Abstract
Saccharomyces cerevisiae Sir2 is an NAD+-dependent histone deacetylase that links chromatin silencing to genomic stability, cellular metabolism, and lifespan regulation. In mice, deficiency for the Sir2 family member SIRT6 leads to genomic instability, metabolic defects, and degenerative pathologies associated with aging. Until recently, SIRT6 was an orphan enzyme whose catalytic activity and substrates were unclear. However, new mechanistic insights have come from the discovery that SIRT6 is a highly substrate-specific histone deacetylase that promotes proper chromatin function in several physiologic contexts, including telomere and genome stabilization, gene expression, and DNA repair. By maintaining both the integrity and the expression of the mammalian genome, SIRT6 thus serves several roles that parallel Sir2 function. Here, we review recent advances in understanding the mechanisms of SIRT6 action and their implications for human biology and disease.
Sir2: A chromatin-aging connection
S. cerevisiae Sir2 is the founding member of an evolutionarily conserved family of proteins, termed sirtuins, whose functions provided one of the first links between chromatin regulation and aging. Sir2 promotes chromatin silencing at sub-telomeric DNA 1, 2, silent mating-type loci 2, 3, and ribosomal DNA (rDNA) repeats 4–6. These effects of Sir2 on chromatin are mediated by its NAD+-dependent histone deacetylase activity: Sir2 catalyzes the deacetylation of several lysine residues on the amino terminal tails of histones H3 and H4 and on the globular core of histone H3 7–11. Among these, deacetylation of H4 lysine 16 (H4K16) and H3 lysine 56 (H3K56) play prominent roles in mediating the silencing effects of Sir2 10, 12, 13.
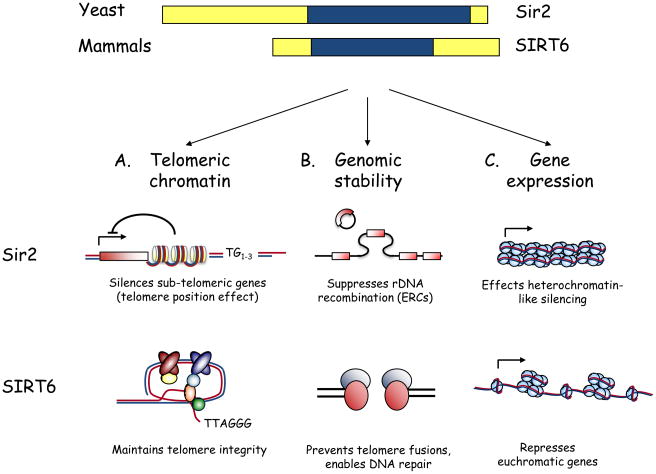
In budding yeast, Sir2 regulates replicative lifespan—a model for the cellular aging of mitotically active cells—through two chromatin-silencing activities. First, by suppressing recombination between rDNA repeats, Sir2 prevents genomic instability and the excision and accumulation of senescence-inducing extrachromosomal rDNA circles (ERCs) 14. Second, recent work has shown that H4K16 acetylation levels increase at telomeres with replicative age, owing to an age-associated decrease in Sir2 protein levels. The resulting chromatin changes lead to defects in telomere position-dependent transcriptional silencing and trigger replicative senescence 12. This finding is particularly interesting for mammalian biology, because maintenance of proper telomere function has an evolutionarily conserved role in preventing genomic instability and cellular senescence, processes that are important in mammalian aging and cancer. (By contrast, ERC accumulation appears to be specific to yeast.) As described below, the human sirtuin SIRT6 parallels yeast Sir2 in linking telomeric chromatin regulation to the prevention of cellular senescence (Fig. 1).
Figure 1.
From yeast to mammals: Parallel functions for Sir2 and SIRT6. A) Schematic of yeast Sir2 and mammalian SIRT6, highlighting the conserved central sirtuin domain (blue). B) Overlapping functions for Sir2 and SIRT6 in regulating telomeric chromatin, genomic stability, and gene expression. i. At yeast telomeres, histone deacetylation by Sir2 is required for telomere position effect, the reversible silencing of sub-telomeric gene expression. SIRT6 is required for generating a proper chromatin structure at human telomeres; this structure allows the binding of the telomere-processing factor WRN and prevents aberrant telomere metabolism. Whether SIRT6 is important for gene silencing due to telomere position effect in human cells is not currently known. ii. Yeast Sir2 maintains genomic stability by preventing recombination at the rDNA locus, thus suppressing formation of senescence-inducing extrachromosomal rDNA circles (ERCs). In mammalian cells, SIRT6 contributes to genomic stability by preventing chromosomal fusions between dysfunctional telomeres, by enabling efficient DNA repair, and perhaps by regulating H3K56 acetylation. iii. Sir2 is required for the transcriptional silencing of three heterchromatin-like domains in yeast: inactive mating-type loci, sub-telomeric DNA, and rDNA repeats. Although SIRT6 also represses gene expression, its reported effects thus far are at euchromatic regions of the genome (NF-κB and HIF1α target genes).
Interestingly, several studies have revealed aging-related Sir2 functions that might be chromatin-independent, suggesting additional layers of complexity in the link between Sir2 and lifespan regulation. For example, Sir2 promotes asymmetric segregation of damaged proteins to the yeast mother cell during cell division, and this asymmetry could contribute to mother cell aging by generating toxic protein aggregates 15. In addition, in a yeast model of chronological aging (in which cells are not dividing), Sir2 blocks lifespan extension in response to nutrient deprivation or mutations in nutrient-sensing pathways 16, in contrast to its effects on replicative lifespan. Sir2 proteins also promote longevity in the post-mitotic organisms Caenorhabditis elegans 17 and Drosophila melanogaster 18, although the molecular pathways and biochemical activities of these sirtuin proteins appear to have diverged from those of yeast Sir2. Notably, some studies have concluded that Sir2 proteins are important for the lifespan extension induced by dietary restriction 18, 19; however, these effects depend on the specific dietary regimens and genetic backgrounds used 20, 21. Together, these data indicate that sirtuin proteins have complex effects on aging-related processes in yeast and other model organisms.
Mammalian sirtuin proteins: Venturing out from chromatin
Mammalian genomes encode seven SIR2 family members, SIRT1-SIRT7 22, 23, and SIRT1 is most closely related to yeast Sir2. However, whereas Sir2 appears to deacetylate histones exclusively, SIRT1 is a robust and promiscuous deacetylase with more than 40 reported substrates. SIRT1 deacetylates several histone residues to regulate facultative heterochromatin 24, and can also modulate chromatin structure indirectly, by deacetylating chromatin-regulatory enzymes such as TIP60 25 and SUV39H1 26. In addition, SIRT1 deacetylates many non-histone proteins and impacts on numerous physiologic processes, including apoptosis, metabolism, and stress resistance 27.
Although SIRT1 remains the best characterized mammalian sirtuin, studies of the other six family members have also revealed links to aging and metabolism 28, and as is the case for SIRT1, some of the biochemical and physiologic functions of these proteins have evolved to be quite different from yeast Sir2. In this context, the mammalian SIRTs vary widely in sub-cellular localization: SIRT1, SIRT6, and SIRT7 are concentrated in different sub-nuclear patterns; SIRT2 is predominantly cytoplasmic; and SIRT3, SIRT4, and SIRT5 appear to reside in the mitochondria 29. However, some controversy exists regarding the localizations of certain mammalian SIRTs, and several of these proteins shuttle between cellular compartments, suggesting cell type- and context-dependent regulation of sub-cellular localization 30–32.
Mammalian SIRTs have also diverged significantly from yeast Sir2 with respect to their enzymatic activity and substrates, and several have been proposed to lack deacetylase activity entirely. Indeed, despite intensive investigation by numerous groups 29, 33, 34, no deacetylase activity was detected for SIRT6 for several years after its discovery (and deacetylase activity for SIRT4 and SIRT7 remain elusive). Recently, this barrier was overcome with the discovery of site-specific histone deacetylase activity for SIRT6, which has allowed new links to be drawn between SIRT6 and several important molecular and cellular pathways that impact on aging, metabolism, cancer, and other diseases.
In this review, we explore the evolution of the SIRT6 field, highlighting the complementary insights that have come both from mouse genetics and whole organism physiology, and—more recently—from biochemical and mechanistic studies of human SIRT6.
SIRTching for a function through knockout mice
The first clues to the in vivo function of SIRT6 came from gene-targeted inactivation of Sirt6 in mice 35. SIRT6-deficient mice, though small, appear relatively normal after birth. Beginning at around two weeks of age, however, these mice develop striking degenerative phenotypes, some of which overlap with pathologies observed in aging. These phenotypes include spinal curvature abnormalities, osteoporosis, loss of subcutaneous fat, colitis, and severe lymphocyte apoptosis due to systemic (non-cell-autonomous) causes 35. The mice also experience dramatic metabolic defects, with a precipitous drop in serum glucose levels despite low levels of the insulin-like protein IGF-1, and all Sirt6−/− mice die by one month of age. Although the functional deterioration and early demise of SIRT6-deficient mice occur earlier and more abruptly than is generally observed in classical premature aging models, the phenotype of SIRT6-deficient mice is reminiscent of these progeroid syndromes, and SIRT6 is clearly important for organismal homeostasis and normal health span.
On the cellular level, SIRT6 deficiency leads to marked genomic instability and hypersensitivity to ionizing radiation (IR), methylmethanesulfonate (MMS), and hydrogen peroxide (H2O2), cellular phenotypes consistent with potential defects in base excision repair (BER) or DNA double-strand break (DSB) repair. Notably, several mouse models of defective DNA repair are associated with defects in insulin/IGF-1 signaling and metabolism 36, and it has been suggested that SIRT6 might influence metabolism through its role in protecting genome integrity 37, 38.
Together, the phenotypes of Sirt6−/− mice and cells demonstrate that SIRT6 is important for many aspects of organismal health, and that SIRT6 loss confers multiple phenotypes relevant for aging, metabolism, and cancer.
An orphan enzyme finds its substrates: SIRT6 deacetylates H3K9 and H3K56
Despite the profound cellular and organismal consequences of SIRT6 deficiency in mice, the function of SIRT6 at the molecular level remained unclear. SIRT6 was found to associate with chromatin 29, 35, but a specific chromatin-regulatory function remained elusive, and multiple studies failed to identify histone deacetylase activity for SIRT6 29, 33, 34. Instead, early in vitro analyses revealed that SIRT6 can promote mono-ADP-ribosylation 34, 35, an alternative NAD+-dependent reaction that has been observed for some sirtuins 22, 39. Indeed, individual SIRT6 molecules undergo intramolecular mono-ADP-ribosylation, leading to the suggestion that auto-ribosylation might contribute to the regulation of SIRT6 activity 34. So far, weak 29, 35 or no 34 ADP-ribosylation activity has been detected for SIRT6 on substrates other than itself, and it remains to be determined whether there are physiologic ADP-ribosylation trans-substrates of SIRT6.
A breakthrough in understanding the molecular mechanisms of SIRT6 function came with the discovery of the enzymatic activity and first known substrate of SIRT6: NAD+-dependent deacetylation of histone H3 lysine 9 (H3K9Ac) 40. An in vitro screen of acetylated histone tail peptides found that SIRT6 specifically deacetylates H3K9, but lacks activity on at least a dozen other histone tail residues 40. Such a high degree of intrinsic substrate specificity is unusual for a mammalian histone deacetylase and likely explains the prior difficulties in detecting deacetylase activity for SIRT6. Because histone deacetylation is associated with decreased chromatin accessibility, the discovery of histone deacetylase activity for SIRT6 prompted a series of studies that demonstrated roles for SIRT6 in regulating telomeric chromatin, gene expression, and the dynamic chromatin association of DNA repair factors.
Following the discovery of the H3K9 deacetylase activity of SIRT6, two groups independently identified a second substrate for SIRT6: lysine 56 of histone H3 (H3K56Ac) 41, 42, a non-tail acetylation mark on the globular core of histone H3. A relatively unstudied chromatin mark only recently demonstrated to exist in mammalian cells 43, 44, H3K56Ac has garnered interest because of its links to genomic stability and DNA damage sensitivity in yeast 45–47. Moreover, several yeast sirtuin proteins deacetylate H3K56 10, 48. Sparked by these observations, as well as by similarities between the sequence contexts of H3K56 and H3K9, one group showed that human SIRT6 deacetylates H3K56 and contributes to dynamic changes in H3K56Ac levels at telomeres during the cell cycle 41. At the same time, a second study focused on murine SIRT6 and demonstrated that global levels of H3K56Ac are dramatically elevated in SIRT6-deficient mouse cells and tissues 42. Together, these findings suggest that one mechanism by which SIRT6 might impact on genomic stability is through its activity as an H3K56 deacetylase. Whether this occurs in genomic contexts in which SIRT6 is already known to function—such as at telomeric chromatin, DNA DSBs, or sites of transcriptional regulation—or at additional sites throughout the genome is an exciting area of future study.
To the core and beyond: Biochemical dissection of SIRT6 function
Sirtuin proteins consist of a conserved central sirtuin domain’—thought to comprise an enzymatic core—flanked by variable N- and C-terminal extensions (Fig. 1). Understanding the mechanistic and regulatory functions of these domains can provide important insights into the physiologic regulation of sirtuin proteins and potentially aid in the design of pharmacologic modulators. A recent study provided such an analysis of SIRT6, revealing that the N- and C-terminal domains of SIRT6 play important roles in regulating SIRT6 function: the C terminus is required for proper nuclear localization but is dispensable for enzymatic activity, whereas the N terminus is important for chromatin association and the intrinsic catalytic activity of SIRT6 49. Furthermore, substitution of a conserved catalytic residue within the core domain of SIRT6 not only abrogates SIRT6 enzymatic activity but also leads to impaired chromatin association in cells despite proficient nucleosome binding in vitro 49, raising the intriguing question of why catalytic activity is required for chromatin association in cells. One possibility is that histone deacetylation by SIRT6 might stabilize SIRT6 occupancy at chromatin or promote propagation of SIRT6 molecules along chromatin, in a manner analogous to the spreading of the Sir2 complex to generate silent chromatin in yeast 50–52. Alternatively, auto-ADP-ribosylation of SIRT6 34 might generate a novel interface for chromatin binding, and such interactions could be important for stabilizing or propagating SIRT6 at chromatin. Indeed, ADP-ribose binding activity has been reported for the macro domain, which is present on macroH2A, a histone variant enriched at silent chromatin 53. Together, these biochemical studies suggest that the N- and C-terminal extensions of SIRT6 could serve as sites of regulation, in both physiologic and pharmacologic contexts.
At the ends of chromosomes: SIRT6 regulates telomeric chromatin
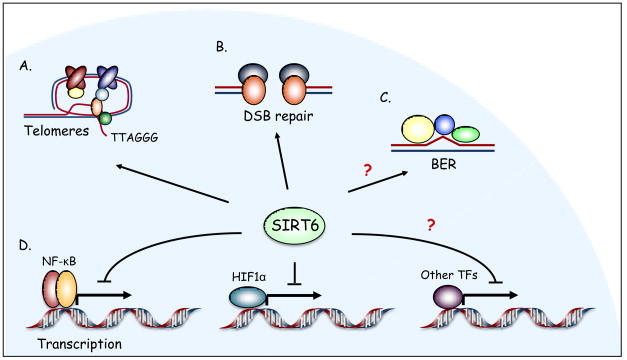
The discovery of H3K9 and H3K56 deacetylase activity for SIRT6 suggested new hypotheses and testable models for SIRT6 functions at the molecular, cellular, and organismal levels. The first physiologic context in which SIRT6 was shown to play an important chromatin-regulatory role was in maintaining the integrity of telomeric chromatin (Fig. 2). Telomeres are specialized DNA–protein structures that protect linear chromosome ends from aberrant DNA repair, degradation, and fusion 54. Mammalian telomeres are packaged into heterochromatin-like structures 55, but relatively little is understood about the role of specific chromatin modifications in telomere metabolism, especially in human cells. In concert with the discovery of its histone deacetylase activity, SIRT6 was shown to associate with telomeric chromatin, where it deacetylates H3K9 40 and H3K56 41. Depletion of SIRT6 from primary human fibroblasts and human cancer cells leads to hyperacetylation of H3K9 and H3K56 at telomeres specifically in S-phase 40, 41. In primary human cells, the resulting abnormalities in telomeric chromatin lead to stochastic replication-associated telomere sequence loss, accumulation of telomeric DNA damage foci, and genomic instability with chromosomal end-to-end fusions 40. Ultimately, this telomere dysfunction triggers premature cellular senescence 40. Mechanistically, site-specific histone deacetylation by SIRT6 at telomeres is required for stable telomere association of WRN, a telomere-processing factor mutated in Werner Syndrome, a disorder of premature aging and cancer susceptibility 40. Together, these observations suggest that SIRT6-mediated histone deacetylation promotes formation of a specialized chromatin state at telomeres that prevents aberrant telomere metabolism.
Figure 2.
Chromatin-bound: SIRT6 regulates multiple chromatin-templated processes. A. SIRT6 deacetylates H3K9 and H3K56 at telomeres, and SIRT6 depletion triggers aberrant telomere metabolism. B. SIRT6 enables efficient DSB repair. C. Although the role of SIRT6 in BER has not been clearly defined, the spectrum of DNA damage sensitivities of Sirt6−/− mouse cells and the results of functional complementation assays suggest a potential chromatin-regulatory function for SIRT6 in this repair pathway. D. SIRT6 represses NF-κB and HIF1α target gene expression, and might act in conjunction with other transcription factors (TFs) to regulate additional gene expression programs. Question marks indicate hypothesized or uncharacterized roles for SIRT6.
Notably, Sirt6−/− mouse cells also display hyperacetylated telomeric chromatin 40. As expected, however, these cells do not exhibit the functional consequences of this hyperacetylation—such as chromosomal end-to-end fusions—because mouse telomeres are long and have considerable functional reserve. Indeed, deficiency in mice for a number of telomere maintenance factors does not lead to severe functional defects unless the mouse telomeres are allowed to shorten or be —humanized through several generations in a telomerase-negative genetic background 56, 57. One would predict that such a background would unmask functional telomere defects in SIRT6-deficient mouse cells.
The discovery that SIRT6 plays a critical role at human telomeres has important implications for cancer and aging. First, proper telomere structure is required to maintain genomic stability, and chromosomal instability is a hallmark of human cancer cells 58. In addition, telomere length decreases with cellular, and possibly organismal, age 59, 60, suggesting that SIRT6’s role at telomeres—like the role of its yeast counterpart Sir2 (Fig. 1)—might be important in the biology of aging. Future studies examining the effect of SIRT6 loss on telomeric gene silencing and on the biology of late-generation telomerase-deficient mice harboring short telomeres should provide further insight into such roles.
Linking SIRT6 to DNA repair
Several observations have linked SIRT6 to DNA repair and suggested models in which SIRT6 impacts on specific DNA repair pathways through chromatin regulation (Fig. 2). SIRT6 was initially hypothesized to play a role in BER, because the DNA damage hypersensitivity of SIRT6-deficient cells could be rescued by over-expression of the isolated dRP lyase domain of DNA polymerase β, which catalyzes the rate-limiting step in BER 35. Previous work had shown that BER is hampered on chromatinized substrates in vitro, thus leading to the speculation that SIRT6 might regulate chromatin to increase the accessibility of DNA to BER factors. In this context, the dRP lyase domain of polymerase β might rescue BER defects in SIRT6-deficient cells because it is small enough to access DNA even in the presence of chromatin constraints. However, a definitive role for SIRT6 in BER has not been demonstrated, as SIRT6 was not found to physically interact with BER factors or to colocalize with these factors at sites of DNA damage 35, and BER defects in cell-based assays of SIRT6-deficient cells have not been reported.
Recently, biochemical and functional studies of SIRT6 in human cells have revealed an unexpected role for SIRT6 in enabling efficient DNA DSB repair 61. An unbiased search for SIRT6-associated factors found that SIRT6 interacts with proteins involved in the non-homologous end-joining (NHEJ) pathway of DSB repair, including DNA-PKcs and Ku70/80. Consistent with this finding, SIRT6 was shown to associate dynamically with chromatin in response to DSBs, promote a global decrease in H3K9Ac levels following DNA damage, and stabilize the DNA damage-dependent association of DNA-PKcs with chromatin. Moreover, in a system for studying biochemical events at defined DSBs (using site-specific endonucleases such as I-SceI and I-PpoI), SIRT6 was detected at chromatin flanking the DSBs and was required for stabilization of DNA-PKcs at these breaks. In SIRT6-depleted cells, accumulation of unrepaired DSBs was observed in both the site-specific DSB system and in comet assays that monitor DNA breaks at the single cell level.
The discovery that SIRT6 modulates DSB repair in human cells was somewhat unexpected given previous results in SIRT6-deficient mice 35, but there are several potential explanations for this discrepancy. For example, Sirt6−/− mice do not appear to have dramatic defects in V(D)J recombination, a process of programmed rearrangement of immunoglobulin genes that is abrogated by inactivation of NHEJ 62; in this context, it is possible that the requirement for SIRT6 in chromatin regulation during NHEJ might be assumed by lymphocyte-specific V(D)J recombination factors 62, 63. In addition, differences in assay sensitivity or compensatory mechanisms in the mouse knockout might have prevented detection of DSB repair defects.
Regardless of the mechanism, it appears that SIRT6 is required for changes in chromatin structure at DSBs that allow for efficient association of DSB repair proteins, and that impaired DSB repair might contribute to the genomic instability of SIRT6-deficient cells. This model provides an interesting parallel with the genome-stabilizing role of yeast Sir2 (Fig. 1) and suggests another link between SIRT6 and the genomic instability observed in cancer cells and aged cells. An important question is whether SIRT6 influences other DNA repair pathways—such as BER —through a similar mechanism, a possibility that can be directly tested in the future.
Sounds of silence: SIRT6 modulates gene expression programs
Another important physiologic context for site-specific histone deacetylation by SIRT6 is in transcriptional repression (Fig. 2). The first study linking SIRT6 to transcriptional regulation centered on nuclear factor-kappa B (NF-κB), a transcription factor that plays pivotal roles in regulating aging-, proliferation-, and inflammation-related gene expression programs 64. Upon NF-κB activation by tumor necrosis factor alpha (TNFα), SIRT6 is recruited to promoters of a subset of NF-κB target genes through a physical interaction with the NF-κB subunit RELA (also known as p65) 65. SIRT6 deacetylates H3K9 at these promoters, which leads to destabilization of RELA promoter occupancy and attenuation of NF-κB signaling. In addition, genomic analyses revealed hyperactivation of certain NF-κB-dependent gene expression programs in SIRT6-deficient human and mouse cells, as well as in multiple SIRT6-deficient mouse tissues, leading to increased expression of NF-κB target genes, including genes involved in inflammation, metabolism, and aging-related processes. Strikingly, RelA heterozygosity partially rescued the premature lethality of SIRT6-deficient mice, attenuated some of their degenerative and metabolic defects, and reversed the excessive levels of NF-κB-driven gene expression observed in these mice, indicating that hyperactive NF-κB signaling contributes to the phenotypes of Sirt6 knockout mice 65. Together, these observations suggest a feedback mechanism wherein SIRT6 regulates the termination of NF-κB-driven gene expression, which is essential for tight control of NF-κB signaling.
Recently, a second role for SIRT6 in transcriptional regulation was reported, this time in the context of the hypoxia-inducible transcription factor HIF1α, an important regulator of glucose homeostasis that has been linked to lifespan regulation 66 and cancer 67. SIRT6 was shown to interact physically with HIF1α and associate with the promoters of a subset of glucose-regulatory HIF1α target genes in a HIF1α-dependent manner. Consistent with the previously discovered function of SIRT6 as an H3K9 deacetylase, SIRT6 deficiency leads to hyperacetylated H3K9 at these promoters, and increased expression of these genes in Sirt6−/− mice causes an increase in glucose uptake and glycolysis in several tissues 68. These data suggest that under normal nutrient conditions, SIRT6 might compete with HIF1α to direct glucose flux away from glycolysis and towards oxidative phosphorylation, perhaps by inhibiting HIF1α–promoter association or blocking HIF1α activity at its target genes. An interesting question is how the interaction between SIRT6 and HIF1α would fit with such a model, and why the association of SIRT6 with its target promoters is dependent on HIF1α.
The newly discovered roles for SIRT6 as a transcriptional regulator suggest that modulation of NF-κB and HIF1α gene expression programs might be only the tip of the SIRT6 iceberg. Indeed, hundreds of genes are differentially expressed in Sirt6−/− mouse cells compared to wild-type controls 65. Moreover, several different cis-regulatory motifs are associated with these gene expression changes, suggesting that SIRT6 regulates numerous gene expression programs, perhaps in tissue- and/or condition-specific manners. Interestingly, in contrast to yeast Sir2, which silences the expression of heterochromatin-like loci, the transcriptional regulatory activity of SIRT6 has thus far been linked to the repression of euchromatic genes (Fig. 1). Whether SIRT6 also regulates the expression of heterochromatic loci remains to be determined.
SIRT6 goes metabolic
As NAD+-dependent histone deacetylases, sirtuin proteins are intrinsically linked to cellular metabolism, given that NAD+ levels reflect cellular redox and energy states 7, 9. The striking metabolic phenotype of Sirt6−/− mice—including an apparently lethal hypoglycemia despite low levels of IGF-1 35—is consistent with an important metabolic role for SIRT6. Indeed, SIRT6-overexpressing mice fed a high-fat diet were found to exhibit improved lipid profiles and increased glucose tolerance compared to wild-type controls 69. The role of SIRT6’s histone deacetylase activity in this mouse model, however, remains to be clarified.
Because the dramatic drop in serum glucose levels in SIRT6-deficient mice is thought to be a primary contributor to their early lethality, understanding the mechanisms underlying the de-regulation of glucose homeostasis is an area of notable interest. Some mechanistic insight into this metabolic phenotype has come from the observation that isolated SIRT6-deficient mouse embryonic stem (ES) cells and mouse embryonic fibroblasts (MEFs) exhibit increased glucose uptake in culture and increased expression of glycolytic genes 68. These data suggest that the hypoglycemia of Sirt6−/− mice is due to a cell-autonomous defect in glucose metabolism 68. In vivo, the increase in glucose uptake appears to be tissue-specific: it was observed in brown adipose tissue and muscle but not in liver, brain, or heart. Importantly, inactivation of HIF1α either pharmacologically or through RNA interference can temporarily rescue the glucose uptake defects of SIRT6-deficient cells and the hypoglycemia of SIRT6-deficient mice 68, suggesting that increased activity of HIF1α contributes to the glucose metabolism defects of these mice.
Although these studies reveal interesting roles for the SIRT6–HIF1α interaction in ES cells and MEFs, HIF1α is best known for its role in cancer cell metabolism and the cellular response to hypoxia 67. Thus, it will be important to determine how SIRT6 deficiency affects the hypoxic response of mouse and human cancer cells. In addition, the observed tissue specificity of the glucose metabolism defects suggests that SIRT6 might play different roles in different physiologic contexts. Studies aimed at identifying upstream regulators of SIRT6 should provide insight into these differences.
Concluding remarks and future directions
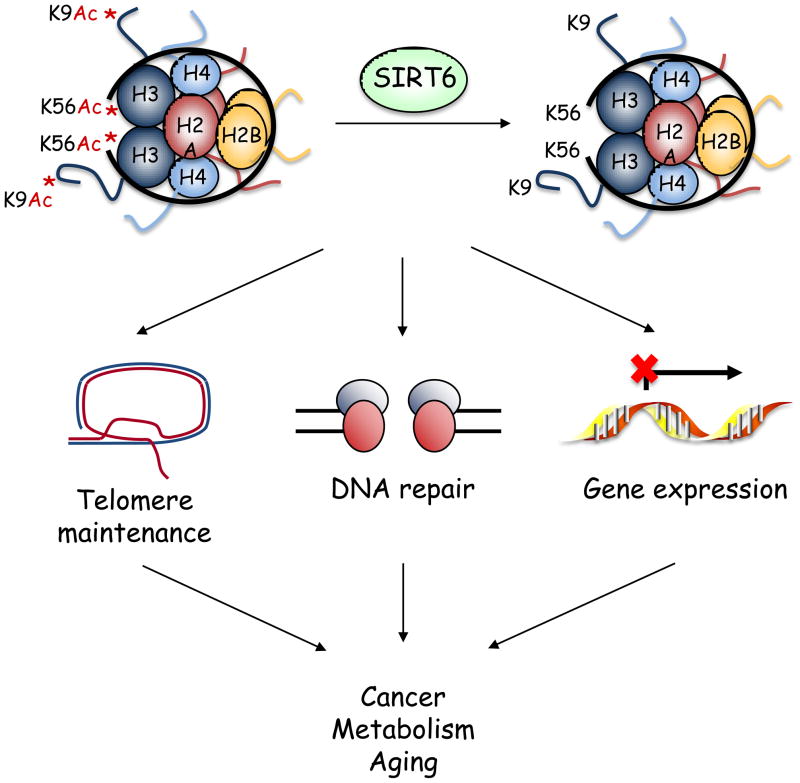
Work over the past several years has demonstrated that SIRT6 is a site-specific histone deacetylase that plays critical roles in maintaining telomere integrity, fine-tuning aging-associated gene expression programs, preventing genomic instability, and maintaining metabolic homeostasis, thus impacting on several pathways relevant for cancer, metabolism, and aging (Fig. 3). These new findings raise many questions, both mechanistic and physiologic.
Figure 3.
From orphan enzyme to site-specific deacetylase. By deacetylating lysines 9 and 56 on histone H3, SIRT6 modulates chromatin structure to regulate telomere integrity, DNA repair, and gene expression, thus impacting on processes important for cancer, metabolism, and aging.
First, SIRT6 appears to function at specific sites in the genome, including telomeres, DNA DSBs, and NF-κB and HIF1α target promoters. Does SIRT6 play a role in other genomic contexts? Based on the available genome-wide data 65, it seems likely that SIRT6 binds to additional gene promoters. Given its role in maintaining proper chromatin structure at telomeres, might SIRT6 associate with other regions of silent chromatin such as centromeric heterochromatin or the inactive X chromosome? Apart from transcription and DNA repair, does SIRT6 function in other chromatin-templated processes such as DNA replication? And how is SIRT6 recruited to these varied genomic loci? We also speculate that SIRT6 might have additional histone- and non-histone substrates that contribute to its roles in regulating genome integrity and gene expression. Genome-wide occupancy studies and the identification of SIRT6-interacting proteins will be critical in defining the role of SIRT6 in these and other cellular contexts.
Second, might there be functional interactions between SIRT6 and other mammalian sirtuin proteins? Crosstalk between different histone modifications is an important aspect of chromatin regulation 70, 71, suggesting that site-specific deacetylation of H3K9 or H3K56 by SIRT6 and deacetylation of other histone residues by SIRT1 24, SIRT2 72, and/or other sirtuins might cooperate to effect proper chromatin regulation.
Third, is there interplay between SIRT6’s roles in regulating genomic stability and maintaining metabolic homeostasis? Or does SIRT6 affect DNA repair and metabolism independently, through modification of histones at unique genomic loci? For example, altered metabolism in the absence of SIRT6 might lead to increased accumulation of DNA damage, or impaired DNA repair could trigger a compensatory metabolic response, as has been observed in mice lacking core nucleotide excision repair (NER) proteins 36.
Fourth, how might SIRT6 impact on cancer? Through its roles in maintaining genome stability and regulating metabolism, SIRT6 might be expected to impact strongly on oncogenic transformation and tumorigenesis. Indeed, several studies have hinted at links between SIRT6 and cancer: the SIRT6 chromosomal locus—19p13.3—is a frequent site of breakage in human acute myeloid leukemia 73; SIRT6 was identified in a genome-wide screen for genes required for K-ras-mediated upregulation of Fas in NIH-3T3 cells 74; the shift from aerobic respiration to glycolysis observed in SIRT6-deficient mouse cells 68 is reminiscent of the Warburg effect, in which cancer cells switch from oxidative phosphorylation to aerobic glycolysis 67; and a recent report suggests that global H3K56 acetylation is increased in many types of human cancer (including skin, thyroid, breast, liver, and colon cancers) and correlates with tumor grade 44, raising the intriguing possibility that aberrant H3K56 acetylation at specific genomic sites in the absence of SIRT6 might contribute to tumorigenesis. Studies examining the role of SIRT6 in cellular transformation and in xenograft tumor models should provide insight into the role of SIRT6 in cancer, as will mice in which Sirt6 is deleted or overexpressed in a tissue-specific and temporally regulated manner.
Finally, can we use recent insights into SIRT6 function—including functional dissection of SIRT6 and the growing list of physiologic contexts in which SIRT6 plays a crucial role—to inform the development of pharmacologic modulators of SIRT6 as treatments for diseases of premature aging, metabolic disorders, and cancer? Future research on SIRT6 will undoubtedly provide exciting new insights into chromatin regulation at both the biochemical and physiologic levels.
Acknowledgments
We apologize to our colleagues whose work could not be cited due to space constraints. We thank Or Gozani and members of the Chua and Gozani labs for helpful discussions and comments on the manuscript. R.I.T. is funded by a National Science Foundation Graduate Research Fellowship and an NIH training grant (1018438-142-PABCA). K.F.C. is a Paul Beeson Scholar and an Ellison Medical Foundation New Scholar in Aging, and is funded by grants from the NIH (R01AG028867 and K08AG028961) and Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gottschling DE, et al. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio OM, et al. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 3.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryk M, et al. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 5.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 6.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 7.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu F, et al. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onishi M, et al. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilaniu H, et al. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 16.Fabrizio P, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 17.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 18.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin SJ, et al. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 20.Kaeberlein M, et al. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 23.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 24.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 27.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 29.Michishita E, et al. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanno M, et al. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 32.Scher MB, et al. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North BJ, et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 34.Liszt G, et al. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 35.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 36.Monnat RJ., Jr From broken to old: DNA damage, IGF1 endocrine suppression and aging. DNA Repair (Amst) 2007;6:1386–1390. doi: 10.1016/j.dnarep.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombard DB, et al. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostoslavsky R. DNA repair, insulin signaling and sirtuins: at the crossroads between cancer and aging. Front Biosci. 2008;13:6966–6990. doi: 10.2741/3203. [DOI] [PubMed] [Google Scholar]
- 39.Tanny JC, et al. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 40.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009:8. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B, et al. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009:8. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan J, et al. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das C, et al. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 47.Han J, et al. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 48.Celic I, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 49.Tennen RI, et al. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hecht A, et al. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 51.Hoppe GJ, et al. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rusche LN, et al. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karras GI, et al. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 55.Blasco MA. The epigenetic regulation of mammalian telomeres. Nature reviews. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 56.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 57.Wong KK, et al. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 58.Negrini S, et al. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 59.Harley CB, et al. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 60.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCord RA, et al. SIRT6 stabilizes DNA-dependent Protein Kinase at chromatin for DNA double-strand break repair. Aging. 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassing CH, et al. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 63.Lieber MR, et al. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 64.Li Q, et al. Inflammation-associated cancer: NF-kappaB is the lynchpin. Trends Immunol. 2005;26:318–325. doi: 10.1016/j.it.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaeberlein M, Kapahi P. The hypoxic response and aging. Cell Cycle. 2009;8:2324. doi: 10.4161/cc.8.15.9126. [DOI] [PubMed] [Google Scholar]
- 67.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 68.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 70.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 72.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahlknecht U, et al. Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int J Oncol. 2006;28:447–456. [PubMed] [Google Scholar]
- 74.Gazin C, et al. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]