Abstract
The Psychiatric GWAS consortium was founded with the aim of conducting statistically rigorous and comprehensive GWAS meta-analyses for five major psychiatric disorders, ADHD, autism, bipolar disorder, major depressive disorder and schizophrenia. In the era of GWAS and high throughput genomics, a major trend has been the emergence of collaborative, consortia approaches. Taking advantage of the scale that collaborative, consortia approaches can bring to a problem, the PGC has been a major driver in psychiatric genetics and provides a model for how similar approaches may be applied to other disease communities.
Introduction
Genome-wide association studies (GWAS) have yielded an extraordinary and unprecedented trove of new knowledge about the genetic causes of human disease. Since 2005, well over 600 human GWAS have been published yielding genetic associations meeting stringent statistical significance (Pe'er et al. 2008) relevant to the etiology of 92 diseases and 117 other traits (Table 1) (Hindorff et al. 2009).Many of these GWAS findings have been surprising, and have engendered new ideas about disease etiology. Because exposure to genetic variation begins at the earliest stage of development, we can generally be confident that genetic risk factors are at the beginning of the causal chain that leads to disease perhaps decades later. Thus, each association is a starting point, a hard clue about disease etiology. As an example from outside of neuroscience, in Crohn's disease, GWAS implicated genes involved in macroautophagy which has provided important insights into pathogenesis (Klionsky 2009). The notably strong association of complement factor H (CFH) with age-related macular degeneration (Klein et al. 2005) has engendered renewed interest in the role of CFH role in initiating the disease process rather than as epiphenomenon associated with the disease. In schizophrenia, multiple independent studies have implicated the major histocompatibility region (International Schizophrenia Consortium 2009; Shi et al. 2009; Stefansson et al. 2009) raising the intriguing possibility of an etiological role for an immune, autoimmune, or infectious process. In addition, some studies have highlighted the role of alternative ways in which genetic variation might be etiological, including the importance of copy number variation (Sebat et al. 2009) and even compelling empirical data that schizophrenia results from the cumulative effects of thousands of different genetic variants in an as yet unknown biological pathway (International Schizophrenia Consortium 2009).
Table 1.
Published descriptions of the GWAS method.
| Citation | Comment |
|---|---|
| (Corvin et al. 2009) | A “primer” on GWAS, covers the major topics with annotations suggesting additional readings |
| (Attia et al. 2009) (Pearson and Manolio 2008) (Attia et al. 2009) |
JAMA series for users of the GWAS literature |
| (Altshuler and Daly 2007) (Hardy and Singleton 2009) (McCarthy and Hirschhorn 2008) |
Notable reviews of the GWAS method |
| (Chanock et al. 2007) | Replication in human genetics |
| (Manolio et al. 2007) | Establishing consortia in human genetics |
| (Neale and Purcell 2008) | Quality control of GWAS data |
| (de Bakker et al. 2008) | Meta-analysis of GWAS data |
| (Psychiatric GWAS Consortium 2009) (Psychiatric GWAS Consortium 2009) (Cross Disorder Phenotype Group of the Psychiatric GWAS Consortium 2009) |
Series of papers describing the PGC |
Even the most cursory review of the GWAS literature reveals that the critical ingredient to success is large samples for initial discovery and replication. Sample size requirements can easily exceed 10,000 cases and 10,000 controls. Obtaining such historically massive sample sizes is beyond the reach of any single group. Therefore, close cooperation among groups has become essential to progress in human genetics.
The purpose of this Neuron NeuroView is to describe the Psychiatric Genome-Wide Association Consortium (PGC) which was created in an attempt to conduct large-scale mega-analyses of GWAS data for psychiatric disorders. Psychiatric diseases are compelling targets for genetics research – they are mostly idiopathic, first-rank public health problems, and cause enormous morbidity, mortality, and personal/societal cost. Moreover, despite considerable research, little is known for certain about disease etiology. The fact that disease definition in psychiatry is descriptive poses particular problems. The diagnostic process relies heavily on signs and symptoms without recourse to biological means of distinguishing affected from unaffected individuals. This poses unique challenges for genetic studies. Although the PGC's focus has been in the area of psychiatric genetics, the organization and consortia approach which the PGC exemplifies could be a model for other disorders and more broadly, even outside of genetics, for other research communities applying high-throughput analytic approaches to biological problems.
GWAS background
As the GWAS method has been reviewed extensively, only a brief description is given here and Table 1 provides ample opportunities for further reading. A GWAS for a human disease is usually a variant of a cross-sectional case-control study, the familiar workhorse in biomedicine. Cases meet lifetime criteria for a disease (e.g., schizophrenia) and controls should have never met criteria and, ideally, be through the period of risk. Each individual in the sample is genotyped for a pre-defined set of a million or more genetic markers spaced across the genome. The genetic markers are single nucleotide polymorphisms (SNPs, “snips”) which are relatively straight-forward to assay in a highly multiplexed fashion. After careful quality control, each SNP is tested for association with disease. In effect, these tests compare the allele frequencies in cases versus controls, and a large case-control difference suggests an etiological role for a particular SNP or its genomic region. Because of the large numbers of statistical comparisons, the laws of probability mandate correction for multiple comparisons. A typical type 1 error threshold for genome-wide significance is often taken to be 5×10-8 (akin to a Bonferroni correction of 0.05 divided by 1 million tests) (Pe'er et al. 2008).
Psychiatric GWAS Consortium (PGC): Background & Science
As increasing numbers of GWAS were published in 2005-2006, it became apparent that typical sample sizes (e.g., 1,000 cases and 1,000 controls) did not usually lead to associations that exceeded chance levels of significance. Such findings provided, for the first time, evidence that the genetic effect sizes for common variation were considerably smaller than had been appreciated. For example, in the pre-GWAS era, many investigators powered their studies to detect genotypic relative risks of °1.54 (1000 cases/1000 controls, α=5×10-8, and 90% power). Using the NHGRI GWAS catalog (Hindorff, 2009)}, the typical genotypic relative in a GWAS is far smaller than appreciated previously (median of 1.28) which necessitates a sample size of over 3,000 cases and 3,000 controls. To identify the 25th percentile genotypic relative risk of 1.18 requires nearly 7,000 cases and 7,000 controls.
Thus, assumptions about power and sample size required revision; studies which seemed well-powered when they began were too small. It became obvious that larger sample sizes were needed and groups working together were the only practical way to achieve this end. Consortia were thus an immediate solution. The concept of working together was influenced by the experiences of other biomedical disorders. For example, the initial three GWAS for type 2 diabetes mellitus were only modestly successful but joint analysis revealed many more strongly significant associations (Zeggini et al. 2008).
The PGC began on a teleconference in March 2007 between principal investigators who had GWAS funded for Attention Deficit Hyperactivity Disorder (ADHD), bipolar disorder, major depressive disorder, and schizophrenia as part of a Foundation for the NIH initiative (Manolio et al. 2007). Even though, at the time, this project was at early stages and as no psychiatric GWAS had yet been published, we were already concerned about power and hence initiated plans for joint analysis of our results. This effort rapidly expanded to include autism, , the other major psychiatric disorder with a considerable body of GWAS data. Subsequently, all investigators in the field with data for these five disorders (ADHD, AUT, BIP, MDD, and SCZ) were invited to join the PGC. All but one group invited has joined.
The over-arching purpose of the PGC is to conduct high-quality GWAS mega-analyses in order to foster rapid progress in what has been a complex and uncertain scientific area. These results are meant to inform research into ADHD, autism, bipolar disorder, major depressive disorder, and schizophrenia along with searches for genetic loci that predispose to more than one disorder. The initial iteration of the PGC had four scientific aims, which were designed to facilitate the over-arching scientific goal of attempting to identify secure associations of comprehensive assessment of common genetic variation with five critically important psychiatric diseases.
The first aim involved dataset harmonization. Experience has taught us that unless this is conducted with expertise and great care, inference is not secure. Harmonization and quality control apply to each step of the GWAS process – ascertainment of subjects, diagnostic procedures, genotyping, removal of subjects and SNPs with unconfident data, and with extensive searches for bias. [pfs1]For the PGC, raw individual-level and de-identified phenotype and genotype data from each study were uploaded to a high performance computing cluster and processed through a robust and comprehensive quality control pipeline conforming to best-practice protocols in order to minimize chances of false positive results (e.g., due to population stratification). As the individual studies used different genotyping platforms, the cleaned data were imputed against a widely used panel of data from European subjects (HapMap3) (Altshuler et al. 2010) so that all studies had a common set of genotypes. In addition, considerable efforts were made to harmonize phenotype data by ensuring that all studies used comparable diagnostic constructs and to database item-level data.
The second aim entailed within-disorder meta-analyses – five different mega-analyses of all available GWAS data for ADHD, autism, bipolar disorder, major depressive disorder and schizophrenia, to attempt to identify convincing genotype-phenotype associations.
The point of the third aim is specific to psychiatry. Throughout the history of psychiatry, diagnoses have been made based on signs and symptoms accrued in conversations between physician and patient. Although test-retest reliability is generally acceptable, these are fundamentally descriptive syndromes and their validity is unknown. Moreover, there is considerable overlap between disorders. For example, people with autism often have ADHD. Cases with schizophrenia frequently have symptoms highly similar to those with BIP and major depressive disorder. Indeed, major depressive disorder and BIP are alike in that both include major depressive episodes whereas BIP additionally has manic episodes. Given that clinically-derived definitions of illness may not have “carved nature at the joint” with respect to the fundamental genetic architecture (Kendell and Brockington 1980; Kendell 1989), This aim attempts to identify convincing genetic associations that are common to two or more of ADHD, autism, bipolar disorder, major depressive disorder, and schizophrenia. This work could provide critical insight into how these disorders are similar and different.
The fourth aim is related to data sharing. Consistent with the goal of rapid progress, we have been communicating pre-publication results widely. Where informed consent and Ethical Committee rulings allow, de-identified phenotype, genotype, and mega-analysis results will be deposited into controlled-access repositories (e.g., dbGaP, NCBI database of Genotypes and Phenotypes) (Mailman et al. 2007) in order to make these data available for future use by the international scientific community.
Since the formation of the PGC, as we move forward, we plan for a new set of aims to continue the work of the PGC. The new aims include a comprehensive assessment of copy number variation and extension of the analytic pipeline to encompass next-generation sequencing data.
The practical side of the PGC
By virtue of the numbers of investigators, subjects, and data points, the PGC is the largest consortium and biological experiment in the history of psychiatry. The PGC currently has over 160 investigators from 65 institutions in 19 countries. Membership has been extended to groups with high-quality GWAS data and usually include the study principal investigator and key collaborators. Joining the PGC entails reading and agreeing to the rules of behavior detailed in a memorandum of understanding. Assent is indicated by email and effectively constitutes a pledge to behave with integrity. The PGC sponsored a series of papers outlining the history of genetic inquiries in psychiatry, a framework for interpretation of GWAS, and issues pertaining to comorbidity between disorders (Table 1).
Participation in the PGC is driven by varying combinations of altruism and enlightened self-interest. Some investigators are inherently collegial and enjoy consortia whereas others would prefer to work independently but have come to believe cooperation is essential for progress. Others are motivated by different imperatives and we are aware that some in the field have chosen not to join given difficulties in functioning comfortably and effectively in a group context.
The PGC consists of a coordinating committee, five disease working groups (ADHD, autism, bipolar, major depressive disorder, and schizophrenia), the cross-disorder working group, and a statistical analysis group that has a CNV subgroup. Working group chairs are: ADHD Dr Stephen Faraone; autism Drs Bernie Devlin and Mark Daly; bipolar disorder Drs John Kelsoe and Pamela Sklar; major depressive disorder Dr Patrick Sullivan; schizophrenia; Dr Pablo Gejman; Statistical analysis Dr Mark Daly; cross-disorder Drs Nick Craddock, Jordan Smoller, and Ken Kendler; and CNV Drs Mark Daly, Steven Scherer, and Jonathan Sebat. Dr Sullivan also chairs the coordinating committee. Additional GWAS for anorexia nervosa and obsessive-compulsive disorder are becoming available and will become part of the PGC in the near future.There are notable computational demands for a project of this scale. We are deeply indebted to Dr Danielle Posthuma (Vrije University Amsterdam) for facilitating the use of a cluster farm in the Netherlands for data warehousing and analysis. The use of this cluster has provided a neutral platform for analyses.
From the beginning, the overall philosophy of the PGC has been to be as inclusive, democratic, transparent, and rapid as possible. No single individual or group dominates. The role of the coordinating committee is to adjudicate procedural issues of relevance to the whole consortium (e.g., to integrate efforts of the working groups and to secure the needed resources). A metaphor for the relationship between the coordinating committee and the working groups is the recurrent theme of United States history, the tension between Federalism and “states’ rights”. The belief is that the best science will emerge if the balance is decidedly shifted towards “states’ rights”. The “federal” coordinating committee has a non-intrusive and facilitating role and all other decisions are delegated to the scientists who understand the issues best. There are often differences of opinion. These are almost always resolved by discussion. Rarely, discussion did not lead to resolution and necessitated a vote (simple majority, one vote per group contributing data).
A key principle has been that groups participate in the PGC at a time appropriate for their group. In practice, this was usually after their GWAS primary manuscript was accepted for publication. Participation in the PGC does not preclude any other academic effort (for courtesy, however, investigators inform their colleagues about any competing activities). An early decision adopted by all working groups was to publish under a consortium byline with all members of that working group listed as “collaborators” in PubMed. (See http://www.ncbi.nlm.nih.gov/pubmed/17554300 for an example) This practical decision allowed the focus to remain on collaborative science and negated the otherwise inevitable jockeying for priority authorship positions. As a result of the clear and consistent application of these basic principles, the PGC has been running smoothly for several years despite its large membership. We note that no one who joined the PGC has quit.
The goal of the PGC is rapid and unfettered progress. As part of the Memorandum of Understanding, PGC members agreed that all genotype and phenotype data should be kept strictly confidential within that working group. Moreover, when the analyses for a specific aim were completed, the results could be freely discussed and participants were free to initiate follow-up experiments. In the interests of maximal progress, we encouraged pre-publication sharing of follow-up experiments. However, the results could not be used in presentations or publications without prior approval.
The PGC encourages a responsible approach to management of intellectual property derived from downstream discoveries that is consistent with the recommendations of the NIH's Best Practices for the Licensing of Genomic Inventions and Research Tools Policy. (http://www.ott.nih.gov/policy/genomic_invention.html and http://ott.od.nih.gov/policy/research_tool.html)In particular, management of patent applications in a manner that restricts use of any findings or that might diminish the value and public benefit provided by these resources is discouraged.
Finally, all PGS members were required to share the commitment to protect the confidentiality of data and to protect the joint analysis activity by insuring that no data were released or published in advance of an agreed-upon group publication and/or data release.
How can genetics inform neurobiology?
The fundamental goal of the PGC is to derive “maps” of the genetic architecture for the major psychiatric disorders, ADHD, autism, bipolar disorder, major depressive disorder and schizophrenia. What does this mean for neuroscientists working on understanding the mechanisms of these disorders? Or to clinicians and patients looking for therapies or at least a better understanding of these disorders and their causes?
For scientists who study processes fundamental to the development of the central nervous system and its function in health and disease, these results are likely to be highly relevant. Genetic risk factors will include a spectrum of variation, from rare variants of strong effect to common variants of more subtle effect. Moreover, these data are likely to uncover novel similarities between currently distinctive disorders and new ways in which genetic changes can lead to disease (e.g., copy number variation and highly polygenic models).
It has now been widely observed that GWAS findings only infrequently implicate the “usual suspects.” In other words, when GWAS identifies a high-confidence and replicated finding, the loci implicated often point in a novel direction and these new leads can then become targeted priorities for more mechanistically oriented experimental work. While the holy grail of GWAS may be the identification of a strongly associated risk allele, as more associations emerge from GWAS and other genomic approaches and these findings are replicated, even apparently modest risk alleles may point us towards relevant biological pathways and networks.
Historically, there has been a gap between psychiatric genetics and neuroscience. In an idealized universe, psychiatric genetics and neuroscience would have rather symbiotic relations. In this way, we may well find that a genetic, molecular, or neuronal process being studied in a lab for one set of reasons ends up emerging as a potentially critical factor for a psychiatric disorder, based on genetic data. Ultimately, it's this kind of synergy, between genetics and biology, which will pave the path to true understanding of how genotype confers risk for phenotype and gives us the best chance of really understanding these disorders and paving the way for more effective therapies.
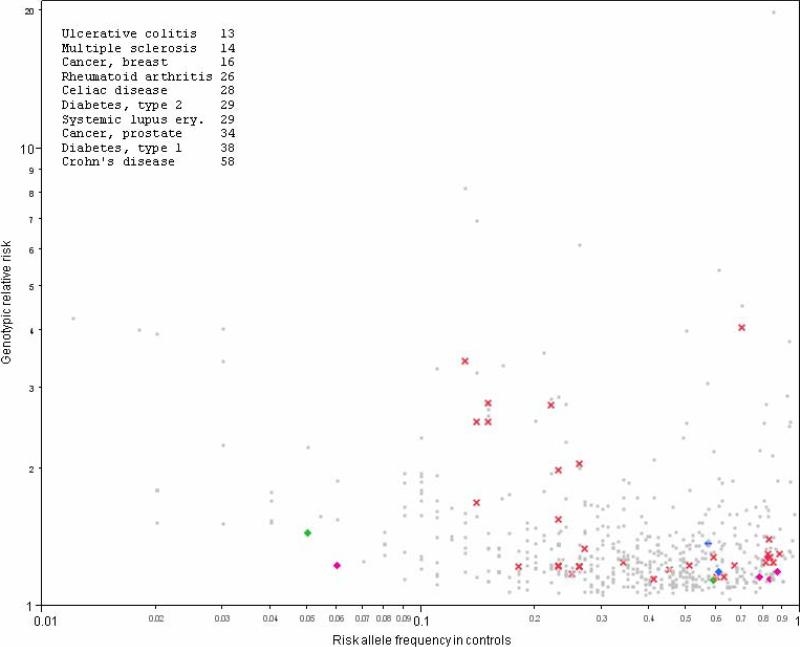
Figure 1.
Depicted are 587 associations for 76 human diseases. Each association is plotted as its genotypic relative risk by the risk allele frequency in controls (both on log10 scale). The insert shows the 10 diseases with the greatest numbers of associations. Red crosses show findings for neurological disorders (Alzheimer disease, amyotrophic lateral sclerosis, essential tremor, multiple sclerosis, narcolepsy, and Parkinson disease). Psychiatric disorders are indicated with plus marks including autism (blue), mood disorders (green), and schizophrenia (fuchsia). All other biomedical disorders are shown by grey dots.
Acknowledgements
Dr. Sullivan reports no conflicts of interest. This project was funded by MH085520. Funding for this project was from the US National Institutes of Health who had no role in manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat Genet. 2007;39(7):813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- Attia J, Ioannidis JP, Thakkinstian A, McEvoy M, Scott RJ, Minelli C, Thompson J, Infante-Rivard C, Guyatt G. How to use an article about genetic association: A: Background concepts. Jama. 2009;301(1):74–81. doi: 10.1001/jama.2008.901. [DOI] [PubMed] [Google Scholar]
- Attia J, Ioannidis JP, Thakkinstian A, McEvoy M, Scott RJ, Minelli C, Thompson J, Infante-Rivard C, Guyatt G. How to use an article about genetic association: C: What are the results and will they help me in caring for my patients? Jama. 2009;301(3):304–308. doi: 10.1001/jama.2008.993. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69(2):428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr., Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Corvin A, Craddock N, Sullivan PF. Genome-wide association studies: a primer. Psychologal Medicine. 2009 doi: 10.1017/S0033291709991723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross Disorder Phenotype Group of the Psychiatric GWAS Consortium Dissecting the phenotype in genome-wide association studies of psychiatric illness. British Journal of Psychiatry. 2009;195:97–99. doi: 10.1192/bjp.bp.108.063156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(R2):R122–128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360(17):1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendell RE. Clinical validity. British Journal of Psychiatry. 1989;19:45–55. doi: 10.1017/s0033291700011016. [DOI] [PubMed] [Google Scholar]
- Kendell RE, Brockington IF. The identification of disease entities and the relationship between schizophrenic and affective psychoses. British Journal of Psychiatry. 1980;137:324–331. doi: 10.1192/bjp.137.4.324. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Crohn's disease, autophagy, and the Paneth cell. N Engl J Med. 2009;360(17):1785–1786. doi: 10.1056/NEJMcibr0810347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, Donnelly P, Faraone SV, Frazer K, Gabriel S, Gejman P, Guttmacher A, Harris EL, Insel T, Kelsoe JR, Lander E, McCowin N, Mailman MD, Nabel E, Ostell J, Pugh E, Sherry S, Sullivan PF, Thompson JF, Warram J, Wholley D, Milos PM, Collins FS. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Purcell S. The positives, protocols, and perils of genome-wide association. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1288–1294. doi: 10.1002/ajmg.b.30747. [DOI] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Pearson TA, Manolio TA. How to interpret a genome-wide association study. Jama. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium A framework for interpreting genomewide association studies of psychiatric disorders. Molecular Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Genome-wide association studies: history, rationale, and prospects for psychiatric disorders. American Journal of Psychiatry. 2009;166:540–546. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25(12):528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]