Abstract
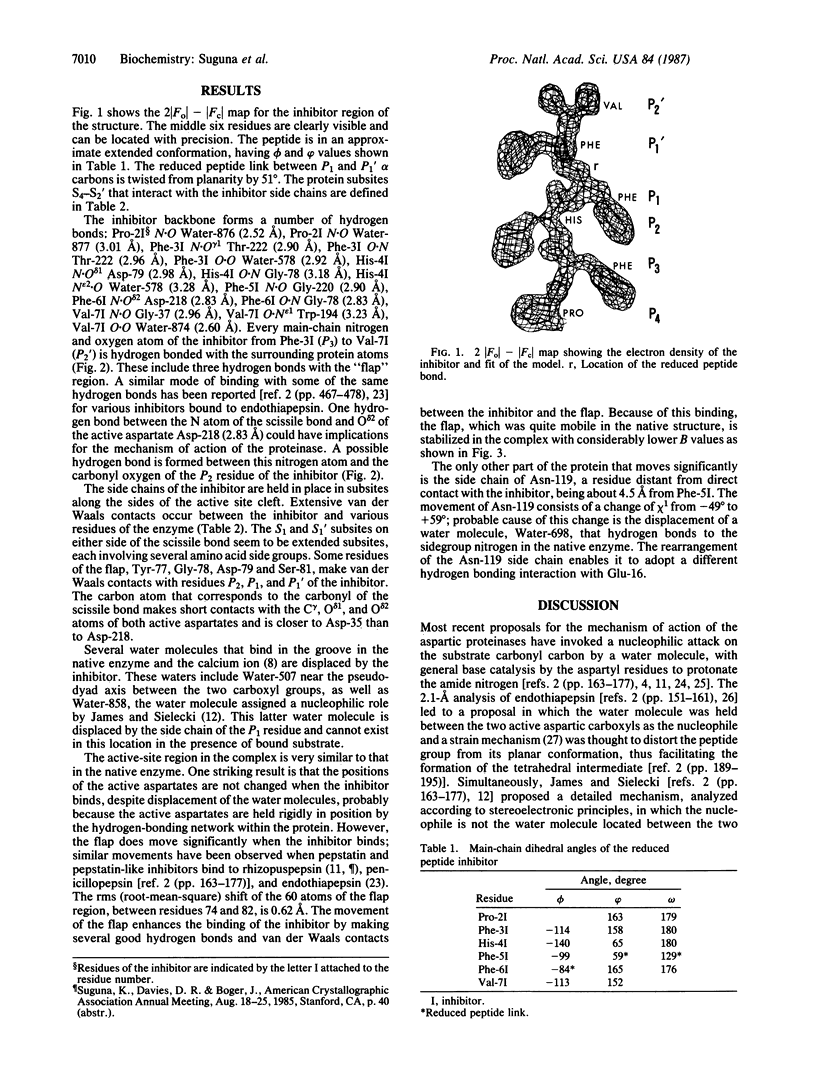
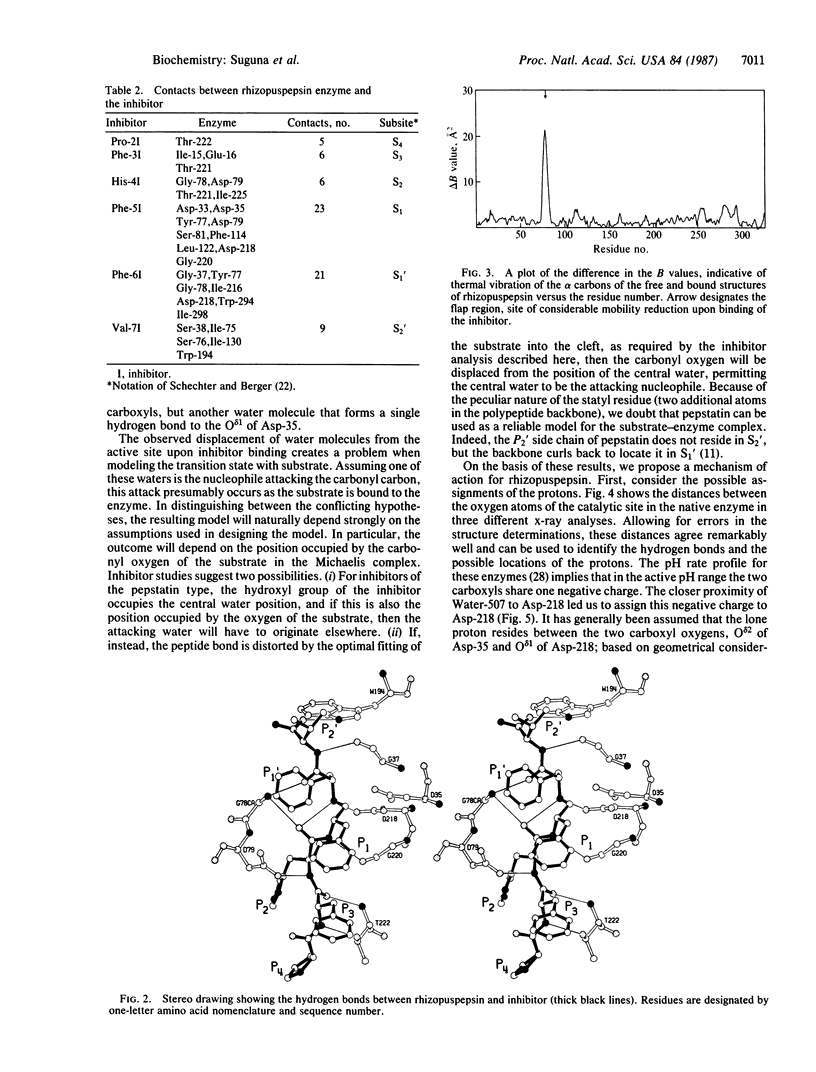
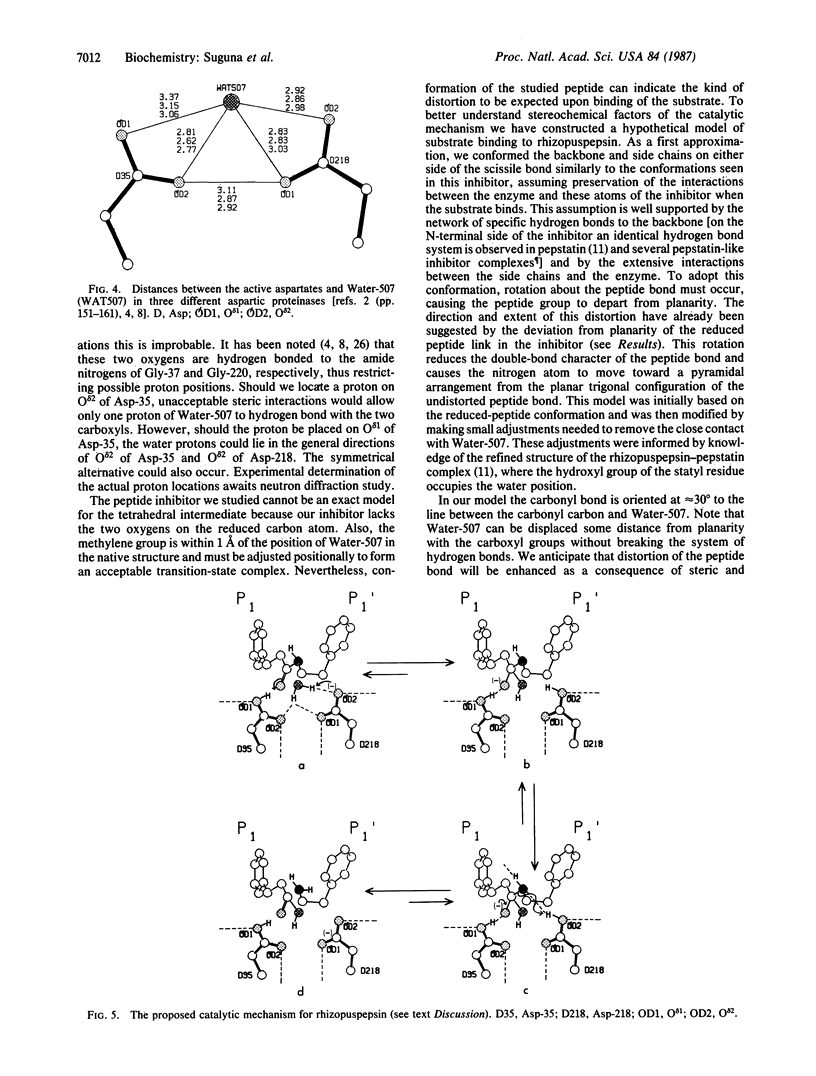
A peptide inhibitor, having the sequence D-His-Pro-Phe-His-Phe psi [CH2-NH]Phe-Val-Tyr, with a reduced bond between the two adjacent phenylalanines, has been diffused into crystals of the aspartic proteinase from Rhizopus chinensis (rhizopuspepsin, EC 3.4.23.6). X-ray diffraction data to 1.8-A resolution have been collected on the complex, which has been subjected to restrained least-squares refinement to an R-factor (R equals the sum of the absolute value of the difference between the observed and calculated structure factor amplitudes divided by the sum of the observed structure factor amplitudes) of 14.7%. The inhibitor lies within the major groove of the enzyme and is clearly defined with the exception of the amino-terminal D-histidine and the carboxyl-terminal tyrosine. The reduced peptide bond is located in the active site with close contacts to the two catalytic aspartyl groups. The active-site water molecule that is held between the two carboxyl groups is displaced by the inhibitor, as are a number of other water molecules seen in the binding groove of the native enzyme. A mechanism of action for this class of enzymes is proposed from these results.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreeva N. S., Gustchina A. E., Fedorov A. A., Shutzkever N. E., Volnova T. V. X-ray crystallographic studies of pepsin. Adv Exp Med Biol. 1977;95:23–31. doi: 10.1007/978-1-4757-0719-9_2. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bott R., Subramanian E., Davies D. R. Three-dimensional structure of the complex of the Rhizopus chinensis carboxyl proteinase and pepstatin at 2.5-A resolution. Biochemistry. 1982 Dec 21;21(26):6956–6962. doi: 10.1021/bi00269a052. [DOI] [PubMed] [Google Scholar]
- Foundling S. I., Cooper J., Watson F. E., Cleasby A., Pearl L. H., Sibanda B. L., Hemmings A., Wood S. P., Blundell T. L., Valler M. J. High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes. 1987 May 28-Jun 3Nature. 327(6120):349–352. doi: 10.1038/327349a0. [DOI] [PubMed] [Google Scholar]
- Fruton J. S. The mechanism of the catalytic action of pepsin and related acid proteinases. Adv Enzymol Relat Areas Mol Biol. 1976;44:1–36. doi: 10.1002/9780470122891.ch1. [DOI] [PubMed] [Google Scholar]
- Hofmann T., Fink A. L., Dunn B. M. Cryoenzymology of penicillopepsin; with an appendix: mechanism of action of aspartyl proteinases. Biochemistry. 1984 Oct 23;23(22):5247–5256. doi: 10.1021/bi00317a024. [DOI] [PubMed] [Google Scholar]
- Hofmann T., Hodges R. S., James M. N. Effect of pH on the activities of penicillopepsin and Rhizopus pepsin and a proposal for the productive substrate binding mode in penicillopepsin. Biochemistry. 1984 Feb 14;23(4):635–643. doi: 10.1021/bi00299a008. [DOI] [PubMed] [Google Scholar]
- Howard A. J., Nielsen C., Xuong N. H. Software for a diffractometer with multiwire area detector. Methods Enzymol. 1985;114:452–472. doi: 10.1016/0076-6879(85)14030-9. [DOI] [PubMed] [Google Scholar]
- James M. N., Hsu I. N., Delbaere L. T. Mechanism of acid protease catalysis based on the crystal structure of penicillopepsin. Nature. 1977 Jun 30;267(5614):808–813. doi: 10.1038/267808a0. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Stereochemical analysis of peptide bond hydrolysis catalyzed by the aspartic proteinase penicillopepsin. Biochemistry. 1985 Jul 2;24(14):3701–3713. doi: 10.1021/bi00335a045. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Structure and refinement of penicillopepsin at 1.8 A resolution. J Mol Biol. 1983 Jan 15;163(2):299–361. doi: 10.1016/0022-2836(83)90008-6. [DOI] [PubMed] [Google Scholar]
- Jenkins J., Tickle I., Sewell T., Ungaretti L., Wollmer A., Blundell T. X-ray analysis and circular dichroism of the acid protease from Endothia parasitica and chymosin. Adv Exp Med Biol. 1977;95:43–60. doi: 10.1007/978-1-4757-0719-9_4. [DOI] [PubMed] [Google Scholar]
- Pearl L. H. The catalytic mechanism of aspartic proteinases. FEBS Lett. 1987 Apr 6;214(1):8–12. doi: 10.1016/0014-5793(87)80003-0. [DOI] [PubMed] [Google Scholar]
- Pearl L., Blundell T. The active site of aspartic proteinases. FEBS Lett. 1984 Aug 20;174(1):96–101. doi: 10.1016/0014-5793(84)81085-6. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967 Apr 20;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Subramanian E., Liu M., Swan I. D., Davies D. R. The crystal structure of an acid protease from Rhizopus chinensis at 2.5 A resolution. Adv Exp Med Biol. 1977;95:33–41. doi: 10.1007/978-1-4757-0719-9_3. [DOI] [PubMed] [Google Scholar]
- Subramanian E., Swan I. D., Liu M., Davies D. R., Jenkins J. A., Tickle I. J., Blundell T. L. Homology among acid proteases: comparison of crystal structures at 3A resolution of acid proteases from Rhizopus chinensis and Endothia parasitica. Proc Natl Acad Sci U S A. 1977 Feb;74(2):556–559. doi: 10.1073/pnas.74.2.556. [DOI] [PMC free article] [PubMed] [Google Scholar]


