatment of human diseases and might help extend the healthy lifespan. However, hyperactivation of the antioxidant response is detrimental in model organisms and has been linked to chemoresistant cancers in humans. These observations caution that sustained induction of Cnc factors can be deleterious, and indicate that the antioxidant response system must be tightly controlled; moreover, they imply that compounds inhibiting Nrf2 may also be useful therapeutics as chemotherapy sensitizers. This Review with 3 figures, 2 tables, and 321 citations, describes the stress-activated Cnc transcription factors and their regulatory mechanisms, and highlights their roles in aging and human disease
Acknowledgments
Funding: Support was provided by JP Wilmot Cancer Research Fellowship (G.P.S.) and NIH grant RO3 CA123591 (D.B).
Footnotes
Competing interests: The authors declare no competing interests.
References
- 1.Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 2.Mohler J, Vani K, Leung S, Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech Dev. 1991;34:3–9. doi: 10.1016/0925-4773(91)90086-l. [DOI] [PubMed] [Google Scholar]
- 3.Chan JY, Cheung MC, Moi P, Chan K, Kan YW. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum Genet. 1995;95:265–269. doi: 10.1007/BF00225191. [DOI] [PubMed] [Google Scholar]
- 4.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 5.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ney PA, Andrews NC, Jane SM, Safer B, Purucker ME, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JY, Han XL, Kan YW. Isolation of cDNA encoding the human NF-E2 protein. Proc Natl Acad Sci U S A. 1993;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap’n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 11.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 13.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Kwok AM, Chan JY. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J Biol Chem. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- 15.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtsubo T, Kamada S, Mikami T, Murakami H, Tsujimoto Y. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. 1999;6:865–872. doi: 10.1038/sj.cdd.4400566. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 20.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 21.Lecine P, Villeval JL, Vyas P, Swencki B, Xu Y, Shivdasani RA. Mice lacking transcription factor NF-E2 provide in vivo validation of the proplatelet model of thrombocytopoiesis and show a platelet production defect that is intrinsic to megakaryocytes. Blood. 1998;92:1608–1616. [PubMed] [Google Scholar]
- 22.Derjuga A, Gourley TS, Holm TM, Heng HH, Shivdasani RA, Ahmed R, Andrews NC, Blank V. Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Mol Cell Biol. 2004;24:3286–3294. doi: 10.1128/MCB.24.8.3286-3294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sies H. Oxidative stress: oxidants and antioxidants. Exp Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 24.Halliwell B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007;35:1147–1150. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- 25.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 26.Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004:157–176. doi: 10.1042/bss0710157. [DOI] [PubMed] [Google Scholar]
- 27.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. Nrf2, a multi-organ protector? Faseb J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 29.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol. 2005;25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 31.Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prestera T, Zhang Y, Spencer SR, Wilczak CA, Talalay P. The electrophile counterattack response: protection against neoplasia and toxicity. Adv Enzyme Regul. 1993;33:281–296. doi: 10.1016/0065-2571(93)90024-8. [DOI] [PubMed] [Google Scholar]
- 33.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79:1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 36.Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Liew C, Chan JY, Kong AN. Toxicogenomics of endoplasmic reticulum stress inducer tunicamycin in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Toxicol Lett. 2007;168:21–39. doi: 10.1016/j.toxlet.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 38.Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 39.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2(Keap1) Signaling in Oxidative Stress. Free Radic Biol Med. 2009 doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhakshinamoorthy S, Jaiswal AK. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2001;20:3906–3917. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 45.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci U S A. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 53.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem. 2006;281:37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 63.Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Jain MR, Chen C, Yue X, Hebbar V, Zhou R, Kong AN. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J Biol Chem. 2005;280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 66.Theodore M, Kawai Y, Yang J, Kleshchenko Y, Reddy SP, Villalta F, Arinze IJ. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J Biol Chem. 2008;283:8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 68.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watai Y, Kobayashi A, Nagase H, Mizukami M, McEvoy J, Singer JD, Itoh K, Yamamoto M. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells. 2007;12:1163–1178. doi: 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- 71.Tashiro S, Muto A, Tanimoto K, Tsuchiya H, Suzuki H, Hoshino H, Yoshida M, Walter J, Igarashi K. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol Cell Biol. 2004;24:3473–3484. doi: 10.1128/MCB.24.8.3473-3484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niture SK, Jaiswal AK. Prothymosin-alpha mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. J Biol Chem. 2009;284:13856–13868. doi: 10.1074/jbc.M808084200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Yuan X, Xu C, Pan Z, Keum YS, Kim JH, Shen G, Yu S, Oo KT, Ma J, Kong AN. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog. 2006;45:841–850. doi: 10.1002/mc.20234. [DOI] [PubMed] [Google Scholar]
- 74.Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- 75.Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, Kong AN. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 76.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, Han J, Agarwal A, Kong AN. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 77.Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, Khor TO, Li W, Ma J, Kong AN. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 78.Chen C, Pung D, Leong V, Hebbar V, Shen G, Nair S, Li W, Kong AN. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radic Biol Med. 2004;37:1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 79.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J Biol Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 80.Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm Res. 2006;23:2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- 81.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 82.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 83.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285:C334–342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 84.Cho MK, Kim WD, Ki SH, Hwang JI, Choi S, Lee CH, Kim SG. Role of Galpha12 and Galpha13 as novel switches for the activity of Nrf2, a key antioxidative transcription factor. Mol Cell Biol. 2007;27:6195–6208. doi: 10.1128/MCB.02065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J Biol Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- 86.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pi J, Bai Y, Reece JM, Williams J, Liu D, Freeman ML, Fahl WE, Shugar D, Liu J, Qu W, Collins S, Waalkes MP. Molecular mechanism of human Nrf2 activation and degradation: role of sequential phosphorylation by protein kinase CK2. Free Radic Biol Med. 2007;42:1797–1806. doi: 10.1016/j.freeradbiomed.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem. 2006;281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 89.Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem. 2007;282:16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- 90.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 91.Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, Keum YS, Han J, Gallo MA, Kong AN. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem. 2004;279:23052–23060. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- 92.Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Min KJ, Kim JH, Jou I, Joe EH. Adenosine induces hemeoxygenase-1 expression in microglia through the activation of phosphatidylinositol 3-kinase and nuclear factor E2-related factor 2. Glia. 2008;56:1028–1037. doi: 10.1002/glia.20676. [DOI] [PubMed] [Google Scholar]
- 94.Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol. 2008;22:63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- 95.Naidu S, Vijayan V, Santoso S, Kietzmann T, Immenschuh S. Inhibition and genetic deficiency of p38 MAPK up-regulates heme oxygenase-1 gene expression via Nrf2. J Immunol. 2009;182:7048–7057. doi: 10.4049/jimmunol.0900006. [DOI] [PubMed] [Google Scholar]
- 96.Kocanova S, Buytaert E, Matroule JY, Piette J, Golab J, de Witte P, Agostinis P. Induction of heme-oxygenase 1 requires the p38MAPK and PI3K pathways and suppresses apoptotic cell death following hypericin-mediated photodynamic therapy. Apoptosis. 2007;12:731–741. doi: 10.1007/s10495-006-0016-x. [DOI] [PubMed] [Google Scholar]
- 97.Hwang YP, Jeong HG. The coffee diterpene kahweol induces heme oxygenase-1 via the PI3K and p38/Nrf2 pathway to protect human dopaminergic neurons from 6-hydroxydopamine-derived oxidative stress. FEBS Lett. 2008;582:2655–2662. doi: 10.1016/j.febslet.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 98.Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007 doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 99.Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J Biol Chem. 2005;280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- 101.Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 102.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of nrf2 protein in activation of antioxidant response by oxidants. Mol Pharmacol. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 103.Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem. 2003;278:49246–49253. doi: 10.1074/jbc.M306764200. [DOI] [PubMed] [Google Scholar]
- 104.Kaspar JW, Jaiswal AK. Antioxidant-induced phosphorylation of tyrosine 486 leads to rapid nuclear export of Bach1 that allows Nrf2 to bind to the antioxidant response element and activate defensive gene expression. J Biol Chem. 285:153–162. doi: 10.1074/jbc.M109.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, Hayashi N, Yamamoto M, Igarashi K. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275:15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 106.Muto A, Tashiro S, Tsuchiya H, Kume A, Kanno M, Ito E, Yamamoto M, Igarashi K. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J Biol Chem. 2002;277:20724–20733. doi: 10.1074/jbc.M112003200. [DOI] [PubMed] [Google Scholar]
- 107.Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol Cell Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ki SH, Cho IJ, Choi DW, Kim SG. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol Cell Biol. 2005;25:4150–4165. doi: 10.1128/MCB.25.10.4150-4165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iwasaki K, Hailemariam K, Tsuji Y. PIAS3 interacts with ATF1 and regulates the human ferritin H gene through an antioxidant-responsive element. J Biol Chem. 2007;282:22335–22343. doi: 10.1074/jbc.M701477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 113.Novotny V, Prieschl EE, Csonga R, Fabjani G, Baumruker T. Nrf1 in a complex with fosB, c-jun, junD and ATF2 forms the AP1 component at the TNF alpha promoter in stimulated mast cells. Nucleic Acids Res. 1998;26:5480–5485. doi: 10.1093/nar/26.23.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levy S, Jaiswal AK, Forman HJ. The Role of c-Jun Phosphorylation in EpRE Activation of Phase II Genes. Free Radic Biol Med. 2009 doi: 10.1016/j.freeradbiomed.2009.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim YC, Masutani H, Yamaguchi Y, Itoh K, Yamamoto M, Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 116.Ikeda Y, Sugawara A, Taniyama Y, Uruno A, Igarashi K, Arima S, Ito S, Takeuchi K. Suppression of rat thromboxane synthase gene transcription by peroxisome proliferator-activated receptor gamma in macrophages via an interaction with NRF2. J Biol Chem. 2000;275:33142–33150. doi: 10.1074/jbc.M002319200. [DOI] [PubMed] [Google Scholar]
- 117.Ansell PJ, Lo SC, Newton LG, Espinosa-Nicholas C, Zhang DD, Liu JH, Hannink M, Lubahn DB. Repression of cancer protective genes by 17beta-estradiol: Ligand-dependent interaction between human Nrf2 and estrogen receptor alpha. Mol Cell Endocrinol. 2005;243:27–34. doi: 10.1016/j.mce.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 118.Zhou W, Lo SC, Liu JH, Hannink M, Lubahn DB. ERRbeta: a potent inhibitor of Nrf2 transcriptional activity. Mol Cell Endocrinol. 2007;278:52–62. doi: 10.1016/j.mce.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 119.Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci U S A. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brown SL, Sekhar KR, Rachakonda G, Sasi S, Freeman ML. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res. 2008;68:364–368. doi: 10.1158/0008-5472.CAN-07-2170. [DOI] [PubMed] [Google Scholar]
- 121.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 122.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:Quinone oxidoreductase1 gene. J Biol Chem. 2000;275:40134–40141. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 123.Dhakshinamoorthy S, Jaiswal AK. c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction. Oncogene. 2002;21:5301–5312. doi: 10.1038/sj.onc.1205642. [DOI] [PubMed] [Google Scholar]
- 124.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 125.Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 127.Chenais B, Derjuga A, Massrieh W, Red-Horse K, Bellingard V, Fisher SJ, Blank V. Functional and placental expression analysis of the human NRF3 transcription factor. Mol Endocrinol. 2005;19:125–137. doi: 10.1210/me.2003-0379. [DOI] [PubMed] [Google Scholar]
- 128.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hubbs AF, Benkovic SA, Miller DB, O’Callaghan JP, Battelli L, Schwegler-Berry D, Ma Q. Vacuolar leukoencephalopathy with widespread astrogliosis in mice lacking transcription factor Nrf2. Am J Pathol. 2007;170:2068–2076. doi: 10.2353/ajpath.2007.060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yoh K, Itoh K, Enomoto A, Hirayama A, Yamaguchi N, Kobayashi M, Morito N, Koyama A, Yamamoto M, Takahashi S. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60:1343–1353. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 133.Li J, Stein TD, Johnson JA. Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiol Genomics. 2004;18:261–272. doi: 10.1152/physiolgenomics.00209.2003. [DOI] [PubMed] [Google Scholar]
- 134.Ma Q, Battelli L, Hubbs AF. Multiorgan autoimmune inflammation, enhanced lymphoproliferation, and impaired homeostasis of reactive oxygen species in mice lacking the antioxidant-activated transcription factor Nrf2. Am J Pathol. 2006;168:1960–1974. doi: 10.2353/ajpath.2006.051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY. Nrf1 is critical for redox balance and survival of liver cells during development. Mol Cell Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 137.Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc Natl Acad Sci U S A. 1995;92:8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kuroha T, Takahashi S, Komeno T, Itoh K, Nagasawa T, Yamamoto M. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J Biochem (Tokyo) 1998;123:376–379. doi: 10.1093/oxfordjournals.jbchem.a021947. [DOI] [PubMed] [Google Scholar]
- 139.Martin F, van Deursen JM, Shivdasani RA, Jackson CW, Troutman AG, Ney PA. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood. 1998;91:3459–3466. [PubMed] [Google Scholar]
- 140.Motohashi H, Kimura M, Fujita R, Inoue A, Pan X, Takayama M, Katsuoka F, Aburatani H, Bresnick EH, Yamamoto M. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood. 115:677–686. doi: 10.1182/blood-2009-05-223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Y, Lucocq JM, Yamamoto M, Hayes JD. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem J. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nouhi Z, Chevillard G, Derjuga A, Blank V. Endoplasmic reticulum association and N-linked glycosylation of the human Nrf3 transcription factor. FEBS Lett. 2007;581:5401–5406. doi: 10.1016/j.febslet.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 145.Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 Transcription Factor Is a Membrane-bound Glycoprotein Targeted to the Endoplasmic Reticulum through Its N-terminal Homology Box 1 Sequence. J Biol Chem. 2009;284:3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Y, Lucocq JM, Hayes JD. The Nrf1 CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem J. 2009;418:293–310. doi: 10.1042/BJ20081575. [DOI] [PubMed] [Google Scholar]
- 147.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 148.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pepe AE, Xiao Q, Zampetaki A, Zhang Z, Kobayashi A, Hu Y, Xu Q. Crucial Role of Nrf3 in Smooth Muscle Cell Differentiation From Stem Cells. Circ Res. doi: 10.1161/CIRCRESAHA.109.211417. [DOI] [PubMed] [Google Scholar]
- 150.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 151.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 153.Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- 154.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kell A, Ventura N, Kahn N, Johnson TE. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic Biol Med. 2007;43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. A cap ‘n’ collar protein isoform contains a selective Hox repressor function. Development. 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- 161.Mohler J, Mahaffey JW, Deutsch E, Vani K. Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development. 1995;121:237–247. doi: 10.1242/dev.121.1.237. [DOI] [PubMed] [Google Scholar]
- 162.Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- 163.Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat Res. 2001;480–481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Y, Gordon GB. A strategy for cancer prevention: stimulation of the Nrf2-ARE signaling pathway. Mol Cancer Ther. 2004;3:885–893. [PubMed] [Google Scholar]
- 165.Sporn MB, Liby KT. Cancer chemoprevention: scientific promise, clinical uncertainty. Nat Clin Pract Oncol. 2005;2:518–525. doi: 10.1038/ncponc0319. [DOI] [PubMed] [Google Scholar]
- 166.Zhang Y, Munday R. Dithiolethiones for cancer chemoprevention: where do we stand? Mol Cancer Ther. 2008;7:3470–3479. doi: 10.1158/1535-7163.MCT-08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, Gribble GW, Johnson DA, Johnson JA, Burton NC, Guilarte TR, Yamamoto M, Sporn MB, Kensler TW. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 168.Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, Huang MT, Chan JY, Kong AN. Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5:39–51. doi: 10.1158/1535-7163.MCT-05-0293. [DOI] [PubMed] [Google Scholar]
- 169.Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell Mol Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 171.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yates MS, Kwak MK, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, Baumgartner KJ, Roebuck BD, Liby KT, Yore MM, Honda T, Gribble GW, Sporn MB, Kensler TW. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 173.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O’Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 174.Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24:461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 175.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 176.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. Faseb J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 178.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 179.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Burton NC, Kensler TW, Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–1100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 181.Kanninen K, Malm TM, Jyrkkanen HK, Goldsteins G, Keksa-Goldsteine V, Tanila H, Yamamoto M, Yla-Herttuala S, Levonen AL, Koistinaho J. Nuclear factor erythroid 2-related factor 2 protects against beta amyloid. Mol Cell Neurosci. 2008;39:302–313. doi: 10.1016/j.mcn.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 182.Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J Neurosci. 2008;28:13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580–11584. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- 184.Xu W, Hellerbrand C, Kohler UA, Bugnon P, Kan YW, Werner S, Beyer TA. The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab Invest. 2008;88:1068–1078. doi: 10.1038/labinvest.2008.75. [DOI] [PubMed] [Google Scholar]
- 185.Lamle J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 186.Collins AR, Lyon CJ, Xia X, Liu JZ, Tangirala RK, Yin F, Boyadjian R, Bikineyeva A, Pratico D, Harrison DG, Hsueh WA. Age-accelerated atherosclerosis correlates with failure to upregulate antioxidant genes. Circ Res. 2009;104:e42–54. doi: 10.1161/CIRCRESAHA.108.188771. [DOI] [PubMed] [Google Scholar]
- 187.Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 189.van Muiswinkel FL, Kuiperij HB. The Nrf2-ARE Signalling pathway: promising drug target to combat oxidative stress in neurodegenerative disorders. Curr Drug Targets CNS Neurol Disord. 2005;4:267–281. doi: 10.2174/1568007054038238. [DOI] [PubMed] [Google Scholar]
- 190.Kern JT, Hannink M, Hess JF. Disruption of the Keap1-containing ubiquitination complex as an antioxidant therapy. Curr Top Med Chem. 2007;7:972–978. doi: 10.2174/156802607780906825. [DOI] [PubMed] [Google Scholar]
- 191.Yamamoto T, Yoh K, Kobayashi A, Ishii Y, Kure S, Koyama A, Sakamoto T, Sekizawa K, Motohashi H, Yamamoto M. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun. 2004;321:72–79. doi: 10.1016/j.bbrc.2004.06.112. [DOI] [PubMed] [Google Scholar]
- 192.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. Faseb J. 2007 doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 193.Guan CP, Zhou MN, Xu AE, Kang KF, Liu JF, Wei XD, Li YW, Zhao DK, Hong WS. The susceptibility to vitiligo is associated with NF-E2-related factor2 (Nrf2) gene polymorphisms: a study on Chinese Han population. Exp Dermatol. 2008;17:1059–1062. doi: 10.1111/j.1600-0625.2008.00752.x. [DOI] [PubMed] [Google Scholar]
- 194.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Hasegawa S, Takagi T, Wang FY, Hirata I, Nakano H. The relationship between Helicobacter pylori infection and promoter polymorphism of the Nrf2 gene in chronic gastritis. Int J Mol Med. 2007;19:143–148. [PubMed] [Google Scholar]
- 195.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Yoshioka D, Arima Y, Okubo M, Hirata I, Nakano H. Association between promoter polymorphisms of nuclear factor-erythroid 2-related factor 2 gene and peptic ulcer diseases. Int J Mol Med. 2007;20:849–853. [PubMed] [Google Scholar]
- 196.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Yoshioka D, Okubo M, Sakata M, Wang FY, Hirata I, Nakano H. Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population. Hepatogastroenterology. 2008;55:394–397. [PubMed] [Google Scholar]
- 197.Calverley PM, Walker P. Chronic obstructive pulmonary disease. Lancet. 2003;362:1053–1061. doi: 10.1016/s0140-6736(03)14416-9. [DOI] [PubMed] [Google Scholar]
- 198.Mannino DM, Buist AS. Global burden of COPD: risk factors, prevalence, and future trends. Lancet. 2007;370:765–773. doi: 10.1016/S0140-6736(07)61380-4. [DOI] [PubMed] [Google Scholar]
- 199.Barnes PJ. Mediators of chronic obstructive pulmonary disease. Pharmacol Rev. 2004;56:515–548. doi: 10.1124/pr.56.4.2. [DOI] [PubMed] [Google Scholar]
- 200.Yoshida T, Tuder RM. Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol Rev. 2007;87:1047–1082. doi: 10.1152/physrev.00048.2006. [DOI] [PubMed] [Google Scholar]
- 201.Ishii Y, Itoh K, Morishima Y, Kimura T, Kiwamoto T, Iizuka T, Hegab AE, Hosoya T, Nomura A, Sakamoto T, Yamamoto M, Sekizawa K. Transcription Factor Nrf2 Plays a Pivotal Role in Protection against Elastase-Induced Pulmonary Inflammation and Emphysema. J Immunol. 2005;175:6968–6975. doi: 10.4049/jimmunol.175.10.6968. [DOI] [PubMed] [Google Scholar]
- 202.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, Sporn MB, Gabrielson KL, Champion HC, Tuder RM, Kensler TW, Biswal S. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci U S A. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, Crestani B, Fournier M, Leseche G, Soler P, Boczkowski J, Bonay M. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–924. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 204.Malhotra D, Thimmulappa R, Navas-Acien A, Sandford A, Elliott M, Singh A, Chen L, Zhuang X, Hogg J, Pare P, Tuder RM, Biswal S. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am J Respir Crit Care Med. 2008;178:592–604. doi: 10.1164/rccm.200803-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 205.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc Natl Acad Sci U S A. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ooe H, Maita C, Maita H, Iguchi-Ariga SM, Ariga H. Specific cleavage of DJ-1 under an oxidative condition. Neurosci Lett. 2006;406:165–168. doi: 10.1016/j.neulet.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 208.Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, Kondo S, Nishimura M. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2008;39:673–682. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- 209.de Vries HE, Witte M, Hondius D, Rozemuller AJ, Drukarch B, Hoozemans J, van Horssen J. Nrf2-induced antioxidant protection: a promising target to counteract ROS-mediated damage in neurodegenerative disease? Free Radic Biol Med. 2008;45:1375–1383. doi: 10.1016/j.freeradbiomed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 210.Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, Chen PC. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 212.Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc Natl Acad Sci U S A. 2005;102:244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jakel RJ, Townsend JA, Kraft AD, Johnson JA. Nrf2-mediated protection against 6-hydroxydopamine. Brain Res. 2007;1144:192–201. doi: 10.1016/j.brainres.2007.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yang L, Calingasan NY, Thomas B, Chaturvedi RK, Kiaei M, Wille EJ, Liby KT, Williams C, Royce D, Risingsong R, Musiek ES, Morrow JD, Sporn M, Beal MF. Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One. 2009;4:e5757. doi: 10.1371/journal.pone.0005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kraft AD, Resch JM, Johnson DA, Johnson JA. Activation of the Nrf2-ARE pathway in muscle and spinal cord during ALS-like pathology in mice expressing mutant SOD1. Exp Neurol. 2007;207:107–117. doi: 10.1016/j.expneurol.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dumont M, Wille E, Calingasan NY, Tampellini D, Williams C, Gouras GK, Liby K, Sporn M, Nathan C, Flint Beal M, Lin MT. Triterpenoid CDDO-methylamide improves memory and decreases amyloid plaques in a transgenic mouse model of Alzheimer’s disease. J Neurochem. 2009;109:502–512. doi: 10.1111/j.1471-4159.2009.05970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lim JH, Kim KM, Kim SW, Hwang O, Choi HJ. Bromocriptine activates NQO1 via Nrf2-PI3K/Akt signaling: novel cytoprotective mechanism against oxidative damage. Pharmacol Res. 2008;57:325–331. doi: 10.1016/j.phrs.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 221.Nakaso K, Nakamura C, Sato H, Imamura K, Takeshima T, Nakashima K. Novel cytoprotective mechanism of anti-parkinsonian drug deprenyl: PI3K and Nrf2-derived induction of antioxidative proteins. Biochem Biophys Res Commun. 2006;339:915–922. doi: 10.1016/j.bbrc.2005.11.095. [DOI] [PubMed] [Google Scholar]
- 222.Paupe V, Dassa EP, Goncalves S, Auchere F, Lonn M, Holmgren A, Rustin P. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PLoS One. 2009;4:e4253. doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, Hamilton RL, Chu CT, Jordan-Sciutto KL. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66:75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sarlette A, Krampfl K, Grothe C, Neuhoff N, Dengler R, Petri S. Nuclear erythroid 2-related factor 2-antioxidative response element signaling pathway in motor cortex and spinal cord in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2008;67:1055–1062. doi: 10.1097/NEN.0b013e31818b4906. [DOI] [PubMed] [Google Scholar]
- 225.Bacskai BJ, Hyman BT. Alzheimer’s disease: what multiphoton microscopy teaches us. Neuroscientist. 2002;8:386–390. doi: 10.1177/107385802236963. [DOI] [PubMed] [Google Scholar]
- 226.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 227.Mitchell JD, Borasio GD. Amyotrophic lateral sclerosis. Lancet. 2007;369:2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]
- 228.Eisen A. Amyotrophic lateral sclerosis: A 40-year personal perspective. J Clin Neurosci. 2009;16:505–512. doi: 10.1016/j.jocn.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 229.Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9:198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- 230.Pehar M, Vargas MR, Robinson KM, Cassina P, Diaz-Amarilla PJ, Hagen TM, Radi R, Barbeito L, Beckman JS. Mitochondrial superoxide production and nuclear factor erythroid 2-related factor 2 activation in p75 neurotrophin receptor-induced motor neuron apoptosis. J Neurosci. 2007;27:7777–7785. doi: 10.1523/JNEUROSCI.0823-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kirby J, Halligan E, Baptista MJ, Allen S, Heath PR, Holden H, Barber SC, Loynes CA, Wood-Allum CA, Lunec J, Shaw PJ. Mutant SOD1 alters the motor neuronal transcriptome: implications for familial ALS. Brain. 2005;128:1686–1706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- 232.Vargas MR, Pehar M, Cassina P, Martinez-Palma L, Thompson JA, Beckman JS, Barbeito L. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes: consequences for motor neuron survival. J Biol Chem. 2005;280:25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- 233.Liu YL, Guo YS, Xu L, Wu SY, Wu DX, Yang C, Li CY. Ultrastructural evidence of neurofilament involvement in immune-mediated motor neuron injury. Neurol Res. 2008;30:990–994. doi: 10.1179/016164108X323780. [DOI] [PubMed] [Google Scholar]
- 234.Guo Y, Liu Y, Xu L, Wu D, Wu H, Li CY. Reduced Nrf2 and Phase II enzymes expression in immune-mediated spinal cord motor neuron injury. Neurol Res. 2009 doi: 10.1179/174313209X385563. [DOI] [PubMed] [Google Scholar]
- 235.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 237.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 239.Ishii T, Itoh K, Sato H, Bannai S. Oxidative stress-inducible proteins in macrophages. Free Radic Res. 1999;31:351–355. doi: 10.1080/10715769900300921. [DOI] [PubMed] [Google Scholar]
- 240.Aono J, Yanagawa T, Itoh K, Li B, Yoshida H, Kumagai Y, Yamamoto M, Ishii T. Activation of Nrf2 and accumulation of ubiquitinated A170 by arsenic in osteoblasts. Biochem Biophys Res Commun. 2003;305:271–277. doi: 10.1016/s0006-291x(03)00728-9. [DOI] [PubMed] [Google Scholar]
- 241.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res. 2004;94:609–616. doi: 10.1161/01.RES.0000119171.44657.45. [DOI] [PubMed] [Google Scholar]
- 242.Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci U S A. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Du Y, Wooten MC, Gearing M, Wooten MW. Age-associated oxidative damage to the p62 promoter: implications for Alzheimer disease. Free Radic Biol Med. 2009;46:492–501. doi: 10.1016/j.freeradbiomed.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Du Y, Wooten MC, Wooten MW. Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiol Dis. 2009;35:302–310. doi: 10.1016/j.nbd.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.van Duijn CM, Dekker MC, Bonifati V, Galjaard RJ, Houwing-Duistermaat JJ, Snijders PJ, Testers L, Breedveld GJ, Horstink M, Sandkuijl LA, van Swieten JC, Oostra BA, Heutink P. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am J Hum Genet. 2001;69:629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 247.Lev N, Ickowicz D, Barhum Y, Melamed E, Offen D. DJ-1 changes in G93A-SOD1 transgenic mice: implications for oxidative stress in ALS. J Mol Neurosci. 2009;38:94–102. doi: 10.1007/s12031-008-9138-7. [DOI] [PubMed] [Google Scholar]
- 248.Gal J, Strom AL, Kilty R, Zhang F, Zhu H. p62 accumulates and enhances aggregate formation in model systems of familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282:11068–11077. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- 249.Mizuno Y, Amari M, Takatama M, Aizawa H, Mihara B, Okamoto K. Immunoreactivities of p62, an ubiqutin-binding protein, in the spinal anterior horn cells of patients with amyotrophic lateral sclerosis. J Neurol Sci. 2006;249:13–18. doi: 10.1016/j.jns.2006.05.060. [DOI] [PubMed] [Google Scholar]
- 250.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 251.Harman D. Free radical theory of aging: an update: increasing the functional life span. Ann N Y Acad Sci. 2006;1067:10–21. doi: 10.1196/annals.1354.003. [DOI] [PubMed] [Google Scholar]
- 252.Dugan LL, Quick KL. Reactive oxygen species and aging: evolving questions. Sci Aging Knowledge Environ. 2005;2005:pe20. doi: 10.1126/sageke.2005.26.pe20. [DOI] [PubMed] [Google Scholar]
- 253.Luchak JM, Prabhudesai L, Sohal RS, Radyuk SN, Orr WC. Modulating longevity in Drosophila by over- and underexpression of glutamate-cysteine ligase. Ann N Y Acad Sci. 2007;1119:260–273. doi: 10.1196/annals.1404.000. [DOI] [PubMed] [Google Scholar]
- 254.Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 255.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 257.Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, Poosala S, Csiszar A, Ungvari Z, Kensler TW, Yamamoto M, Egan JM, Longo DL, Ingram DK, Navas P, de Cabo R. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci U S A. 2008;105:2325–2330. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hyun DH, Emerson SS, Jo DG, Mattson MP, de Cabo R. Calorie restriction up-regulates the plasma membrane redox system in brain cells and suppresses oxidative stress during aging. Proc Natl Acad Sci U S A. 2006;103:19908–19912. doi: 10.1073/pnas.0608008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kritchevsky D. Caloric restriction and experimental carcinogenesis. Hybrid Hybridomics. 2002;21:147–151. doi: 10.1089/153685902317401753. [DOI] [PubMed] [Google Scholar]
- 261.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 262.Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 263.Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 264.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 265.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Suh JH, Shenvi SV, Dixon BM, Liu H, Jaiswal AK, Liu RM, Hagen TM. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci U S A. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shih PH, Yen GC. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signaling pathway. Biogerontology. 2007;8:71–80. doi: 10.1007/s10522-006-9033-y. [DOI] [PubMed] [Google Scholar]
- 269.Noyan-Ashraf MH, Sadeghinejad Z, Davies GF, Ross AR, Saucier D, Harkness TA, Juurlink BH. Phase 2 protein inducers in the diet promote healthier aging. J Gerontol A Biol Sci Med Sci. 2008;63:1168–1176. doi: 10.1093/gerona/63.11.1168. [DOI] [PubMed] [Google Scholar]
- 270.Kim HJ, Barajas B, Wang M, Nel AE. Nrf2 activation by sulforaphane restores the age-related decrease of T(H)1 immunity: role of dendritic cells. J Allergy Clin Immunol. 2008;121:1255–1261. e1257. doi: 10.1016/j.jaci.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Duan W, Zhang R, Guo Y, Jiang Y, Huang Y, Jiang H, Li C. Nrf2 activity is lost in the spinal cord and its astrocytes of aged mice. In Vitro Cell Dev Biol Anim. 2009;45:388–397. doi: 10.1007/s11626-009-9194-5. [DOI] [PubMed] [Google Scholar]
- 272.Habeos IG, Ziros PG, Chartoumpekis D, Psyrogiannis A, Kyriazopoulou V, Papavassiliou AG. Simvastatin activates Keap1/Nrf2 signaling in rat liver. J Mol Med. 2008;86:1279–1285. doi: 10.1007/s00109-008-0393-4. [DOI] [PubMed] [Google Scholar]
- 273.Ali F, Zakkar M, Karu K, Lidington EA, Hamdulay SS, Boyle JJ, Zloh M, Bauer A, Haskard DO, Evans PC, Mason JC. Induction of the cytoprotective enzyme heme oxygenase-1 by statins is enhanced in vascular endothelium exposed to laminar shear stress and impaired by disturbed flow. J Biol Chem. 2009;284:18882–18892. doi: 10.1074/jbc.M109.009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Przybysz AJ, Choe KP, Roberts LJ, Strange K. Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech Ageing Dev. 2009;130:357–369. doi: 10.1016/j.mad.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zaman MB, Leonard MO, Ryan EJ, Nolan NP, Hoti E, Maguire D, Mulcahy H, Traynor O, Taylor CT, Hegarty JE, Geoghegan JG, O’Farrelly C. Lower expression of Nrf2 mRNA in older donor livers: a possible contributor to increased ischemia-reperfusion injury? Transplantation. 2007;84:1272–1278. doi: 10.1097/01.tp.0000288229.53064.e2. [DOI] [PubMed] [Google Scholar]
- 276.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 277.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hayes JD, McMahon M. The double-edged sword of Nrf2: subversion of redox homeostasis during the evolution of cancer. Mol Cell. 2006;21:732–734. doi: 10.1016/j.molcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 279.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 281.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 282.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. 1368, e1351–1354. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 283.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 284.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett. 2008;260:96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 286.Kim SK, Yang JW, Kim MR, Roh SH, Kim HG, Lee KY, Jeong HG, Kang KW. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 287.Tarumoto T, Nagai T, Ohmine K, Miyoshi T, Nakamura M, Kondo T, Mitsugi K, Nakano S, Muroi K, Komatsu N, Ozawa K. Ascorbic acid restores sensitivity to imatinib via suppression of Nrf2-dependent gene expression in the imatinib-resistant cell line. Exp Hematol. 2004;32:375–381. doi: 10.1016/j.exphem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 288.Pi J, Diwan BA, Sun Y, Liu J, Qu W, He Y, Styblo M, Waalkes MP. Arsenic-induced malignant transformation of human keratinocytes: involvement of Nrf2. Free Radic Biol Med. 2008;45:651–658. doi: 10.1016/j.freeradbiomed.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N, Itoh K, Yamamoto M. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 290.Loignon M, Miao W, Hu L, Bier A, Bismar TA, Scrivens PJ, Mann K, Basik M, Bouchard A, Fiset PO, Batist Z, Batist G. Cul3 overexpression depletes Nrf2 in breast cancer and is associated with sensitivity to carcinogens, to oxidative stress, and to chemotherapy. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-1186. [DOI] [PubMed] [Google Scholar]
- 291.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson E, Feinstein E, Biswal S. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Devling TW, Lindsay CD, McLellan LI, McMahon M, Hayes JD. Utility of siRNA against Keap1 as a strategy to stimulate a cancer chemopreventive phenotype. Proc Natl Acad Sci U S A. 2005;102:7280–7285A. doi: 10.1073/pnas.0501475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jain AK, Mahajan S, Jaiswal AK. Phosphorylation and dephosphorylation of tyrosine 141 regulate stability and degradation of INrf2: a novel mechanism in Nrf2 activation. J Biol Chem. 2008;283:17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 294.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 295.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 296.Strachan GD, Morgan KL, Otis LL, Caltagarone J, Gittis A, Bowser R, Jordan-Sciutto KL. Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein. Biochemistry. 2004;43:12113–12122. doi: 10.1021/bi0494166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 299.Ito N, Fukushima S, Tsuda H. Carcinogenicity and modification of the carcinogenic response by BHA, BHT, and other antioxidants. Crit Rev Toxicol. 1985;15:109–150. doi: 10.3109/10408448509029322. [DOI] [PubMed] [Google Scholar]
- 300.Wang XJ, Hayes JD, Wolf CR. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 301.Wondrak GT, Cabello CM, Villeneuve NF, Zhang S, Ley S, Li Y, Sun Z, Zhang DD. Cinnamoyl-based Nrf2-activators targeting human skin cell photo-oxidative stress. Free Radic Biol Med. 2008;45:385–395. doi: 10.1016/j.freeradbiomed.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Reisman SA, Csanaky IL, Aleksunes LM, Klaassen CD. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol Sci. 2009;109:31–40. doi: 10.1093/toxsci/kfp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Reisman SA, Csanaky IL, Yeager RL, Klaassen CD. Nrf2 activation enhances biliary excretion of sulfobromophthalein by inducing glutathione-S-transferase activity. Toxicol Sci. 2009;109:24–30. doi: 10.1093/toxsci/kfp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Primiano T, Egner PA, Sutter TR, Kelloff GJ, Roebuck BD, Kensler TW. Intermittent dosing with oltipraz: relationship between chemoprevention of aflatoxin-induced tumorigenesis and induction of glutathione S-transferases. Cancer Res. 1995;55:4319–4324. [PubMed] [Google Scholar]
- 305.Bae SK, Lee SJ, Kim T, Kim JW, Lee I, Kim SG, Lee MG. Pharmacokinetics and therapeutic effects of oltipraz after consecutive or intermittent oral administration in rats with liver cirrhosis induced by dimethylnitrosamine. J Pharm Sci. 2006;95:985–997. doi: 10.1002/jps.20597. [DOI] [PubMed] [Google Scholar]
- 306.Rojo AI, Sagarra MR, Cuadrado A. GSK-3beta down-regulates the transcription factor Nrf2 after oxidant damage: relevance to exposure of neuronal cells to oxidative stress. J Neurochem. 2008;105:192–202. doi: 10.1111/j.1471-4159.2007.05124.x. [DOI] [PubMed] [Google Scholar]
- 307.Cuadrado A, Moreno-Murciano P, Pedraza-Chaverri J. The transcription factor Nrf2 as a new therapeutic target in Parkinson’s disease. Expert Opin Ther Targets. 2009;13:319–329. doi: 10.1517/13543780802716501. [DOI] [PubMed] [Google Scholar]
- 308.Wruck CJ, Huppertz B, Bose P, Brandenburg LO, Pufe T, Kadyrov M. Role of a fetal defence mechanism against oxidative stress in the aetiology of preeclampsia. Histopathology. 2009;55:102–106. doi: 10.1111/j.1365-2559.2009.03339.x. [DOI] [PubMed] [Google Scholar]
- 309.Kruidenier L, Kuiper I, Van Duijn W, Mieremet-Ooms MA, van Hogezand RA, Lamers CB, Verspaget HW. Imbalanced secondary mucosal antioxidant response in inflammatory bowel disease. J Pathol. 2003;201:17–27. doi: 10.1002/path.1408. [DOI] [PubMed] [Google Scholar]
- 310.Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121:1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- 311.Theiss AL, Vijay-Kumar M, Obertone TS, Jones DP, Hansen JM, Gewirtz AT, Merlin D, Sitaraman SV. Prohibitin is a novel regulator of antioxidant response that attenuates colonic inflammation in mice. Gastroenterology. 2009;137:199–208. e191–196. doi: 10.1053/j.gastro.2009.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Siedlinski M, Postma DS, Boer JM, van der Steege G, Schouten JP, Smit HA, Boezen HM. Level and course of FEV1 in relation to polymorphisms in NFE2L2 and KEAP1 in the general population. Respir Res. 2009;10:73. doi: 10.1186/1465-9921-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Reddy NM, Suryanaraya V, Yates MS, Kleeberger SR, Hassoun PM, Yamamoto M, Liby KT, Sporn MB, Kensler TW, Reddy SP. The Triterpenoid CDDO-Imidazolide Confers Potent Protection Against Hyperoxic Acute Lung Injury in Mice. Am J Respir Crit Care Med. 2009 doi: 10.1164/rccm.200905-0670OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhao J, Moore AN, Redell JB, Dash PK. Enhancing expression of Nrf2-driven genes protects the blood brain barrier after brain injury. J Neurosci. 2007;27:10240–10248. doi: 10.1523/JNEUROSCI.1683-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Dash PK, Zhao J, Orsi SA, Zhang M, Moore AN. Sulforaphane improves cognitive function administered following traumatic brain injury. Neurosci Lett. 2009;460:103–107. doi: 10.1016/j.neulet.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zhao J, Kobori N, Aronowski J, Dash PK. Sulforaphane reduces infarct volume following focal cerebral ischemia in rodents. Neurosci Lett. 2006;393:108–112. doi: 10.1016/j.neulet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 318.Zhao X, Sun G, Zhang J, Strong R, Dash PK, Kan YW, Grotta JC, Aronowski J. Transcription factor Nrf2 protects the brain from damage produced by intracerebral hemorrhage. Stroke. 2007;38:3280–3286. doi: 10.1161/STROKEAHA.107.486506. [DOI] [PubMed] [Google Scholar]
- 319.Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26:817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 320.So HS, Kim HJ, Lee JH, Park SY, Park C, Kim YH, Kim JK, Lee KM, Kim KS, Chung SY, Jang WC, Moon SK, Chung HT, Park RK. Flunarizine induces Nrf2-mediated transcriptional activation of heme oxygenase-1 in protection of auditory cells from cisplatin. Cell Death Differ. 2006;13:1763–1775. doi: 10.1038/sj.cdd.4401863. [DOI] [PubMed] [Google Scholar]
- 321.Park HM, Cho JM, Lee HR, Shim GS, Kwak MK. Renal protection by 3H-1,2-dithiole-3-thione against cisplatin through the Nrf2-antioxidant pathway. Biochem Pharmacol. 2008;76:597–607. doi: 10.1016/j.bcp.2008.06.021. [DOI] [PubMed] [Google Scholar]