Abstract
Most metazoans have at least some ability to regenerate damaged cells and tissues, although the regenerative capacity varies depending on the species, organ, or developmental stage. Cell replacement and regeneration occur in two contexts: renewal of spent cells during tissue homeostasis (homeostatic growth), and in response to external injury, wounding, or amputation (epimorphic regeneration). Model organisms that display remarkable regenerative capacity include amphibians, planarians, Hydra, and the vertebrate liver. In addition, several mammalian organs—including the skin, gut, kidney, muscle, and even the human nervous system—have some ability to replace spent or damaged cells. Although the regenerative response is complex, it typically involves the induction of new cell proliferation through formation of a blastema, followed by cell specification, differentiation, and patterning. Stem cells and undifferentiated progenitor cells play an important role in both tissue homeostasis and tissue regeneration. Stem cells are typically quiescent or passing slowly through the cell cycle in adult tissues, but they can be activated in response to cell loss and wounding. A series of studies, mostly performed in Drosophila as well as in Hydra, Xenopus, and mouse, has revealed an unexpected role of apoptotic caspases in the production of mitogenic signals that stimulate the proliferation of stem and progenitor cells to aid in tissue regeneration. This Review summarizes some of the key findings and discusses links to stem cell biology and cancer.
Regulation of Apoptosis
Most animal cells have the ability to self-destruct by undergoing apoptosis, a morphologically distinct form of programmed cell death (1). The proper regulation of apoptosis is critical for both development and tissue homeostasis, and inhibition of apoptosis contributes to the development and progression of cancer (2, 3). A central step for the execution of apoptosis is the activation of caspases, a family of cysteine proteases that are present as weakly active zymogens in virtually all cells (4). Caspases are activated by proteolytic cleavage of the zymogen in response to different stimuli, including developmental signals, as well as various forms of persistent cellular stress or injury, such as DNA damage, viral infection, hypoxia, increased presence of reactive oxygen species, loss of cellular adhesion, accumulation of unfolded proteins, excitotoxicity, shear stress, cytoskeletal damage, and other insults (1). Apoptosis can also be induced indirectly by cells that undergo necrosis in response to overwhelming physical injury. In all these cases, apoptosis appears to serve as an efficient cellular quality control mechanism that removes dysfunctional, unwanted, and potentially dangerous cells from the organism.
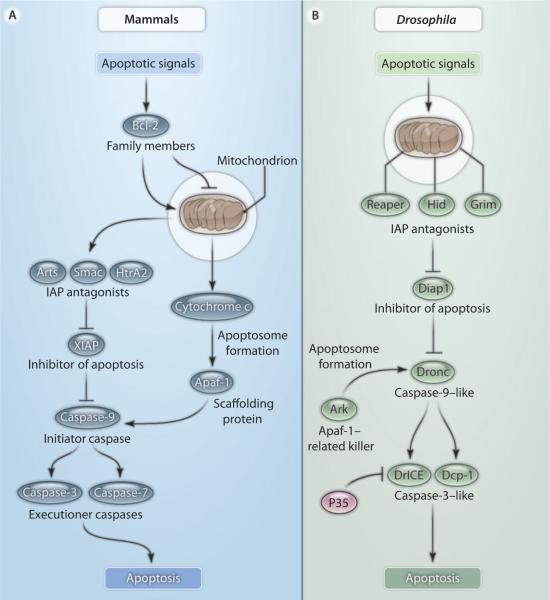
Caspases are classified according to the length of their prodomains. Initiator caspases have long prodomains, whereas executioner caspases have short prodomains. The prodomain of the initiator caspases contains protein-protein interaction motifs for binding to upstream adaptor molecules. For example, mammalian caspase-9 is activated upon recruitment into the apoptosome complex with the adaptor protein Apaf-1 and cytochrome c (Fig. 1A) (5, 6). An important event for apoptosome formation is the release of cytochrome c from mitochondria, a step that is regulated by Bcl-2 family proteins (Fig. 1A) (7–9). Once initiator caspases have been activated, they promote cleavage and activation of executioner caspases, such as caspase-3 and caspase-7 (Fig. 1A), which then induce the demise of the cell. Caspases are well conserved in the animal kingdom. In Drosophila, they are represented by one caspase-9 ortholog, termed Dronc, and two caspase-3–like proteins, Dcp-1 and DrICE (Fig. 1B) (10–12).
Fig. 1.
The apoptosis pathways in mammals and Drosophila. (A) In mammals, apoptotic signals control the balance of pro- and antiapoptotic Bcl-2 family members and thereby the release of cytochrome c and IAP antagonists from mitochondria. Binding of cytochrome c to Apaf-1 promotes apoptosome formation. The apoptosome recruits the initiator caspase-9, which leads to apoptosome activation. At the same time, IAP antagonists release caspases from inhibition by IAPs, most notably XIAP (X-linked inhibitor of apoptosis protein). Activated caspase-9 cleaves and in turn activates executioner caspases such as caspase-3 and caspase-7. (B) In Drosophila, apoptotic signals control the abundance and activity of the IAP antagonists Reaper, Hid, and Grim and recruit them to mitochondria, where these proteins promote ubiquitin-mediated degradation of Drosophila IAP1 (Diap1) (22–27). Release of Dronc from inhibition by Diap1 enables it to associate with the scaffolding protein Ark, which creates an apoptosome-like structure that activates the executioner caspases DrICE and Dcp-1. The baculovirus protein P35 specifically inhibits the activity of DrICE and Dcp-1 in Drosophila.
In addition to zymogen processing, an equally important layer of cell death control involves the inhibition of caspases (13, 14). One important family of caspase inhibitors are the IAP (inhibitor of apoptosis proteins), which can bind to and inhibit caspases (Fig. 1) (15, 16). IAPs were originally discovered in insect viruses (17), but a family of related proteins was subsequently described in both insect and mammalian genomes. IAPs are characterized by the presence of at least one BIR (baculovirus inhibitory repeat) domain, which can directly bind to and inhibit caspases (15, 16). Several IAPs also carry a RING (really interesting new gene) E3 ubiquitin ligase domain that promotes ubiquitination of key cell death regulators (16, 18, 19). In cells that are committed to die, IAPs are inhibited by a family of IAP antagonists, such as Reaper, Hid, and Grim, that were initially discovered in Drosophila (Fig. 1B) (20, 21). Reaper, Hid, and Grim must localize to mitochondria to inhibit the apoptotic activity of IAPs (Fig. 1B) (22–27). IAP antagonists contain an N-terminal IAP-binding motif (IBM) that is necessary for binding to the BIR domains of and inhibiting Drosophila IAP1 (Diap1), thus releasing the caspases Dronc and DrICE from Diap1 inhibition (Fig. 1B). A similar IBM motif has been identified in mammalian IAP antagonists, including Smac (known as Diablo in mouse) and HtrA2 (also known as Omi) (Fig. 1A) (28–30). In summary, caspase regulation is under dual control by both activating factors (Apaf-1 and cytochrome c) and inhibitory factors (IAPs), whose activity is in turn regulated by a complex network of upstream signaling pathways (Fig. 1).
Compensatory Proliferation Triggered by Cells That Have Initiated But Not Executed Apoptosis
Many tissues can tolerate a surprising extent of cell death and compensate for the loss of cells through increased cell proliferation and regeneration (31). For example, a full-sized mammalian liver can be regenerated after 75% of the organ has been removed, and developing Drosophila imaginal discs—the larval precursor structures of adult legs, wings, and eyes—can form a normal-sized and patterned organ even after more than 50% of their cells have been killed (32–34). Depending on the type of tissue damage, these regeneration processes involve several steps, including wound healing, the formation of proliferative blastema cells, differentiation, and patterning (35). As proposed in 1988 (36), work in several model organisms—including Drosophila, Hydra, planarians, Xenopus, newt, and mouse—has revealed that, unexpectedly, apoptosis may be the driving force for cell proliferation during tissue regeneration (37–46). Specifically, the proliferation component of regeneration, including the formation of blastema cells, is under the control of apoptotic cells. This phenomenon has been termed “apoptosis-induced compensatory proliferation” (47).
Studies in Hydra have illustrated the link between apoptosis and cellular proliferation in regeneration (39). There are three distinct types of regeneration after amputation in this organism: foot regeneration, apical head regeneration after decapitation, and basal head regeneration after mid-gastric section. However, apoptosis and cellular proliferation were observed only during basal head regeneration. When apoptosis was experimentally induced in the foot-regenerating tip, cell proliferation was observed and the regeneration process was transformed into a head-regenerating one (39). These observations demonstrate that cell proliferation is triggered by apoptosis during regeneration. They also illustrate that although apoptotic cells sooner or later are removed from the organism by phagocytosis (Fig. 2A), they are to some extent still metabolically active and appear to be able to stimulate proliferation and regeneration.
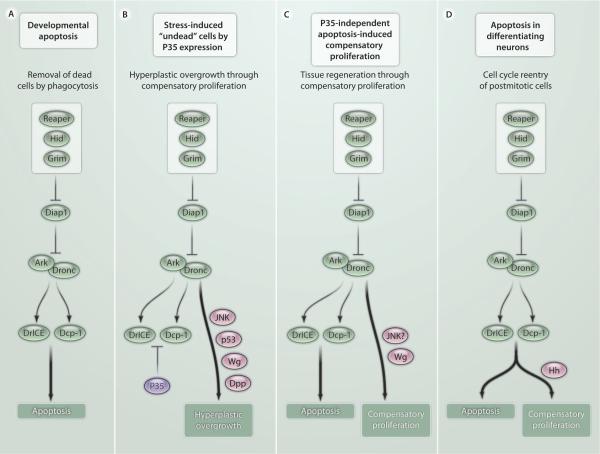
Fig. 2.
Models of apoptosis-induced compensatory proliferation in Drosophila. (A) Normal developmental apoptosis results in removal of dead cells by phagocytosis. An example of a situation in which compensatory proliferation occurs after normal developmental apoptosis is when dying enterocytes in the Drosophila midgut stimulate intestinal stem cell proliferation. (B) P35 expression renders apoptotic cells unable to complete apoptosis. Dronc-dependent p53 and JNK signaling triggers the release of signaling cytokines such as Wg and Dpp, which promote hyperplastic overgrowth of the affected tissue. (C) Induction of apoptosis in a temporally and spatially restricted manner without P35 expression results in tissue regeneration through compensatory proliferation. The question marks indicate that it is unclear whether JNK signaling operates either in dying cells or regenerating cells. (D) Apoptosis induction in differentiating photoreceptor neurons in the Drosophila retina engages a different pathway of compensatory proliferation. Dying photoreceptor neurons release Hedgehog (Hh) in a manner that requires DrICE and Dcp-1. Hh stimulates cell cycle reentry of postmitotic non-neuronal cells.
The original demonstration that cells that undergo apoptosis in response to stress or injury can stimulate cell proliferation and tissue regeneration came from a series of studies in Drosophila (34, 37, 48). In these studies, massive apoptosis was induced by x-ray radiation, overexpression of IAP antagonists, or loss of Diap1. The key to identifying the mechanisms and signals involved in compensatory proliferation was to keep apoptotic cells alive by expression of P35, a potent inhibitor of executioner caspases (Fig. 2B) (49). Under these conditions, the apoptotic program is induced but cannot be executed because P35 inhibits executioner caspases, thus producing what has been termed “undead” cells. However, P35 inhibits DrICE and Dcp-1, but not the initiator caspase Dronc (Fig. 1B); hence, Dronc is active in these cells. Therefore, although Dronc cannot induce apoptosis, it does promote nonapoptotic functions, such as the induction of compensatory proliferation, which can lead to hyperplastic overgrowth (Fig. 2B) (34, 37, 38, 48, 50, 51). Consistently, mutations in dronc block compensatory proliferation in apoptotic larval wing discs (38, 48, 52). Nonapoptotic substrates of Dronc have not been identified, but indirect evidence for their existence has been provided (53).
The P35-dependent cell system made it possible to identify genes involved in compensatory proliferation and to determine a mechanism. In cells that have initiated but not fully executed apoptosis, Drosophila p53 is required for the induction of compensatory proliferation and is transcriptionally activated by a mechanism that requires Dronc function (Fig. 2B) (52). The function of p53 for compensatory proliferation is independent of the DNA damage–sensing pathway because it does not require the genes atm and chk2, which encode DNA damage checkpoint proteins (52). This study also showed that blastema formation requires Dronc and p53 in Drosophila. Furthermore, because the IAP antagonists Reaper and Hid are direct transcriptional targets of p53 (54, 55), a positive feedback loop involving Dronc, p53, Reaper, and Hid may operate in this system. Similarly, a planarian p53 ortholog has been implicated in stem cell renewal and proliferation (56).
Furthermore, Jun N-terminal kinase (JNK) signaling plays a critical role for compensatory proliferation as well as wound healing in Drosophila (Fig. 2B) (37, 57, 58). Inhibition of JNK activity impairs the ability of the tissue to proliferate and to heal wounds. However, the exact function of JNK in compensatory proliferation is still a subject of debate. Initial work suggested that JNK acts downstream of Dronc in P35-expressing apoptotic cells to promote the expression of mitogens (Fig. 2B) (37). Another study found that JNK induces compensatory proliferation in P35-expressing apoptotic cells independently of the apoptotic program (50). In a different regeneration system that does not involve P35-mediated apoptosis inhibition (see below), JNK activity was observed in the proliferating (regenerating) tissue but not in the dying tissue (59). Finally, JNK signaling can drive oncogenic cooperation through compensatory proliferation and Jak-STAT signaling (60). In particular, Wu et al. (60) used a Drosophila tumor model to show that both wound-induced and stress-induced JNK activity can be propagated to RasV12 cells and trigger the production of Jak-STAT–activating cytokines, thereby promoting tumor development. Therefore, additional work is needed to clarify the multiple and complex roles of the JNK pathway for compensatory proliferation in regeneration and tumor development.
These studies also revealed several mitogens or morphogens that are secreted in a Dronc-dependent manner by P35-expressing cells that cannot execute apoptosis. These include Wingless (Wg), a Wnt family member, and Dpp, a member of the bone morphogenetic protein (BMP)/transforming growth factor–β (TGF-β) superfamily (Fig. 2B) (37). Of note is the initial evidence that Wnt signaling from apoptotic cells is necessary and sufficient for cell proliferation in Drosophila (37), because the Wnt pathway was later found to play an important role in tissue regeneration in planarians, zebrafish, Xenopus, and Hydra (39, 61–70). Likewise, BMP signaling contributes to asymmetric tissue regeneration in planarians (71). Other reports also have implicated Hedgehog, Notch, and Jak-STAT signaling from apoptotic cells in compensatory proliferation (51, 72–74). Thus, the pathways that regulate cell proliferation during normal development are also used for compensatory proliferation in response to injury and apoptosis, and are conserved in evolution.
P35-Independent Regeneration Systems
The inability of P35-expressing apoptotic cells to complete apoptosis may change their cellular behavior (75, 76). Therefore, concern has been raised about the relevance of the signaling mechanisms triggered by these cells for understanding of normal compensatory proliferation and regeneration (50). Consequently, several labs have developed P35-independent proliferation and regeneration systems (59, 77). In these systems, apoptosis is temporally induced in a spatially restricted manner, and the events leading to compensatory proliferation are analyzed. Although only two reports have been published so far, the results confirm the findings obtained in the so-called “undead” Drosophila cells. Compensatory proliferation triggered by apoptotic cells under these conditions requires Wg as a mitogen (77) and JNK signaling (59) (Fig. 2C). However, it is unclear whether JNK acts in apoptotic cells, as indicated by studies using P35-expressing apoptotic cells (37), or in proliferating cells, as suggested by a P35-independent system (59).
Another concern with the use of P35-expressing apoptotic cells was that the experiments were performed in proliferation-competent tissues, such as larval wing imaginal discs. Imaginal disc growth is under the control of an unknown size-sensing mechanism that instructs the cells to stop proliferating when the regular size of the disc has been reached. It was therefore suggested that normal compensatory proliferation (which does not use P35) is not triggered by apoptosis-induced Wg and Dpp expression, but instead is controlled by the developmental size-sensing mechanism of the disc (50). In contrast, if cells are kept unable to complete apoptosis by the presence of P35, Wg and Dpp are induced to stimulate compensatory proliferation, which leads to hyperplastic overgrowth (Fig. 2B) (50). Despite these reasonable concerns, the P35-independent models demonstrate that Wg signaling is involved in regeneration (77).
It is important to identify the effectors of Wg signaling that mediate compensatory proliferation and tissue regeneration. Genetic screens using Drosophila leg imaginal discs have identif ied at least three conserved Wg target genes that function in P35-independent tissue regeneration (78). Mammalian homologs of these genes—augmenter of liver regeneration (alr), regeneration (rgn), and Matrix metalloproteinase-1 (Mmp1)—have characterized functions in regeneration. However, their precise role was unknown. Genetic analysis in Drosophila revealed that rgn controls the timing of blastema formation, whereas alr regulates proliferation of blastema cells and the extent of regeneration. Mmp1 functions to limit regenerative proliferation by controlling cell cycle arrest of non-blastema cells (78). Mmp1 is also a target gene of JNK signaling, which suggests a link between Wg and JNK in tissue regeneration (79).
JNK and cytokine signaling have also been implicated in increased stem cell activity in the Drosophila midgut. Homeostatic turnover and replacement of cells of the entire midgut in healthy flies takes about 3 weeks (80). However, upon epithelial cell loss induced by bacterial infection, experimental injury, stress signaling, or expression of IAP antagonists, compensatory proliferation by intestinal stem cells (ISCs) regenerates the entire midgut within 2 to 3 days (80–83). This increased ISC activity can be at least partially blocked by expression of P35, which inhibits caspase-3–like executioner caspases (Fig. 1B); this suggests that increased ISC activity can be induced by the apoptotic caspases DrICE and Dcp-1. The mechanisms leading to increased compensatory ISC activity are less clear. It has been reported that dying enterocytes in the midgut produce interleukin 6 (IL-6)–like cytokines [Unpaired (Upd), Upd2, Upd3] in a JNK-dependent manner, which stimulate Jak-STAT signaling in ISCs for proliferation, thereby driving renewal of the gut epithelium (80, 82, 83). In addition, insulin signaling has been implicated in promoting ISC proliferation and intestinal tissue repair in response to detergent-induced damage of the gut (81). Therefore, it is possible that different stresses induce different mechanisms for compensatory ISC activity and gut regeneration. Despite these uncertainties, given that IL-6 has been linked to ISC-derived colorectal cancer in humans (84, 85), studies on stress-induced compensatory ISC activity for gut regeneration in Drosophila may provide insights into the mechanisms by which colon cancer arises in humans.
As mentioned above, these studies were performed in proliferation-competent tissue, mostly the proliferating wing and leg imaginal disc, as well as intestinal stem cells. Thus, these models do not mimic the postmitotic state of most cells in an adult organism. In fact, developing tissues in Drosophila (and likely also those in vertebrates) progressively lose the ability to regenerate as development proceeds. For example, wing imaginal discs become unable to induce regenerative growth between day 7 and day 10 after egg laying when kept at 18°C (77). This corresponds to the third larval instar stage, when the disc reaches its final size and cells in the disc cease mitosis. Nevertheless, at least one example has been reported in which apoptotic cell death induces compensatory proliferation in predominantly postmitotic tissue in a P35-independent manner (51). During eye development in the third larval instar stage, a wave of differentiation initiated at the morphogenetic furrow sweeps across the disc from posterior to anterior (86). As part of the differentiation process, all cells exit the cell cycle and enter a quiescent state. They differentiate into photoreceptor neurons, cone cells, and pigment cells in a defined sequence (86). However, hid-induced apoptosis stimulates many cells to reenter the cell cycle in this otherwise postmitotic tissue (Fig. 2D) (51). Given the postmitotic state of this tissue, it is not surprising that this form of apoptosis-induced compensatory proliferation uses a different mechanism relative to proliferation-competent tissue. Signaling through Hedgehog (Hh), but not through Wg and Dpp, is required for this form of compensatory proliferation (Fig. 2D). Furthermore, Hh secretion requires not only the activity of the initiator caspase Dronc, but also that of the executioner caspases DrICE and Dcp-1 (Fig. 2D) (51). Thus, P35 expression blocks compensatory proliferation in this postmitotic model because of the requirement for executioner caspases, but not in the proliferating wing (see above). The Hh signal that is produced by dying photoreceptor neurons stimulates cell cycle reentry of non-neuronal postmitotic cells that have not yet started to differentiate (51). Thus, although these cells have exited the cell cycle, they have not completely lost mitotic potential. However, they do not reenter the cell cycle immediately after hid-induced apoptosis. It takes them about 6 to 12 hours to become cell cycle–competent again (51). This is similar to quiescent mammalian cells, which need about 8 hours for reentry into the cell cycle (87). The reason for this delay is unknown, but understanding the mechanism of cell cycle reentry of quiescent cells may be critical for understanding of cancer initiation.
Apoptosis-Induced Proliferation in Other Regeneration Models
In addition to the studies in Drosophila, links among apoptosis, proliferation, and tissue regeneration have been established in other models, including Hydra, Xenopus, planarians, newts, and mouse (Fig. 3) (37–46). In the freshwater animal Hydra, apoptosis is both necessary and sufficient for head regeneration after mid-gastric amputation (Fig. 3A) (39). Caspases trigger release of Wnt3 from apoptotic cells, promoting proliferation and regeneration (Fig. 3A). In contrast, foot regeneration in Hydra does not require proliferation. Experimental induction of apoptosis at the foot-regenerating tip promotes proliferation and results in head regeneration (39), suggesting a link between apoptosis and regenerative proliferation.
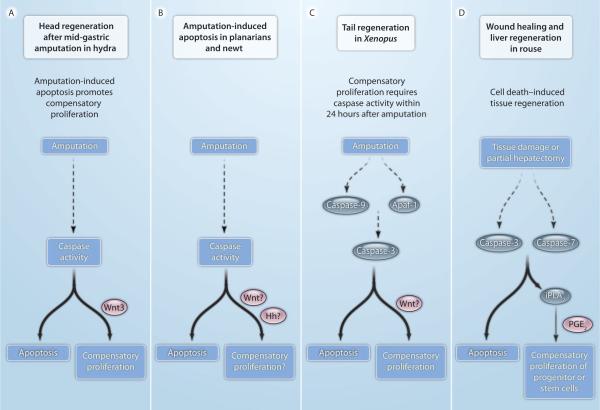
Fig. 3.
Apoptosis-induced compensatory proliferation in other model systems. In several model organisms, apoptosis and caspase activity have been detected in regenerating tissue. Identified intracellular factors are indicated in blue, secreted mitogens in red. Question marks indicate uncertainties. (A) In Hydra, head regeneration after mid-gastric amputation requires caspase activity. Apoptotic cells release Wnt3 to induce compensatory proliferation. (B) In planarians and newts, amputation correlates with localized apoptosis and caspase activity at the cut side. It is unknown whether this apoptotic response is required for the release of Wnt and Hh signals to induce regeneration. (C) In Xenopus, caspase activity during the first 24 hours after amputation is required for tail regeneration. Inhibition of initiator (caspase-9) and executioner (caspase-3) caspases blocks proliferation and regeneration. It has not been documented whether Wnt signals are released from dying cells. (D) In mouse, wound healing and liver regeneration require executioner caspases. Animals lacking caspase-3 and caspase-7 fail to induce these regenerative processes. The activity of calcium-independent iPLA2 is enhanced after proteolytic cleavage by caspase-3. iPLA2 produces arachidonic acid, which is converted to PGE2, a stimulator of stem cell proliferation and regeneration.
Localized massive apoptosis has also been observed at the cut side after amputation in planarians and newts (Fig. 3B) (43, 44, 46). However, it is currently unknown whether apoptosis is required for proliferation and regeneration in these animals. It is also unknown whether Wnt and Hh signals, which have been implicated in planarian and newt regeneration (61–64, 74, 88), are released by apoptotic cells.
In addition to model systems of invertebrate regeneration, similar apoptosis-dependent mechanisms of compensatory proliferation have been observed in vertebrates, most notably in Xenopus and in mouse. In the Xenopus tadpole, a large number of apoptotic cells were detected in the nascent regeneration bud within 12 hours after tail amputation (42). Inhibition of both initiator and executioner caspases completely abolished regeneration (42), indicating a strict requirement of caspases for regeneration (Fig. 3C). Caspases are required for the early phase of regeneration in which proliferation occurs, because caspase inhibition after 24 hours has no effect on regeneration. Although Wnt signaling has been implicated in regeneration in Xenopus, it has not been demonstrated that Wnt ligands are secreted by dying cells in this organism.
In a mouse model, dying mouse embryonic fibroblasts (MEFs) stimulate stem and progenitor cells to proliferate in response to x-ray treatment both in vitro and in vivo (41). Similar to the observations in Drosophila and Xenopus, this activity is directly dependent on the executioner caspases caspase-3 and caspase-7 (41, 42, 51). Mice lacking caspase-3 or caspase-7 have impaired wound healing and defects in liver regeneration after partial hepatectomy, which suggests that these two caspases are required for these regenerative processes in living animals (Fig. 3D) (41). Because prostaglandin E2 (PGE2) and Wnt signaling have been implicated in fin regeneration in zebrafish (68), Li et al. also tested PGE2 release in the mouse model and found that PGE2 concentrations increased in a caspase-dependent manner (41). A known caspase substrate in the PGE2 biosynthetic pathway is calcium-independent phospholipase A2 (iPLA2), which produces arachidonic acid, a precursor of PGE2 (Fig. 3D). Indeed, iPLA2 activity is enhanced after proteolytic cleavage by caspase-3 and is required for tissue regeneration (41).
In summary, there is now overwhelming evidence that both initiator and executioner caspases are required for the release of several cytokines, the most critical being Wnt, Hh, and PGE2 (in vertebrates). In this way, apoptotic cells can induce compensatory proliferation and promote regeneration in invertebrates and vertebrates. It will be important to further identify the mechanisms by which dying cells trigger the release of these factors.
Compensatory Proliferation and Cancer
Mitogenic signaling by apoptotic cells may contribute to the formation of neoplastic tumors. The idea that tumors resemble wounds that do not heal is an old one (89, 90), and many of the pathways involved in tissue regeneration and stem cell self-renewal, including Wnt and Hh, play prominent roles in human cancer (91–96). Therefore, even if wounding, inflammation, tissue stress, or tissue damage promotes apoptosis of cancer cells, these dying cells may release mitogens that promote malignant growth. Even if the release of mitogens by apoptotic cells does not contribute to the overall growth of a tumor, it may stimulate proliferation of cancer stem cells (97) or provide a supportive microenvironment for tumor growth (98, 99). If so, this could contribute to regrowth and relapse in response to cancer therapy, and also to seeding metastases.
This possibility warrants serious consideration for several reasons. First, virtually all cancer cells have acquired at least some degree of resistance toward apoptosis and hence may have features of so-called “undead” cells (3, 37, 100). Second, most existing cancer therapies, including radiation and chemotherapy, kill cancer cells by apoptosis and hence are expected to induce a “compensatory proliferation” response. Third, compensatory growth can explain oncogenic cooperation between genetically distinct cells in a Drosophila tumor model (60). Therefore, the combination of apoptosis resistance and strong, therapy-induced cellular stress and damage may lead to the expansion of cancer stem cells and hence increase the likelihood of tumor regeneration and the formation of secondary tumors.
Compensatory proliferation may play a role during different stages of carcinogenesis (including initiation, tumor promotion, and formation of secondary tumors) as well as in the response to cancer therapy, although the effects are likely to vary considerably in different paradigms. Inflammation is commonly thought to promote tumorigenesis, and a complex relationship exists between inflammation and apoptosis (101–103). For example, hepatocellular carcinoma (HCC) frequently develops in response to chronic liver damage, and agents that induce cell death can promote HCC (104). The chemical carcinogen DEN (diethylnitrosamine) induces hepatocyte death by DNA damage. DEN-induced hepatocyte death can activate the production of mitogens in adjacent myeloid cells, which in turn promotes compensatory proliferation of surviving hepatocytes (105). Activation of both JNK and Stat3 has been implicated in several types of human cancer (106, 107), and Stat3 function is critically required for the development of HCC (108, 109). Because these pathways drive compensatory proliferation in Drosophila, and because caspase-3 and caspase-7 are required for liver regeneration in mouse (41), it is likely that caspases may regulate the production of some tumor-promoting cytokines.
Conclusions
The original view that apoptotic cells only display “eat-me” signals for engulfment has been superseded by the realization that they can release various factors to communicate with their microenvironment. In addition to molecules that promote tissue regeneration, apoptotic cells can also release paracrine “alarm signals,” including microRNAs, to modulate immune cell responses, promote vascular repair, and exert atheroprotective effects (110). Therefore, mutations that affect the kinetics by which apoptosis is executed, or that influence the destruction or clearance of apoptotic corpses, are expected to alter the duration and strength of these signals. This presents new challenges as well as opportunities. For example, much remains to be learned about exactly how caspases control the production of signals, and how cells that undergo developmental apoptosis avoid the induction of compensatory growth. Fortunately, it appears that the underlying pathways have been conserved in evolution, and therefore, it is likely that results obtained from invertebrate models can guide mammalian work. Also, advances in apoptosis research have provided a wealth of tools and targets to further investigate basic mechanisms and to explore how compensatory proliferation may be manipulated for therapeutic purposes. We expect that progress in this area will have many important clinical implications, including the areas of regenerative medicine and cancer treatment.
Acknowledgments
We thank our colleagues in the field, G. Morata, H. D. Ryoo, M. Miura, and Y. Hiromi, for support of this review.
Funding: A.B. is supported by NIH grants GM068016, GM074977, and GM081543, Welch Foundation grant G-1496, and an anonymous donor. H.S. is an Investigator of the Howard Hughes Medical Institute and is supported by NIH grant R01GM60124.
References and Notes
- 1.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Wang J. Initiator caspases in apoptosis signaling pathways. Apoptosis. 2002;7:313–319. doi: 10.1023/a:1016167228059. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 1999;274:1156–1163. doi: 10.1074/jbc.274.2.1156. [DOI] [PubMed] [Google Scholar]
- 8.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 9.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 10.Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 12.Fraser AG, McCarthy NJ, Evan GI. drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J. 1997;16:6192–6199. doi: 10.1093/emboj/16.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornbluth S, White K. Apoptosis in Drosophila: Neither fish nor fowl (nor man, nor worm) J. Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- 14.Steller H. Staying alive: Apoptosome feedback inhibition. Nat. Cell Biol. 2008;10:1387–1388. doi: 10.1038/ncb1208-1387. [DOI] [PubMed] [Google Scholar]
- 15.Salvesen GS, Duckett CS. IAP proteins: Blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 16.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 17.Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schile AJ, García-Fernández M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 20.White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 21.Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- 22.Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4936–4941. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clavería C, Caminero E, Martínez-A C, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson MR, Holley CL, Gan EC, Colón-Ramos DA, Kaplan B, Kornbluth S. A GH3-like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J. Biol. Chem. 2003;278:44758–44768. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- 25.Freel CD, Richardson DA, Thomenius MJ, Gan EC, Horn SR, Olson MR, Kornbluth S. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J. Biol. Chem. 2008;283:367–379. doi: 10.1074/jbc.M708931200. [DOI] [PubMed] [Google Scholar]
- 26.Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Sandu C, Ryoo HD, Steller H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J. Cell Biol. 2010;190:1039–1052. doi: 10.1083/jcb.201004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 30.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 31.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 32.Haynie JL, Bryant PJ. The effects of X-rays on the proliferation dynamics of cells in the imaginal disc of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 1977;183:85–100. doi: 10.1007/BF00848779. [DOI] [PubMed] [Google Scholar]
- 33.Milán M, Campuzano S, García-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5691–5696. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Garijo A, Martín FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 35.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 36.Kondo S. Altruistic cell suicide in relation to radiation hormesis. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1988;53:95–102. doi: 10.1080/09553008814550461. [DOI] [PubMed] [Google Scholar]
- 37.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou JC, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell. 2009;17:279–289. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Hu DY, Chu Q, Wu JH, Gao C, Zhang YQ, Huang YR. Cell apoptosis and regeneration of hepatocellular carcinoma after transarterial chemoembolization. World J. Gastroenterol. 2004;10:1876–1880. doi: 10.3748/wjg.v10.i13.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Huang Q, Chen J, Peng Y, Roop DR, Bedford JS, Li CY. Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci. Signal. 2010;3:ra13. doi: 10.1126/scisignal.2000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev. Biol. 2007;301:62–69. doi: 10.1016/j.ydbio.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang JS, Kobayashi C, Agata K, Ikeo K, Gojobori T. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene. 2004;333:15–25. doi: 10.1016/j.gene.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 44.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: From humans to planarians. Annu. Rev. Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- 46.Vlaskalin T, Wong CJ, Tsilfidis C. Growth and apoptosis during larval forelimb development and adult forelimb regeneration in the newt (Notophthalmus viridescens) Dev. Genes Evol. 2004;214:423–431. doi: 10.1007/s00427-004-0417-1. [DOI] [PubMed] [Google Scholar]
- 47.Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 49.Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg AH, Miller LK, Wong WW. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 50.Pérez-Garijo A, Shlevkov E, Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development. 2009;136:1169–1177. doi: 10.1242/dev.034017. [DOI] [PubMed] [Google Scholar]
- 51.Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev. Cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr. Biol. 2006;16:1606–1615. doi: 10.1016/j.cub2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 55.Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H, Bergmann A. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ. 2010;17:912–921. doi: 10.1038/cdd.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosch M, Serras F, Martín-Blanco E, Baguñà J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev. Biol. 2005;280:73–86. doi: 10.1016/j.ydbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004;2:E239. doi: 10.1371/journal.pbio.0020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergantiños C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development. 2010;137:1169–1179. doi: 10.1242/dev.045559. [DOI] [PubMed] [Google Scholar]
- 60.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen CP, Reddien PW. Smed–catenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 62.Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adell T, Salò E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- 64.Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 66.Sodhi D, Micsenyi A, Bowen WC, Monga DK, Talavera JC, Monga SP. Morpholino oligonucleotide-triggered β-catenin knockdown compromises normal liver regeneration. J. Hepatol. 2005;43:132–141. doi: 10.1016/j.jhep.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 67.Zhong N, Gersch RP, Hadjiargyrou M. Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone. 2006;39:5–16. doi: 10.1016/j.bone.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, Zon LI. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Martí M, Dubova I, Izpisúa Belmonte JC. Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 2009;330:186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 72.Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, Bolduc C, Bergmann A. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martín FA, Herrera SC, Morata G. Cell competition, growth and size control in the Drosophila wing imaginal disc. Development. 2009;136:3747–3756. doi: 10.1242/dev.038406. [DOI] [PubMed] [Google Scholar]
- 76.Martín FA, Peréz-Garijo A, Morata G. Apoptosis in Drosophila: Compensatory proliferation and undead cells. Int. J. Dev. Biol. 2009;53:1341–1347. doi: 10.1387/ijdb.072447fm. [DOI] [PubMed] [Google Scholar]
- 77.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell. 2009;16:797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McClure KD, Sustar A, Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev. Biol. 2008;319:68–77. doi: 10.1016/j.ydbio.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006;25:5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Becker C, Fantini MC, Wirtz S, Nikolaev A, Lehr HA, Galle PR, Rose-John S, Neurath MF. IL-6 signaling promotes tumor growth in colorectal cancer. Cell Cycle. 2005;4:217–220. [PubMed] [Google Scholar]
- 85.Schneider MR, Hoeflich A, Fischer JR, Wolf E, Sordat B, Lahm H. Interleukin-6 stimulates clonogenic growth of primary and metastatic human colon carcinoma cells. Cancer Lett. 2000;151:31–38. doi: 10.1016/s0304-3835(99)00401-2. [DOI] [PubMed] [Google Scholar]
- 86.Voas MG, Rebay I. Signal integration during development: Insights from the Drosophila eye. Dev. Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 87.Coller HA. What's taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 2007;8:667–670. doi: 10.1038/nrm2223. [DOI] [PubMed] [Google Scholar]
- 88.Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005;132:3243–3253. doi: 10.1242/dev.01906. [DOI] [PubMed] [Google Scholar]
- 89.Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 90.Dvorak HF. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 91.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 92.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 93.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: Mediators of oncogenic Hedgehog signalling. Eur. J. Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 94.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 95.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 96.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, Munchhof M, VanArsdale T, Beachy PA, Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 98.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 99.Curran T, Ng JM. Cancer: Hedgehog's other great trick. Nature. 2008;455:293–294. doi: 10.1038/455293a. [DOI] [PubMed] [Google Scholar]
- 100.Fennell DA. Caspase regulation in non-small cell lung cancer and its potential for therapeutic exploitation. Clin. Cancer Res. 2005;11:2097–2105. doi: 10.1158/1078-0432.CCR-04-1482. [DOI] [PubMed] [Google Scholar]
- 101.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 102.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 104.Sarma DS, Rao PM, Rajalakshmi S. Liver tumour promotion by chemicals: Models and mechanisms. Cancer Surv. 1986;5:781–798. [PubMed] [Google Scholar]
- 105.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKα couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 106.Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, Marinkovich MP, Tao S, Lin Q, Kubo Y, Khavari PA. NF-κB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 107.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, Armstrong B, Bebernitz G, Weng S, Wang L, Ye M, McEachern K, Chen H, Morosini D, Bell K, Alimzhanov M, Ioannidis S, McCoon P, Cao ZA, Yu H, Jove R, Zinda M. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, Sieghart W, Peck-Radosavljevic M, Leffert HL, Karin M. Hepatocyte IKKα/NF-κB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]