Abstract
We investigated the hypothesis that a prominent effect of chronic ethanol consumption is mitochondrial DNA (mtDNA) injury and compared this injury in IL-6 knockout (KO) and wild-type (WT) mice. Ethanol feeding for 4 weeks resulted in steatosis and oxidative mtDNA damage (8-OHdG) in both IL-6KO and WT mice. However, the WT mice were able to repair the injury by increased production of mtDNA repair enzymes (OGG-1, Neil 1) and check point (p21, p53) proteins and avoid the mtDNA mutations. By contrast the IL-6 KO mice were unable to repair mtDNA resulting in deletions and diminished transcription of the mtDNA encoded protein cytochrome c oxidase subunit-I (COI). The mitochondrial injury was reflected by decreased membrane potential, reduced levels of ATP, and apoptosis-inducing factor (AIF)-induced apoptosis.
Conclusion
IL-6 plays a critical role in allowing the liver to recover from significant mtDNA oxidation caused by alcohol. The data suggests that IL-6 activates mtDNA repair enzymes and induces cell cycle arrest allowing time for mtDNA repair.
Keywords: ROS, 8-OHdG, Cell cycle protein, NEIL1, OGG1
Acute and chronic ethanol intake has been shown to increase the production of reactive oxygen species (ROS) and lower cellular antioxidant levels in many tissues especially the liver (1–5). A consequence of increased ROS production is oxidation of mtDNA reflected by increased amounts of 8-hydroxy-2-deoxyguanosine (8-OHdG) and mutations of mitochondrial DNA (mtDNA) which have been reported to be involved in the pathogenesis of many chronic conditions (6–12). After acute administration of toxic dosages of alcohol, hepatic mtDNA degradation has been found in mice (13, 14). Similarly, a 4977-base pair common deletion has been detected in the hepatic mtDNA of alcoholic patients with microvesicular steatosis, a lesion ascribed to impaired mitochondrial beta-oxidation (15, 16). Interestingly, clinical studies have shown that moderate drinkers tend to have better health and live longer than those who are either abstainers or heavy drinkers (17–19). In addition to having fewer heart attacks and strokes, moderate consumers are generally less likely to suffer hypertension or high blood pressure, peripheral artery disease, Alzheimer’s disease (20) and the common cold (21). Moderate alcohol intake has beneficial effects on insulin and triglyceride concentrations and insulin sensitivity in non-diabetic postmenopausal women (22). Chronic alcohol consumption in mice only causes moderate liver injury which does not progress to cirrhosis. These findings suggest the presence of important defenses and/or repair systems which compensate for the toxic attributes of moderate alcohol ingestion.
A major candidate for ameliorating liver injury is interleukin-6 (IL-6). Indeed, it has been reported that IL-6 reduces fatty changes caused by alcohol feeding in mice, and that the mechanism is related to improved mitochondrial function (23, 24). We have reported that in vitro IL-6 treatment markedly reduced mortality associated with fatty liver transplants from alcohol-fed rats (25). We propose that alcohol consumption causes mtDNA damage manifested by 8-OHdG production which activates the mtDNA repair system consisting of Neil1 and OGG1. We propose that IL-6 enables these enzymes to accomplish repair by causing cell cycle arrest. To test this hypothesis, wild-type (WT) mice and IL-6 knockout (KO) mice were fed Lieber-DeCarli ethanol-containing diets or pair-fed control diets for 4 weeks. First, we measured ethanol induced liver injury chemically, histologically and by TUNEL reaction. Second, we evaluated mtDNA oxidation by measuring levels of 8-OHdG in mtDNA. Third, we monitored the effects of mtDNA oxidation by determining the frequency of common deletions of mtDNA and the consequence of mtDNA deletion. Finally, we compared the cell cycle protein and DNA repair enzyme levels in ethanol-fed IL-6 KO mice with wild-type mice.
Materials and Methods
Chronic Ethanol Consumption Model
Male C57BL/6J background IL-6 KO mice and control C57BL/6J (WT) mice weighing 20–25g were purchased from the Jackson Laboratory (Bar Harbor, Maine). The mice were divided into 4 groups: 1. IL-6 KO fed alcohol (BioServ, Inc., Frenchtown, NJ), 2. IL-6 KO pair-fed isocalorically with dextrin maltose (BioServ, Inc.), 3. WT mice fed alcohol as above, 4. WT mice pair-fed control diet as above. Both diets were dispensed in glass liquid diet feeding tubes (BioServ, Inc.). The ethanol diet was introduced gradually by increasing the ethanol content of the diet by 1% (v/v) every 2 days until the mice were consuming diets containing 5% (v/v) ethanol. All mice were then fed the liquid diet containing 5% ethanol for 4 weeks. IL-6 KO and WT mice were observed to consume similar volumes of ethanol or control diets and that the ethanol-fed and pair-fed IL-6 KO and WT mice gained weight similarly. The protocol for this study was approved by the National Institute on Alcohol Abuse and Alcoholism Animal Care and Use Committee. Animals were cared for according to NIH guidelines.
Other Methods
The following methods are described in supplemental supporting materials: Analysis of alanine and asparate aminotransaminase activity; Caspase-3 activity and Adenosine Triphosphate assays; Histopathological analysis; Western blot analysis; Preparation of mitochondria; ELISA assay for measurement of 8-OHdG levels in mtDNA; ROS release measurements; Determination of mitochondrial membrane potential (MMP); Detection of mtDNA deletion by polymerase chain reaction; Real-Time PCR Analysis for DNA repair enzyme mRNA.
Statistics
The results were expressed as mean values ± SEM of n-independent experiments. Analyses were performed using the paired Student t test and the analysis of variance (ANOVA) with repeated measures. P less than .05 considered significant difference between KO mice and WT mice and between alcohol-containing diets and pair-fed control diets.
Results
More severe liver injuries in IL-6 KO mice fed ethanol compared to WT mice
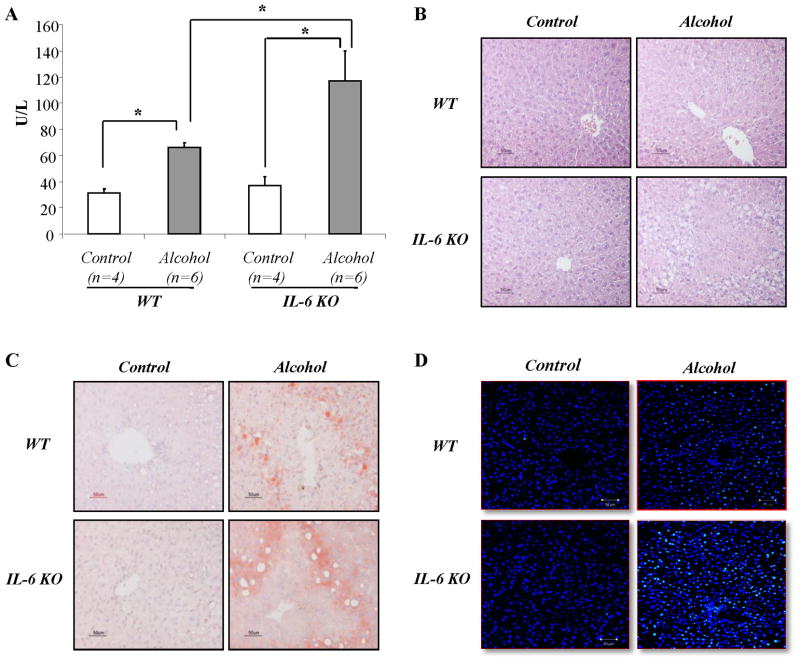
All animals survived the 4 week feeding period, but serum levels of alanine aminotransferase (ALT) were significantly increased in mice fed ethanol-containing diet compared to those pair-fed control diets. Moreover, IL-6 KO mice had significantly higher levels of serum ALT than wild-type mice after feeding ethanol diet (Fig. 1A). Liver histological studies showed that significantly greater hepatic steatosis occurred in the IL-6 KO mice compared to the wild-type mice as demonstrated by both H&E staining (Fig. 1B) and Oil red O staining (Fig. 1C). TUNEL staining of liver tissue sections revealed few TUNEL positive cells in liver tissue sections from mice pairs fed control diets. In contrast, TUNEL positivity was increased in both wild-type and IL-6 KO mice fed with ethanol-containing diets (Fig. 1D). Similar to ALT results, the number of TUNEL positive cells was significantly higher in the IL-6 KO mice compared to the WT mice after feeding ethanol diet.
Figure 1. Liver injury in IL-6 KO mice fed ethanol compared to WT mice.
Liver injuries were evaluated by (A) serum ALT, sections stained with hematoxylin-eosin (B), oil red O (C), and (D) DNA fragmentation staining (TUNEL) in liver sections was carried out using dUTP-FITC (green color), whereas nuclei was stained by Daipi (blue color). *p<0.01
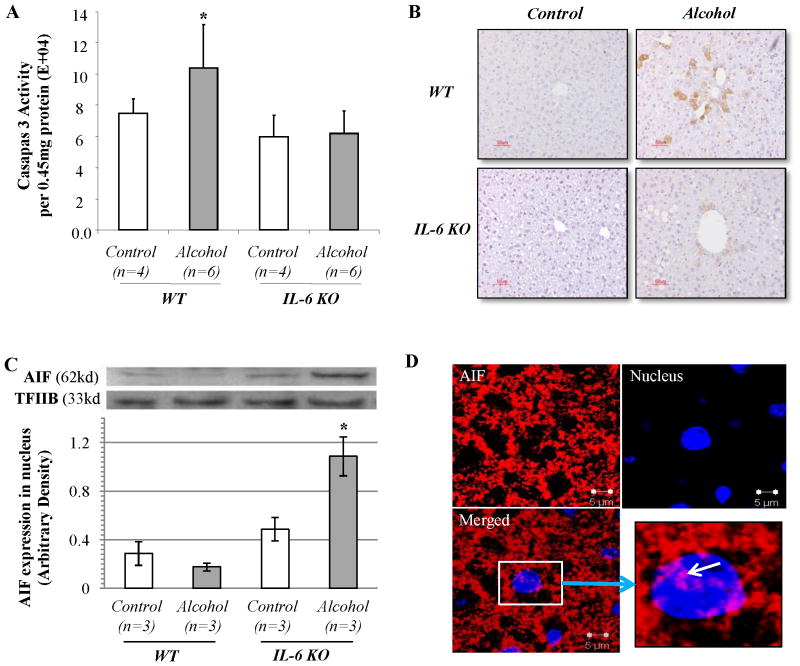
To determine the possible pathway of alcohol induced liver injury, we quantified caspase-3 activity in liver tissue extracts. Caspase-3 activity was increased in wild-type mice fed with ethanol-containing diets for 4 weeks. Surprisingly, caspase-3 activity was not elevated in ethanol-fed IL-6 KO mice compared to pair-fed IL-6 KO mice (Fig. 2A). This finding was confirmed histologically as activated caspase-3 was found in small foci of cells present in the WT mice fed ethanol, but not in IL-6 KO mice (Fig. 2B). To account for the presence of apoptosis, we examined the expression of apoptosis-inducing factor (AIF), a mitochondrial intermembrane protein which translocates to the cytoplasm and nucleus (28), where it binds to DNA triggering DNA fragmentation and nuclear condensation (29, 30). AIF induces apoptosis in a caspase-independent fashion (30). To determine whether there was translocation of AIF to the nucleus, we quantified AIF expression in nuclear proteins by using western blot analysis. Transcription factor IIB (TFIIB) that is localized in the nucleus was used as a loading control for western blot and protein normalization. After ethanol diet feeding, AIF expression in the nucleus was slightly decreased in wild-type mice but markedly increased in IL-6 KO mice (Fig. 2C). The evidence for mitochondria-nuclear translocation of AIF in liver tissue sections from ethanol-fed IL-6 KO mice was strengthened using confocal microscopy (Fig. 2D). In contrast, no nuclear AIF redistribution was observed in wild-type mice fed ethanol or mice fed control diets (Supplemental Fig. S1). These results suggest that the AIF apoptotic pathway may contribute to ethanol induced apoptosis in the livers of IL-6 KO mice.
Figure 2. Caspase-3 activity and AIF translocation in ethanol-fed IL-6 KO mice compared to WT mice.
(A) Caspase-3 activity was measured by Caspase-3 Fluorometric Assay Kit. Data represent mean ± SEM (n=4 or 6). (B) Immunohistochemical staining for activated caspase-3. (C) Western blot analyses of AIF expression in nuclear proteins. TFIIB was used as loading control. (D) Confocal evidence for the mitochondria-nuclear translocation of AIF. Liver tissue sections were stained for AIF (red fluorescence) and nucleus (Daipi-blue). Representative data of three individual samples per group.
Multiple mtDNA deletions in IL-6 KO mice fed ethanol
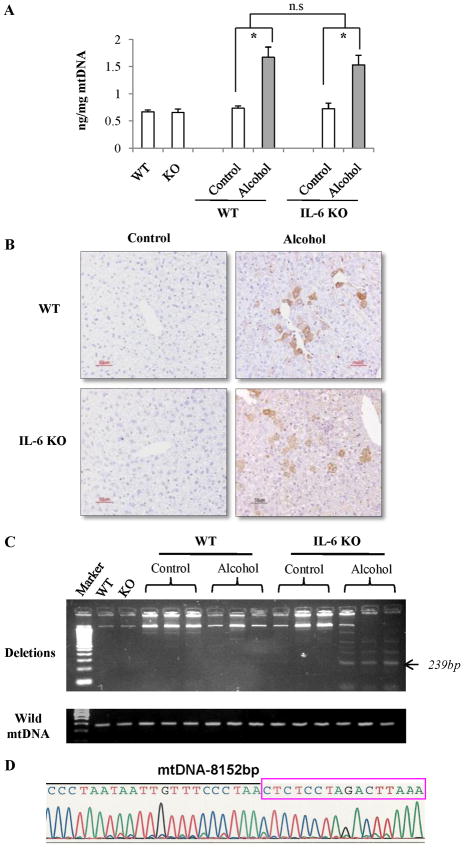
Using a competitive ELISA method, levels of 8-OHdG were determined after 4 weeks of feeding. As illustrated in Fig. 3A, ethanol feeding significantly elevated 8-OHdG levels in the mtDNA of the livers from both ethanol-fed wild-type and IL-6KO mice. Such elevation was comparable between WT and KO mice. Immunohistochemical staining confirmed these findings. As shown in Fig. 3B, pair-fed mice had few 8-OHdG–positive cells in livers, while ethanol-fed mice had strong 8-OHdG staining in the cytoplasm of hepatocytes especially in the area immediately around central veins. Although the method for 8-OHdG detection stains nuclear as well as mtDNA, the staining was primarily localized in the cytoplasm (Supplemental Fig. S2), indicating that this oxidative adduct was present mainly in the mitochondria.
Figure 3. Oxidative mtDNA damage and deletion in IL-6 KO mice fed ethanol compared to WT mice.
(A) Levels of 8-OHdG in mtDNA of hepatocytes recovered from pair-fed and ethanol-fed mice. Data represent mean ± SEM (n= 3). n.s., no significant difference. *p<0.05. (B) Immunohistochemical staining for 8-OHdG. 8-OHdG-positive cells stained with brown color. (C) PCR analysis of mtDNA deletion. Each lane represents mtDNA extracted from one animal in each group. (D) The representative PCR products (239bp) were further analyzed by auto-sequencer. The 239-bp fragment was generated from mtDNA with the 4,834bp deletion which is located between 8,152bp and 12,986bp.
Template mtDNA from the livers of ethanol-fed mice was amplified in a condition of 40 cycles using the primer pair between 7,837 and 13,126 bp. PCR amplification showed that multiple mtDNA deletions of 4,834 bp were found in the livers recovered from IL-6 KO mice at 4 weeks after ethanol feeding. In contrast, mtDNA deletions were undetectable at 4 weeks in pair-fed WT or IL-6 KO mice. Although mtDNA deletions were also detected at low levels in two of three WT mice at 4 weeks after ethanol feeding, the mtDNA recovered from IL-6 KO mice fed with ethanol exhibited higher levels of deletion products (Fig. 3C). In addition to the common mtDNA deletion of 4,834 bp, the representative PCR products were further analyzed by an auto-sequencer. The 239-bp fragment was generated from mtDNA with the 4,834bp deletion which is located between 8,152bp and 12,986bp (Fig. 3D).
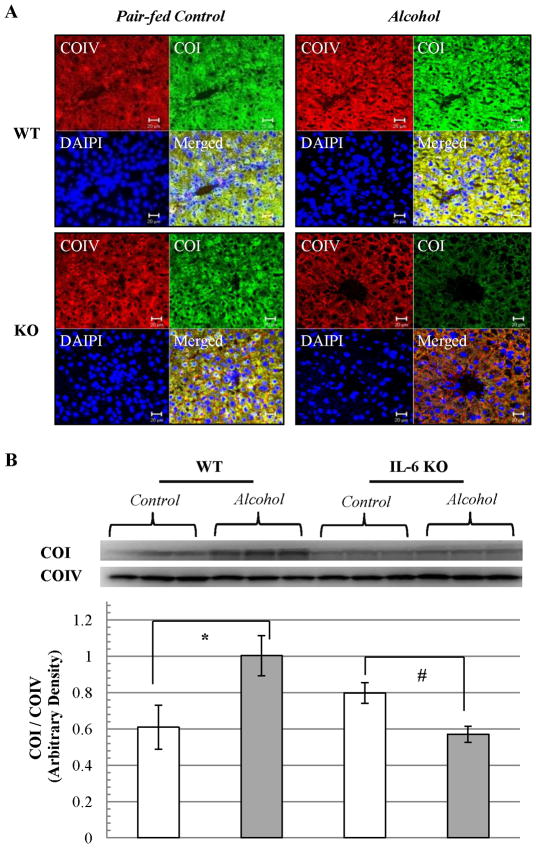
To determine if common deletions in mtDNA influence mtDNA encoded protein expression, the polypeptide of oxidative phosphorylation enzyme cytochrome C oxidase subunit-I (COI) was examined by immunofluorescence staining. Nuclear DNA encoded cytochrome C oxidase subunit-IV (COIV) was used as internal control. Immunofluorescence staining showed that COI was slightly increased in WT mice after ethanol feeding for 4 weeks. However, a decrease occurred in COI fluorescence around central vein areas in liver tissue sections recovered from IL-6 KO mice after ethanol feeding for 4 weeks (Fig. 4A). Western blot analyses of mitochondria fractions (Fig. 4B) confirmed that mtDNA encoded COI expression relative to nuclear encoded COIV (COI/COIV) was significantly increased in WT mice fed ethanol, while COI was decreased in IL-6 KO mice fed ethanol.
Figure 4. mtDNA encoded protein expression in ethanol-fed IL-6 KO mice compared to WT mice.
(A) The polypeptide of oxidative phosphorylation enzyme cytochrome C oxidase subunit-I (COI) was examined by immunofluorescence staining (green color). Nuclear DNA encoded cytochrome C oxidase subunit-IV (COIV) was used as internal control (red color). Nuclei stained with DAPI (blue color). (B) Western blot analyses of COI and COIV expression. Each lane represents mitochondrial protein from one animal. COI expression relative to COIV (ratio of COI/COIV) in each group was calculated as mean ± SEM. *p<0.01 vs pair-fed WT, #p<0.05 vs pair-fed KO.
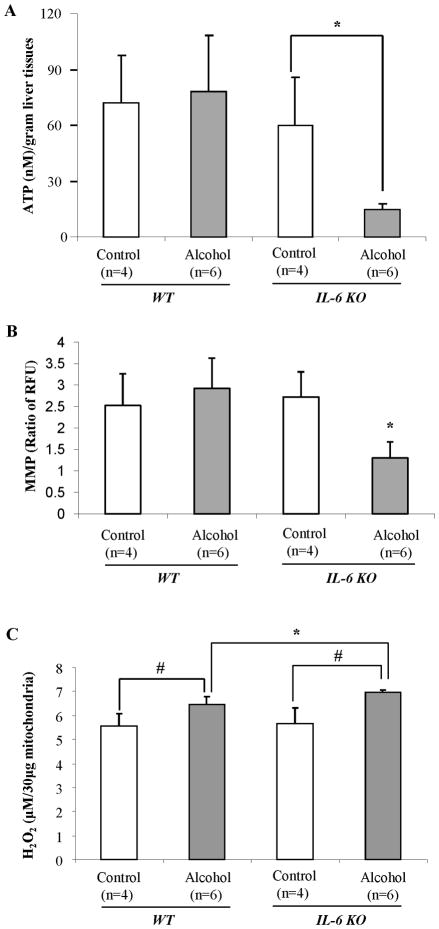
Decreased ATP production and increased mitochondrial ROS generation in IL-6 KO mice fed ethanol
To determine if mtDNA deletions influenced primary mitochondrial functions, we measured ATP content in the liver using the ATP Luciferase Assay Kit. As shown in Fig. 5A, ethanol feeding for 4 weeks did not affect hepatic ATP levels in WT mice, but significantly decreased the level of ATP in the livers of IL-6 KO mice compared to pair-fed IL-6 KO mice. These results suggest that the combination of alcohol feeding and the absence of IL-6 results in depletion of cellular energy reserves.
Figure 5. ATP content, MMP and mitochondrial ROS generation.
(A) ATP content in the liver was measured using the ATP Luciferase Assay Kit. (B) MMP in fresh isolated mitochondria was measured by using JC-1 MMP detection Kit. (C) H2O2 production in isolated mitochondria was measured using the Amplex Red peroxidase assay kit. Values are mean ± SEM of 4 or 6 samples. *p<0.05, #p< 0.01.
Sustaining a strong mitochondrial membrane potential (MMP) is essential for the synthesis of ATP through oxidative phosphorylation. We measured MMP in freshly isolated mitochondria by using JC-1 MMP detection Kit. After four weeks of feeding there was no significant difference in MMP between the ethanol-fed and pair-fed WT mice. However, the MMP was significantly decreased in IL-6 KO mice after ethanol feeding (Fig. 5B).
Further, after 4 weeks of feeding, significantly increased amounts of H2O2 were present in the mitochondrial fraction of livers recovered from animals fed ethanol compared to control diets (Fig. 5C). Such elevation was higher in IL-6 KO mice fed ethanol (Fig. 5C).
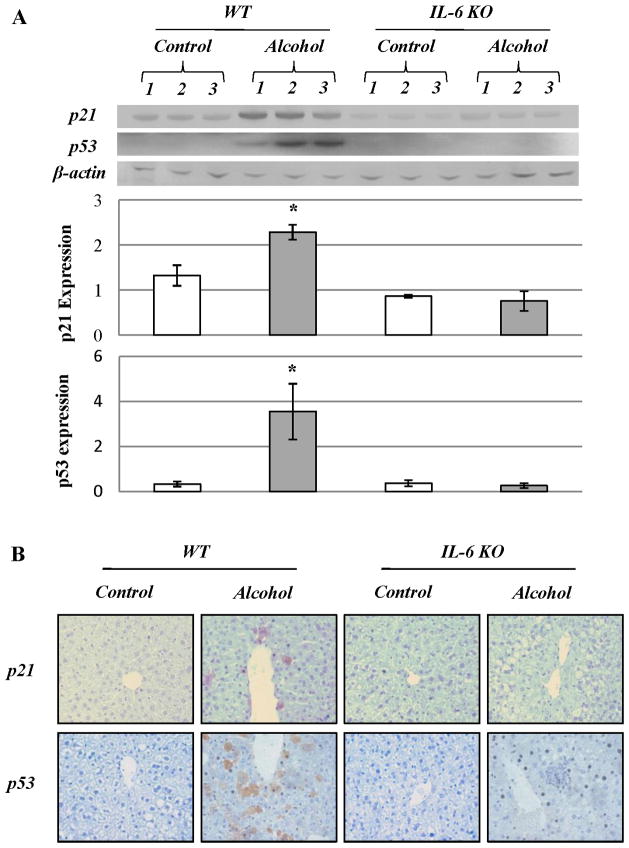
Cell cycle arrest and activation of DNA repair enzymes occurred in ethanol-fed WT mice but not in IL-6 KO mice after 4 weeks of ethanol feeding
Using western blot analysis, we found dramatic increases in proteins, p21 and p53, which are associated with cell cycle inhibition, in wild-type mice fed with ethanol for 4 weeks. By contrast, no significant increase in p21 or p53 expression occurred in IL-6 KO mice fed ethanol (Fig. 6A). Using immunohistochemistry, there were more p21 and p53 positive hepatocytes around central vein areas in the liver tissue sections from ethanol-fed WT mice compared to ethanol-fed IL-6 KO mice (Fig. 6B). Although proliferating cell nuclear antigen (PCNA) positive hepatocytes around central vein areas in liver tissue sections were increased in both WT and IL-6 KO mice fed ethanol compared to pair-fed mice, the number of Ki67 positive hepatocytes was only increased in IL-6 KO mice fed ethanol (Supplemental Fig. S3).
Figure 6. Cell cycle inhibitors, p21 and p53 expression in WT mice fed ethanol.
(A) Western blot analysis of p21 and p53. Each lane represents hepatic protein extracted from one animal in each group. There are three animals per group. p21 or p53 expression relative to β-actin was calculated as mean ± SEM. (B) Immunohistochemical staining for p21 and p53 in liver sections. Representative data of three individual samples per group.
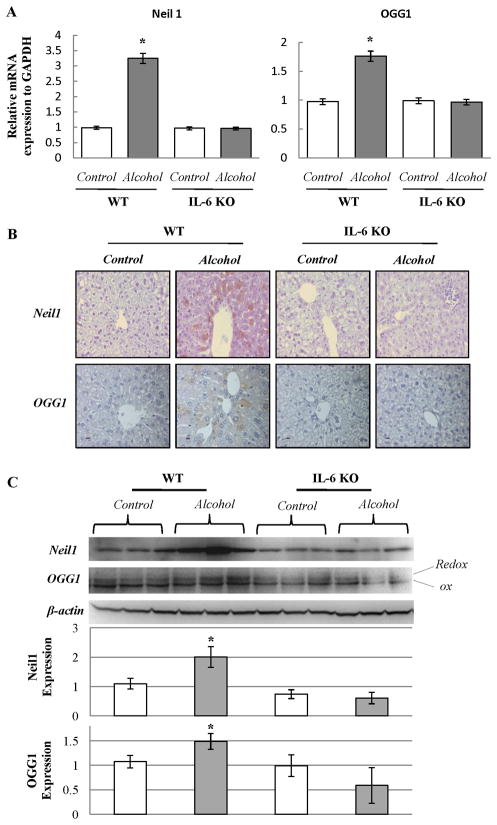
The mRNA expression of Neil1 and OGG1 was determined in WT and IL-6 KO mice fed ethanol for 4 weeks using quantitative real-time PCR. The mRNA of Neil1 and OGG1 was significantly increased in ethanol-fed WT mice compared to pair-fed WT mice. In contrast, there was no increase in mRNA of Neil1 and OGG1 in IL-6 KO mice (Fig. 7A). Neil1 and OGG1 protein levels were also studied by immunohistochemistry and western blot analyses. As shown in Fig. 7B, the number of Neil1 or OGG1 positive hepatocytes was significantly increased in WT mice fed ethanol compared to ethanol-fed IL-6 KO mice. Western blot analysis confirmed the finding that expression of Neil1 and OGG1 was increased only in ethanol-fed WT mice (Fig. 7C).
Figure 7. Detection of Neil1 and OGG1 in livers recovered from mice fed ethanol containing diets or control diets.
(A) Quantitative real-time PCR analysis for mRNA expression of Neil1 and OGG1 in WT mice or IL-6 KO mice fed with ethanol. Relative mRNA expression to GAPDH was calculated and data represent mean ± SEM of n= 4 or 6 animals per group. *p<0.05. (B) Immunohistochemical staining for Neil1 and OGG1. Positive cells show brown. Representative data of 4 or 6 individual samples per group. (C) Western blot analysis. Each lane represents hepatic protein extracted from one animal. OGG1 or Neil1 expression relative to β-actin was calculated as mean ± SEM.
Discussion
Our studies demonstrated that four weeks of alcohol feeding of WT mice resulted in increased ALT, steatosis, mtDNA oxidation, caspase-3 activation, and cell cycle protein and mtDNA repair enzyme expression. In these animals, low levels of mtDNA deletions were detected while the mitochondrial membrane potential and ATP levels remained unchanged compared to that present in pair-fed mice. By contrast, IL-6 KO mice fed alcohol for 4 weeks had more severe liver injury manifested by higher levels of ALT, greater steatosis, and more TUNEL positive cells. Ethanol feeding in IL-6 KO mice induced significant mtDNA deletions that were associated with the loss of the mitochondrial membrane potential, greatly diminished levels of COI synthesized in the mitochondria, along with diminished ATP and mtDNA repair enzymes levels. Thus oxidative injury that was fairly well tolerated in WT mice fed ethanol had highly deleterious effects in IL-6 KO mice.
Elevation of serum IL-6 is generally present in patients with alcoholic liver disease (ALD) and correlates with the severity of ALD (31). However, the findings from animal models suggest that IL-6 plays an important role in protecting against early alcoholic liver injury via multiple mechanisms. IL-6 exerts anti-apoptotic function through the gp130 protein that associates with and activates Janus kinase-signal transducer and transcription factor-3 (JAK-STAT3) (32). Lack of IL-6/gp130/STAT3 in hepatocytes predisposes the liver to steatosis in mice (33). Emerging evidence suggests that the hepatoprotective effect of IL-6 on ALD is mediated via induction of anti-apoptotic factors including Bcl-2 and Bcl-x(L), and anti-oxidative genes including metallothionein (23). Here we have shown in these experiments that IL-6 is necessary to provide cell cycle check-points via induction of p21 and p53, and stimulate the transcription of DNA repair enzymes.
We observed that p21 and p53 proteins are up-regulated in WT but not IL-6 KO mice fed ethanol (Fig. 5). The mechanism resulting in the failure of IL-6 KO mice to increase transcription of p21 may be found in our previous studies (23–25). IL-6 stimulation significantly increases phosphorylated-STAT3, which was then stimulates p21 transcription (34, 35). Over-expression of p21 resulted in G1 arrest. PCNA expression (G1 phase) was increased in both WT and IL-6 KO mice fed ethanol, but failed to increase Ki67 (S phase) in WT mice fed ethanol suggesting cell cycle arrest occurred. Thus, we propose that normal mice have sufficiently high levels of IL-6 during continuous moderate alcohol exposure to increase p21 production via activated STAT3 thereby leading to cell cycle (G1/S) arrest. This hypothesis is supported by the findings that pretreatment of obese animals with rIL-6 normalized proliferating cell nuclear antigen (PCNA) expression in steatotic hepatocytes but failed to increase DNA synthesis (BrdU, S phase) (36).
While the activation of p53 leads to hepatocyte apoptosis its deficiency results in the abrogation of hepatocyte apoptosis and the early appearance of liver dysplasia after ethanol feeding (37). However, the increased TUNEL reactivity in ethanol-fed IL-6 KO mice occurred in the absence of p53 and caspase-3 activation. This suggests that ethanol-induced hepatocyte apoptosis in IL-6 KO mice is independent of p53 and caspase-3. At present, the underlying mechanisms for this remain largely unknown. The facts that significant translocation of the mitochondrial intermembrane protein AIF into the nucleus was observed in the ethanol-fed IL-6 KO mice suggest that AIF translocation may contribute to ethanol-induced hepatocyte apoptosis in IL-6 KO mice as AIF has been shown to contribute to caspase-independent apoptosis (29, 30). Moreover, significant ATP depletion observed in ethanol-fed IL-6 KO mice appears to be one mechanisms leading to AIF translocation to the nucleus in these mice (38).
The OGG1 enzyme is the primary one for the repair of 8-oxoguanine (8-oxoG), a common product of DNA oxidation, in both the nuclear and mtDNA (39, 40). It has been reported that the OGG1 glycosylase plays a more important role in mitochondria than in nuclear DNA repair. Under conditions of oxidative stress the accumulation of 8-oxoG in the mitochondrial DNA from OGG1−/− (knockout) mice was much greater than in nuclear DNA when both were compared with wild type (41). Nei-like homologs (NEILs) are a recently identified group of bi-functional DNA glycosylases, which are mammalian orthologs of the Escherichia coli MutM/Nei. It was initially suggested that NEIL1 (42) and NEIL2 (43) localize exclusively to the nucleus. However, it has been recently reported that NEIL1 is also found in mouse liver mitochondria (44). NEIL1 may initiate base excision repair of ring-fragmented purines and some saturated pyrimidines. Interestingly, NEIL1 knockout (neil1−/−) and heterozygotic (neil1+/−) mice develop severe obesity, dyslipidemia, and fatty liver disease and also have a tendency to develop hyperinsulinemia at age of 4 to 6 months (45). Additionally, mtDNA from neil1−/− mice show increased levels of steady-state DNA damage and deletions relative to wild-type controls. Our results show that NEIL1 and OGG1 were increased in response to ethanol stimulation in wild-type mice and this was associated with much less mtDNA deletion, fatty changes and hepatocyte injury compared to IL-6 knock-out mice. IL-6 may play an essential role in the activation of DNA repair enzymes in response to ethanol stimulation.
We studied the components of cytochrome C oxidase (COI and COIV) for two reasons: first to determine if the deletions in mtDNA had functional significance resulting in a decreased level of COI and second, to compare the transcriptional capability of nuclear DNA (COIV) vs mtDNA (COI). We found that the level of mtDNA encoded COI was significantly increased in wild-type mice fed ethanol (Fig. 4). The increased oxidative stress may be responsible for the elevation of mtDNA encoded COI because oxidative stress has been shown to regulate the expression of COI and COII in hepatocytes (46). The increased production of COI of the mitochondrial respiratory chain in response to ethanol in WT mice is likely to represent an adaptive response of compensatory effect of mitochondrial to oxidative injury and to re-establish ionic balance and to support the synthesis of cellular repair systems in ethanol injured cells. In contrast, oxidative mtDNA damage without repair which occurred in the IL-6 KO mice fed ethanol results in common deletions responsible for the transcription of COI. The finding that nuclear COIV synthesis was relatively unaffected after four weeks of alcohol feeding in both WT and IL-6 KO mice is the strongest argument supporting the concept that the principal target of chronic alcohol toxicity is mtDNA.
In summary, IL-6 is required to activate DNA repair enzymes (Neil1 and OGG1) and cell cycle proteins (p21 and p53) in response to chronic ethanol consumption. These newly assigned functions for IL-6 are likely to be important in other forms of injury and perhaps in other organ systems.
Supplementary Material
Acknowledgments
Financial Support: This work was supported in part by the Bertram M. Bernheim Research Fund.
List of Abbreviations
- AIF
apoptosis-inducing factor
- ATP
Adenosine-5′-triphosphate
- COI
cytochrome c oxidase subunit-I
- COIV
cytochrome C oxidase subunit-IV
- IL-6
interleukin 6
- KO
knockout
- MMP
mitochondrial membrane potential
- mtDNA
mitochondrial DNA
- NEIL
Nei-like homologs
- OGG1
8-oxoguanine glycosylase
- 8-OHdG
8-hydroxy-2 -deoxyguanosine
- PCR
Polymerase chain reaction
- ROS
reactive oxygen species
- STAT3
Signal transducer and activator of transcription 3
- TUNEL
terminal deoxynucleotidyltransferase-mediated nick-end labeling
- WT
wild-type
References
- 1.Bailey SM, Cunningham CC. Acute and chronic ethanol increases reactive oxygen species generation and decreases viability in fresh, isolated rat hepatocytes. Hepatology. 1998;28:1318–1326. doi: 10.1002/hep.510280521. [DOI] [PubMed] [Google Scholar]
- 2.Bailey SM, Pietsch EC, Cunningham CC. Ethanol stimulates the production of reactive oxygen species at mitochondrial complexes I and III. Free Radic Biol Med. 1999;27:891–900. doi: 10.1016/s0891-5849(99)00138-0. [DOI] [PubMed] [Google Scholar]
- 3.Satre J, Minana JB, Alguacil P, Malet G, Gomez-Cambronero L, Marin JA, et al. Chronic ethanol feeding causes oxidative stress in rat liver mitochondria: prevention by S-adenosyl methionine. Free Radic Res. 1999;30:325–337. [Google Scholar]
- 4.Bailey SM, Patel VB, Young TA, Asayama K, Cunningham CC. Chronic ethanol consumption alters the glutathione/glutathione peroxidase-1 system and protein oxidation status in rat liver. Alcohol Clin Exp Res. 2001;25:726–733. [PubMed] [Google Scholar]
- 5.Arteel GE. Oxidants and antioxidants in alcohol-induced liver disease. Gastroenterology. 2003 Mar;124(3):778–90. doi: 10.1053/gast.2003.50087. [DOI] [PubMed] [Google Scholar]
- 6.Cahill A, Wang X, Hoek JB. Increased oxidative damage to mitochondrial DNA following chronic ethanol consumption. Biochem Biophys Res Commun. 1997 Jun 18;235(2):286–90. doi: 10.1006/bbrc.1997.6774. [DOI] [PubMed] [Google Scholar]
- 7.Cahill A, Stabley GJ, Wang X, Hoek JB. Chronic ethanol consumption causes alterations in the structural integrity of mitochondrial DNA in aged rats. Hepatology. 1999;30:881–888. doi: 10.1002/hep.510300434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Monte SM, Luong T, Neely TR, Robinson D, Wands JR. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer’s disease. Lab Invest. 2000;80:1323–1335. doi: 10.1038/labinvest.3780140. [DOI] [PubMed] [Google Scholar]
- 9.Warita H, Hayashi T, Murakami T, Manabe Y, Abe K. Oxidative damage to mitochondrial DNA in spinal motoneurons of transgenic ALS mice. Brain Res Mol Brain Res. 2001;89:147–152. doi: 10.1016/s0169-328x(01)00029-8. [DOI] [PubMed] [Google Scholar]
- 10.Hoek JB, Cahill A, Pastorino JG. Alcohol and mitochondria: a dysfunctional relationship. Gastroenterology. 2002 Jun;122(7):2049–63. doi: 10.1053/gast.2002.33613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakimoto M, Inoguchi T, Sonta T, Yu HY, Imamura M, Etoh T, et al. Accumulation of 8-hydroxy-2 -deoxyguanosine and mitochondrial DNA deletion in kidney of diabetic rats. Diabetes. 2002;51:1588–1595. doi: 10.2337/diabetes.51.5.1588. [DOI] [PubMed] [Google Scholar]
- 12.Lin PH, Lee SH, Su CP, Wei YH. Oxidative damage to mitochondrial DNA in atrial muscle of patients with atrial fibrillation. Free Radic Biol Med. 2003;35:1310–1318. doi: 10.1016/j.freeradbiomed.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Mansouri A, Demeilliers C, Amsellem S, Pessayre D, Fromenty B. Acute ethanol administration oxidatively damages and depletes mitochondrial DNA in mouse liver, brain, heart, and skeletal muscles: protective effects of antioxidants. J Pharmacol Exp Ther. 2001;298:737–743. [PubMed] [Google Scholar]
- 14.Demeilliers C, Maisonneuve C, Grodet A, Mansouri A, Nguyen R, Tinel M, et al. Impaired adaptive resynthesis and prolonged depletion of hepatic mitochondrial DNA after repeated alcohol binges in mice. Gastroenterology. 2002;123:1278–1290. doi: 10.1053/gast.2002.35952. [DOI] [PubMed] [Google Scholar]
- 15.Fromenty B, Grimbert S, Mansouri A, Beaugrand M, Erlinger S, Rotig A, Pessayre D. Hepatic mitochondrial DNA deletion in alcoholics: association with microvesicular steatosis. Gastroenterology. 1995 Jan;108(1):193–200. doi: 10.1016/0016-5085(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 16.Mansouri A, Fromenty B, Berson A, Robin MA, Grimbert S, Beaugrand M, et al. Multiple hepatic mitochondrial DNA deletions suggest premature oxidative aging in alcoholic patients. J Hepatol. 1997;27:96–102. doi: 10.1016/s0168-8278(97)80286-3. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg ML, Amsterdam EA. Alcohol, wine, and cardiovascular health. Clin Cardiol. 2008 Aug;31(8):347–51. doi: 10.1002/clc.20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camargo CA, Jr, Stampfer MJ, Glynn RJ, Grodstein F, Gaziano JM, Manson JE, Buring JE, Hennekens CH. Prospective study of moderate alcohol consumption and mortality in US male physicians. Archives of Internal Medicine. 1997;157:79–85. [PubMed] [Google Scholar]
- 19.Fuchs CS, Stampfer MJ, Colditz GA, Giovannucci EL, Manson JE, Kawachi I, Hunter DJ, Hankinson SE, Hennekens CH, Rosner B. Alcohol consumption and mortality among women. The New England Journal of Medicine. 1995;332(19):1245–1250. doi: 10.1056/NEJM199505113321901. [DOI] [PubMed] [Google Scholar]
- 20.Espeland MA, Coker LH, Wallace R, Rapp SR, Resnick SM, Limacher M, Powell LH, Messina CR. Association between alcohol intake and domain-specific cognitive function in older women. Neuroepidemiology. 2006;27(1):1–12. doi: 10.1159/000093532. [DOI] [PubMed] [Google Scholar]
- 21.Cohen S, Tyrrell DA, Russell MA, Jarvis MJ, Smith AP. Smoking, alcohol consumption and susceptibility to the common cold. American Journal of Public Health. 1993;83(9):1277–1283. doi: 10.2105/ajph.83.9.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies MJ, Baer DJ, Judd JT, Brown ED, Campbell WS, Taylor PR. Effects of moderate alcohol intake on fasting insulin and glucose concentrations and insulin sensitivity in postmenopausal women: a randomized controlled trial. JAMA. 2002 May 15;287(19):2559–62. doi: 10.1001/jama.287.19.2559. [DOI] [PubMed] [Google Scholar]
- 23.Hong F, Kim WH, Tian Z, Jaruga B, Ishac E, Shen X, Gao B. Elevated interleukin-6 during ethanol consumption acts as a potential endogenous protective cytokine against ethanol-induced apoptosis in the liver: involvement of Bcl-2 and Bcl-x(L) proteins. Oncogene. 2002;21:32–43. doi: 10.1038/sj.onc.1205016. [DOI] [PubMed] [Google Scholar]
- 24.Hong F, Radaeva S, Pan H, Tian Z, Veech R, Gao B. Interleukin-6 treatment alleviates steatosis and ischemia/reperfusion injury in mice with fatty liver disease. Hepatology. 2004;40:933–941. doi: 10.1002/hep.20400. [DOI] [PubMed] [Google Scholar]
- 25.Sun Z, Klein AS, Radaeva S, Hong F, El-Assal O, Pan HN, et al. In vitro interleukin-6 treatment prevents mortality associated with fatty liver transplants in rats. Gastroenterology. 2003 Jul;125( 1):202–15. doi: 10.1016/s0016-5085(03)00696-6. [DOI] [PubMed] [Google Scholar]
- 26.Nagakawa Y, Williams GM, Zheng Q, Tsuchida A, Aoki T, Montgomery RA, Klein AS, Sun Z. Oxidative mitochondrial DNA damage and deletion in hepatocytes of rejecting liver allografts in rats: role of TNF-alpha. Hepatology. 2005 Jul;42(1):208–15. doi: 10.1002/hep.20755. [DOI] [PubMed] [Google Scholar]
- 27.Sun Z, Zhang X, Locke JE, Zheng Q, Tachibana S, Diehl AM, Williams GM. Recruitment of host progenitor cells in rat liver transplants. Hepatology. 2009 Feb;49(2):587–97. doi: 10.1002/hep.22653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 29.Cande C, Cohen I, Daugas E, Ravagnan L, Larochette N, et al. Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie. 2002;84:215–222. doi: 10.1016/s0300-9084(02)01374-3. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 31.McClain CJ, Song Z, Barve SS, Hill DB, Deaciuc I. Recent advances in alcoholic liver disease. IV. Dysregulated cytokine metabolism in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2004 Sep;287(3):G497–502. doi: 10.1152/ajpgi.00171.2004. [DOI] [PubMed] [Google Scholar]
- 32.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroy DC, Beraza N, Tschaharganeh DF, Sander LE, Erschfeld S, Giebeler A, et al. Lack of interleukin-6/glycoprotein 130/signal transducers and activators of transcription-3 signaling in hepatocytes predisposes to liver steatosis and injury in mice. Hepatology. 2010 Feb;51(2):463–73. doi: 10.1002/hep.23322. [DOI] [PubMed] [Google Scholar]
- 34.Bienvenu F, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J Biol Chem. 2001 May 18;276(20):16840–7. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]
- 35.Sinibaldi D, Wharton W, Turkson J, Bowman T, Pledger WJ, Jove R. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene. 2000 Nov 16;19(48):5419–27. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 36.Selzner M, Clavien P-A. Failure of regeneration of the steatotic rat liver: disruption at two different levels of the regeneration pathway. Hepatology. 2000;31:35–42. doi: 10.1002/hep.510310108. [DOI] [PubMed] [Google Scholar]
- 37.Pani G, Fusco S, Colavitti R, Borrello S, Maggiano N, Cravero AA, Farré SM, Galeotti T, Koch OR. Abrogation of hepatocyte apoptosis and early appearance of liver dysplasia in ethanol-fed p53-deficient mice. Biochem Biophys Res Commun. 2004 Dec 3;325(1):97–100. doi: 10.1016/j.bbrc.2004.09.213. [DOI] [PubMed] [Google Scholar]
- 38.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, et al. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000 Apr;14(5):729–39. [PubMed] [Google Scholar]
- 39.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 40.Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOGG1 into mitochondria. J Biol Chem. 2002;22; 277(47):44932–7. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 41.Karahalil B, de Souza-Pinto NC, Parsons JL, Elder RH, Bohr VA. Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J Biol Chem. 2003;5;278(36):33701–7. doi: 10.1074/jbc.M301617200. [DOI] [PubMed] [Google Scholar]
- 42.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, et al. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1 and NEIL1 enzymes. J Biol Chem. 2005 Dec 9;280(49):40544–51. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 44.Chen D, Cao G, Hastings T, Feng Y, Pei W, O’Horo C, et al. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J Neurochem. 2002 Jun;81(6):1273–84. doi: 10.1046/j.1471-4159.2002.00916.x. [DOI] [PubMed] [Google Scholar]
- 45.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1864–9. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hutchinson RW, Ing NH, Burghardt RC. Induction of c-fos, and cytochrome c oxidase subunits I and II by gossypol acetic acid in rat liver cells. Cell Biol Toxicol. 1998 Dec;14(6):391–9. doi: 10.1023/a:1007543510337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.