Abstract
The TGF-β and Wnt pathways are involved in cell fate and tumorigenicity. A recent report indicated that a TGF-β target gene, TMEPAI (transmembrane prostate androgen-induced RNA), is possibly also a downstream target of Wnt signaling. Although TMEPAI was believed to be involved in tumorigenicity because of its blockage of TGF-β signaling, how TGF-β and Wnt signals affect the activation of the TMEPAI gene is not well understood. Herein, we show that the TMEPAI promoter is regulated synergistically by TGF-β/Smad and Wnt/β-catenin/T cell factor (TCF) 7L2. The critical cis-element for dual signals, termed TGF-β-responsive TCF7L2-binding element (TTE), is located in intron 1 of the TMEPAI gene. TCF7L2, but not Smad proteins, bound to TTE, whereas the disruption of TTE by mutagenesis remarkably counteracted both TGF-β and TCF7L2 responses. The introduction of mutations in critical Smad-binding elements blocked the activation of the TMEPAI promoter by TCF7L2. Furthermore, our DNA-protein interaction experiments revealed the indirect binding of TCF7L2 to Smad-binding elements via Smad3 upon TGF-β stimulation as well as its TGF-β-dependent association with TTE. We demonstrate that the Wnt/β-catenin/TCF7L2 pathway is preferentially able to alter the transcriptional regulation of the TGF-β-target gene, TMEPAI.
Keywords: β-Catenin, SMAD Transcription Factor, T Cell Factor (TCF), Transcription, Transforming Growth Factor β (TGF-β), Wnt Pathway
Introduction
The TGF-β ligands mediate their signals in cells via specific serine/threonine kinase receptors and intracellular signal transducing molecules, termed Smads (1). Each step of the TGF-β signal transduction pathway appears to be subject to both positive and negative regulation (2, 3). The TGF-β family is implicated in embryogenesis and maintenance of tissue homeostasis during adult life. Thus, aberrant signaling by TGF-β family members is involved in various diseases including cancer, fibrosis, and vascular disorders (4, 5). Although TGF-β acts as a tumor suppressor by inhibiting cell growth, it also promotes tumor progression and metastasis by inducing epithelial-mesenchymal transition, invasion, immunesuppression, and blood vessel intravasation by carcinoma cells (2, 6).
The canonical Wnt cascade is initiated by the binding of Wnt ligands to their cognate receptor complex components such as the Frizzled family and low density lipoprotein receptor-related protein 5/6. In the absence of canonical Wnt signaling, β-catenin is phosphorylated, ubiquitinated, and degraded. Upon the activation of the canonical Wnt pathway, the phosphorylation and degradation of β-catenin are inhibited. Thus, stabilized β-catenin can accumulate to the nucleus, where it makes an active transcription complex with the T cell factor/lymphoid enhancer-binding factor (TCF/LEF)2 family of DNA binding transcription factors (7).
TMEPAI (transmembrane prostate androgen-induced RNA) can interact with either Smad2 or Smad3 via its Smad interaction motif to sequester TGF-β/Smad signaling because of its competition with Smad anchor for receptor activation for binding to Smads. Because TMEPAI is a direct target gene for TGF-β signaling, TMEPAI seems to act as a molecule involved in the negative feedback loop of TGF-β signaling (8). Except for its contribution in TGF-β signaling, TMEPAI is known to be implicated in the degradation of androgen receptor by the recruiting of the E3 ubiquitin ligase to androgen receptor (9) as well as by cell growth inhibition and p53-induced apoptosis in a context-dependent fashion (10, 11). In addition to TGF-β stimulation, TMEPAI has been reported to be induced by treatment with androgen, introduction of mutant p53, or activation of the ERK pathway (11–13). Recently, TMEPAI was reported to be highly expressed in the intestinal polyps of ApcMin/+ mice (8, 14). Thus, these reports supported the theory that the TMEPAI gene might be one of the canonical Wnt target genes. Furthermore, TMEPAI expression was increased in breast cancer, colon cancer, and renal cell carcinoma in humans (13, 15, 16).
It has been reported that the canonical Wnt/β-catenin pathway collaborates with either TGF-β or BMP signaling in an agonistic or antagonistic fashion. In an agonistic manner, the complex of β-catenin and the TCF/LEF family interacts with Smad proteins to coordinate the transcription of target genes (17–20), whereas the transcript of the Id1 gene induced by BMP is antagonistically regulated by Wnt3a, which might inhibit the transcriptional complex formation on the BMP-responsive element of the Id1 gene (21).
Given the recognized role of TMEPAI in the regulation of the TGF-β pathway, we explored the possible role of the canonical Wnt pathway in the modulation of TMEPAI transcription. Our results indicate that the TMEPAI gene is synergistically transactivated by TGF-β/Smad and Wnt/β-catenin/TCF7L2 at the transcriptional level. Furthermore, these results support the notion that TCF7L2 is recruited to a TGF-β-responsive TCF7L2-binding element (TTE) via its indirect binding to Smad-binding elements (SBEs) upon TGF-β stimulation.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Constitutively active activin receptor-like kinase 5 (ALK5ca)/V5, FLAG-Smad2, FLAG-Smad3, FLAG-Smad4, (CAGA)12-luc, FLAG-Smad2Δexon3, HA-TCF7L2, HA-TCF7L2Δ(1–30), and HA-β-catenin were described previously (22–26). Myc-TCF7L2 was kindly gifted by Dr. Watanabe (27). For −1972TMEPAI-luc and −607TMEPAI-luc, the fragments from −1972 to +67 and from −607 to +67 (the sequence information of NC_000068 in NCBI reference sequence is referred) were respectively amplified using TMEPAI gene in BAC mouse genomic library as a template and cloned into pGL3-basic (Promega). pGL3ti-850 was constructed by the ligation of the fragment from +447 to +1294 of mouse TMEPAI gene with pGL3ti (28). For −607TMEPAI-luc-850 and −607TMEPAI-luc-850r, the fragment from +447 to +1294 of mouse TMEPAI gene was put behind the 3′ end of the luciferase gene in −607TMEPAI-luc at both orientations. The other plasmids described were constructed by PCR-based amplification. After generation of all mutants, the sequences in each plasmid were confirmed.
Cell Culture
MDA-MB468, SC3, 293T, and COS7 cells were cultured in DMEM (Invitrogen) containing 10% FCS (Invitrogen) and nonessential amino acids (Sigma). HepG2 cells were maintained in minimum essential medium (Sigma) containing 10% FCS, nonessential amino acids, and sodium pyruvate. For selection of stable transformants with lentiviral vectors in MCF10A1, the cells were cultured in the presence of 1 μg/ml puromycin (Sigma). Smad3-deficient mouse embryonic fibroblasts (Smad3KO-MEFs) were established using Smad3KO mice, which were kindly provided by Dr. A. B. Roberts (29). In parallel, establishment of MEFs from wild-type mice was also carried out. Both MEFs were cultivated in DMEM containing 10% FCS and nonessential amino acids. MCF10A1 cells were maintained by the method described previously except for use of 5 μm forskolin instead of cholera toxin (30).
Transcriptional Reporter Assays
One day before transfection, HepG2 cells were seeded at 1.0 × 105 cells/well in 12-well plates. The cells were transfected using FuGENE 6 (Roche Applied Science). Where indicated, 5 ng/ml TGF-β3 was added into the wells 24 h after transfection. Subsequently, the cells were cultured in the absence of FCS for 18 h. In all of the experiments, β-galactosidase (pCH110; GE Healthcare) activity was measured to normalize for transfection efficiency. Each transfection was carried out in triplicate and repeated at least twice. The transfection into MDA-MB468 cells and MEFs was performed according to the method of the transfection to HepG2 cells except for seeding the cells at 2.5 × 105/well in a 6-well plate.
Immunoprecipitation and Western Blotting
To detect interactions among proteins, the plasmids were transfected into COS7 cells (5 × 105 cells/6 cm dish) using FuGENE 6. Forty hours after transfection, the cells were lysed in 500 μl of TNE buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 100 units/ml aprotinin, 2 mm sodium vanadate, 40 mm NaF, and 20 mm β-glycerophosphate). The cell lysates were precleared with protein G-Sepharose beads (GE Healthcare) for 30 min at 4 °C and then incubated with anti-FLAG M5 antibody (Sigma) for 2 h at 4 °C. Protein complexes were immunoprecipitated by incubation with protein G-Sepharose beads for 30 min at 4 °C followed by three washes with TNE buffer. Immunoprecipitated proteins and aliquots of total cell lysates were boiled for 5 min in sample buffer, separated by SDS-PAGE, and transferred to Hybond-C Extra membrane (GE Healthcare). The membranes were probed with primary antibodies. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and chemiluminescent substrate (Thermo Scientific). Protein expression in total cell lysates was evaluated by Western blotting.
DNA Affinity Precipitation
COS7 cells were seeded at 1.5 × 106 cells/10-cm dish 1 day before transfection. The cells were transfected using FuGENE 6. Forty hours after transfection, the cells were lysed in 1 ml of TNE buffer. The cell lysates were precleared with 12 μg/ml poly(dI·dC) and streptavidin-agarose (Sigma) for 30 min and incubated with 24 μm biotinated (SBE)3(TTE) or biotinated (mSBE)3(TTE) for 2 h at 4 °C. Subsequently, streptavidin-agarose was added to the reaction mixture and incubated for 30 min at 4 °C. After the precipitates were washed with TNE buffer three times, precipitates and aliquots of total lysates were separated by SDS-PAGE. Then proteins were transferred to the membrane. The membrane was incubated with the indicated primary antibodies. Primary antibodies were detected as described above. The sequences of biotinated (SBE)3(TTE) and biotinated (mSBE)3(TTE) are as follows: biotinated (SBE)3(TTE), 5′-biotinated TTTTAGCCAGACAAAAAGCCAGACATTTAGCCAGACATTTTATGAGTCAAAGT-3′ and 3′-AAAATCGGACGTTTTTCGGTCTGTAAATCGGTCTGTAAAATACTCAGTTTCA-5′; and biotinated (mSBE)3(TTE), 5′-biotinated TTTTAGCtacatAAAAAGCtacatATTTAGCtacatATTTTATGAGTCAAAGT-3′ and 3′-AAAATCGatgtaTTTTTCGatgtaTAAATCGatgtaTAAAATACTCAGTTTCA-5′.
Chromatin Immunoprecipitation Assay
HepG2 cells were stimulated with 5 ng/ml TGF-β3 for 1 h and fixed by adding formaldehyde to the medium to a final concentration of 1%. Fifteen minutes after protein-DNA cross-linking at 37 °C, glycine was added to a final concentration of 125 mm. Then the cells were rinsed with PBS once and lysed into nuclei lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS, 10 μg/ml leupeptin, 12.5 μg/ml aprotinin) for 10 min at 4 °C. Chromatin was sonicated until the average length of input DNA became less than 500 bp in size. Then the control IgG, anti-TCF7L2 antibody (Upstate Biotechnology), or anti-RNA polymerase II antibody (clone CTD4H8; Upstate/Millipore, number 05-623) was used for the immunoprecipitation. After purification of immunoprecipitated DNAs, primers specific for detection of the TMEPAI promoter including TTE or the TCF7 promoter including TCF/LEF-binding elements (TBEs) were used for amplification of DNA fragments. The primers used here were 5′-CTCCACTCAACCAAATGTCC-3′ and 5′-TTGGTTCAGTCTGGCTGAGA-3′ for the TMEPAI promoter and 5′-AAGGAAGTCCCTGATTGGCA-3′ and 5′-TGTGAACTGTATCGTGCCCA-3′ for the TCF7 promoter.
ApcMin/+ mice
ApcMin/+ mice that spontaneously show adenomas in the small intestine were sacrificed at 16 weeks of age. Then polyps were isolated to prepare total RNAs. As a control, intestinal mucosa from wild-type mice was collected from total RNA preparation.
RNA Preparation and RT-PCR
Total RNA was extracted using Isogen (Wako). Reverse transcription was carried out by using a high capacity RNA to cDNA kit (Applied Biosystems). PCR was performed using ExTaq polymerase (Takara) as described by the manufacturer. Primer sets to amplify TMEPAI, β-actin and TCF7L2 cDNAs were as follows: for human TMEPAI, 5′-GATCATCATCATCGTGGTGG-3′ and 5′-GATCATCATCATCGTGGTGG-3′; for human β-actin, 5′-CAAGAGATGGCCACGGCTGCT-3′ and 5′-TCCTTCTGCATCCTGTCGGCA-3′; for human TCF7L2, 5′-CAAATCCCGGGAAAGTTTGG-3′ and 5′-GCGTGAAGTGTTCATTGCTG-3′; for mouse TMEPAI, 5′-GTGATGATGGTGATGGTGGT-3′ and 5′-ATCAGACAGTGAGATGGTGG-3′; and for mouse β-actin, 5′-GCTCATAGCTCTTCTCCAGGG-3′ and 5′-TGAACCCTAAGGCCAACCGTG-3′.
Lentiviral shRNAs for TCF7L2
The lentiviral vectors for TCF7L2 shRNA (TRCN0000061894, TRCN0000061895, TRCN0000061896, and TRCN0000061897) and nontargeting shRNA (SHC002) were from Sigma. Lentiviral vectors expressing shTCF7L2 were transfected into 293A cells together with psPAX2 and pMD2.G. Four different lentiviruses were simultaneously incubated in DMEM containing polybrene (8 μg/ml) for 2 h and then added to the dishes. Twelve hours after infection, the cells were washed and cultured in medium. Infected MCF10A1 cells, which became puromycin-resistant, were used for experiments.
RESULTS
Identification of the TGF-β-responsive Region within the First Intron of the TMEPAI Gene
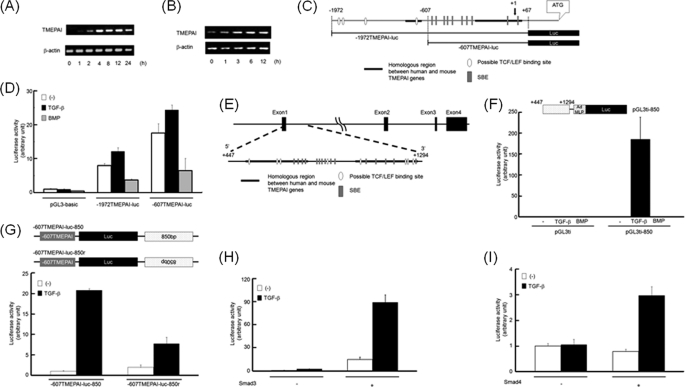
We have previously reported that TMEPAI is one of the early response genes to TGF-β signaling (8, 25). Because we principally used HepG2 cells in the following experiments, we investigated whether TMEPAI mRNA in HepG2 cells could be induced by TGF-β. As seen in Fig. 1A, TMEPAI mRNA was transiently induced by TGF-β. Likewise, SC3 cells, which are capable of responding to androgen, also showed induction of TMEPAI mRNA upon TGF-β stimulation (Fig. 1B). Because the sequences of the TMEPAI promoter from the transcriptional initiation site (+1) to −850 are highly homologous in human and mouse, we cloned the fragment from +67 to −1972 using the mouse BAC clone including the TMEPAI gene and inserted it into the pGL3-basic vector (−1972TMEPAI-luc) (Fig. 1C). In parallel, we made one deletion mutant, termed −607TMEPAI-luc. When we investigated whether these regions in the mouse TMEPAI promoter included TGF-β-responsive element(s), the activity of neither −1972TMEPAI-luc nor −607TMEPAI-luc was induced by TGF-β (Fig. 1D), resulting in the region from +67 to −1972 of the TMEPAI gene failing to respond to TGF-β.
FIGURE 1.
Identification of the TGF-β-responsive region in the TMEPAI gene. A and B, induction of TMEPAI mRNA by TGF-β. HepG2 (A) and SC3 cells (B) were treated with 5 ng/ml TGF-β for indicated times and then analyzed by RT-PCR. C, schematic presentation of deletion mutants for luciferase constructs. The promoter sequences of the mouse TMEPAI gene are shown. The transcriptional initiation site is shown as +1. Luc, luciferase. D, HepG2 cells were transfected with pGL3-basic, −1972TMEPAI-luc, or −607TMEPAI-luc and stimulated with TGF-β or BMP. E, representation of the first intron of the TMEPAI gene from +447 to +1294. F, schematic representation of pGL3ti-850 (upper panel). Ad MLP, adenovirus major late promoter. HepG2 cells were transfected with pGL3ti or pGL3ti-850 and stimulated with TGF-β or BMP (lower panel). G, the enhancer activity of the region consisted of 850 nucleotides in the first intron of the TMEPAI gene. Parts of the first intron in the TMEPAI gene (see E) were inserted below the luciferase gene of −607TMEPAI-luc at both orientations (upper panel). HepG2 cells were transfected with −607TMEPAI-luc-850 or −607TMEPAI-luc-850r and stimulated with TGF-β (lower panel). H and I, Smad-dependent transcriptional activation of the TMEPAI gene. Smad3-deficient MEFs (H) and MDA-MB468 cells (I) were transfected with Smad3 and Smad4 together with pGL3ti-850 in the absence or presence of TGF-β, respectively.
When the first intron was compared between human and mouse, the beginning 850 nucleotide sequences were highly conserved (supplemental Fig. S1). Thus, we made a luciferase reporter construct including the region spanning from +447 to +1294 in the TMEPAI gene (pGL3ti-850) (Fig. 1E). The activity of pGL3ti-850 was drastically potentiated by TGF-β but not by its related ligand BMP (Fig. 1F). In eukaryotes, the enhancer sequences that control mRNA transcription are known to function in both orientations. In addition, the enhancers often mediate their own properties even over a distance (31, 32). In view of these observations, we addressed the question of whether this TGF-β-responsive region of the TMEPAI gene functions over a distance in a manner independent of orientation. Fig. 1G shows that both orientations of the TGF-β-responsive region in the TMEPAI gene could retain the functional capacity as an enhancer, even though this region was connected to the 3′ end of the luciferase gene.
The TGF-β signal can be transduced to the nucleus via the intracellular signaling molecules Smad2, Smad3, and Smad4. Of these molecules, both Smad3 and Smad4 are known to bind to specific DNA elements on the promoter of their target genes (22, 28). To investigate whether the TGF-β-induced reporter activity of pGL3ti-850 depends on Smad3 and Smad4, pGL3ti-850 was transfected into either Smad3KO MEFs (Fig. 1H) or MDA-MB468 cells lacking Smad4 genetically (Fig. 1I). In the absence of either Smad3 or Smad4, the reporter activity of pGL3ti-850 was not potentiated by TGF-β, whereas the TGF-β response was rescued by introduction of Smad3 into Smad3KO MEFs or that of Smad4 into MDA-MB468 cells. In contrast to Smad3, the transfection of Smad2 did not restore the pGL3ti-850 activity upon TGF-β stimulation in Smad3KO MEFs (Fig. 2G). To further investigate whether Smad2 was required for TGF-β-dependent activation of pGL3ti-850, Smad2 was knocked down in MCF10A1 cells. However, we could not observe any differences between cells transfected with control siRNAs and those transfected with Smad2-specific siRNAs (supplemental Fig. S2). These results indicate that both Smad3 and Smad4 appear to be required for TGF-β-induced pGL3ti-850 activity.
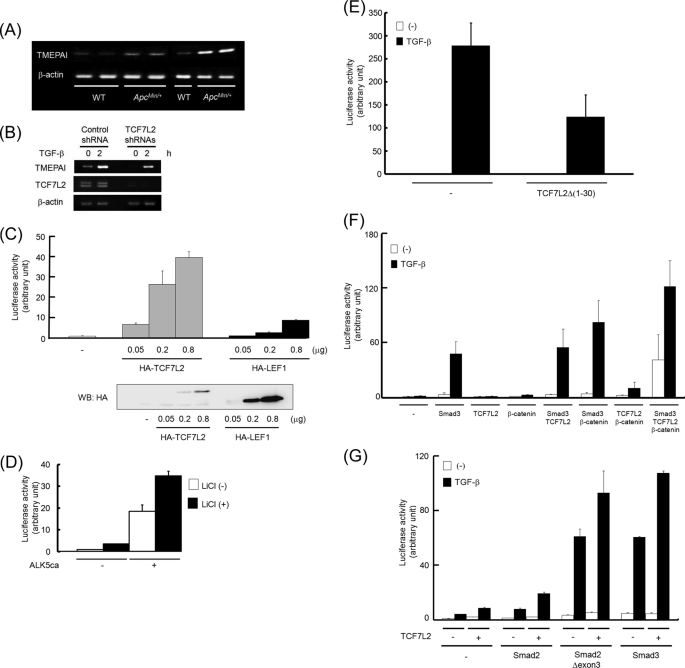
FIGURE 2.
Cross-talk between TGF-β and Wnt signalings to activate the TMEPAI gene. A, expression of TMEPAI mRNA in intestinal adenoma of ApcMin/+ mice. WT, intestinal mucosa of wild-type mice; ApcMin/+, intestinal polyps of ApcMin/+ mice. B, reduction of TGF-β-induced TMEPAI mRNA in TCF7L2 knocked down MCF10A1 cells. MCF10A1 cells carrying nontargeting shRNA (control shRNA) or a mixture of TCF7L2 shRNAs were stimulated with TGF-β for 2 h. Subsequently, the expression of TCF7L2 mRNA was checked using RT-PCR. Because TCF7L2 possesses alternative splicing forms, two PCR products could be seen using primers used here. C, activation of pGL3ti-850 by TCF7L2 or LEF1. HepG2 cells were transfected with a different amount of TCF7L2 or LEF1 together with pGL3ti-850 (upper panel). Simultaneously, the expression of TCF7L2 or LEF1 proteins in HepG2 cells was detected by Western blot (WB) analysis (lower panel). D, synergistic activation of pGL3ti-850 by the combination of ALK5ca with lithium chloride. 293T cells transfected with pGL3ti-850 together with or without ALK5ca were cultured in the presence or absence of 20 mm lithium chloride. E, inhibition of TGF-β-induced pGL3ti-850 activity by TCF7L2Δ(1–30). HepG2 cells transfected with TCF7L2Δ(1–30) were stimulated with TGF-β. F, synergistic activation of pGL-3ti-850. HepG2 cells transfected with Smad3, TCF7L2, β-catenin, or their combinations were stimulated with TGF-β. G, requirement of DNA binding ability in Smad proteins for synergistic activation of pGL3ti-850. Smad3-deficient MEF cells transfected with Smad2, Smad2Δexon3, Smad3, and/or TCF7L2 were stimulated with TGF-β.
Cross-talk between the TGF-β and Wnt Pathways to Activate the Transcription of the TMEPAI Gene
We and others found that the expression of TMEPAI in the intestinal polyps of ApcMin/+ mice is higher than that in the intestinal mucosa of wild-type mice (Fig. 2A) (8, 14). Interestingly, the expression of TMEPAI in the region where β-catenin was expressed at a relatively low level (supplemental Fig. S3, red broken lines) was weaker than that in the region in which β-catenin was highly expressed (supplemental Fig. S3, blue broken lines). Because the Wnt/β-catenin signal in the intestinal polyps of ApcMin/+ mice is constitutively active, we speculated that TMEPAI expression might be regulated by the Wnt/β-catenin pathway as well. To prove this speculation, we reduced the expression of TCF7L2, which is one of the critical DNA-binding transcriptional activators downstream of Wnt/β-catenin signaling, in MCF10A1 cells. Subsequently, these cells were stimulated with TGF-β for 2 h. As expected, the introduction of shRNAs corresponding to TCF7L2 interfered with the basal and TGF-β-induced expressions of TMEPAI mRNA (Fig. 2B). Additionally, the expression of TMEPAI mRNA by TGF-β in cells transfected with both Smad4- and TCF7L2-specific stealth siRNAs was weaker than that using either Smad4- or TCF7L2-specific stealth siRNA (supplemental Fig. S4), which further supported the possibility that both TGF-β and Wnt pathways control TMEPAI mRNA expression. Reciprocally, TMEPAI mRNA induced by TGF-β was further enhanced when cells were simultaneously stimulated with Wnt-3a, although the cooperative effect was marginal (supplemental Fig. S5). Thus, it is possible that the transcription of the TMEPAI gene was regulated by both the Wnt and TGF-β signaling pathways. Indeed, TCF7L2 potentiated the activity of pGL3ti-850 in a dose-dependent manner. However, LEF1, a member of the same protein family as TCF7L2, could only marginally activate the promoter of the TMEPAI gene, although its expressions were relatively higher than those of TCF7L2 when the same amount of DNA was transfected in cells (Fig. 2C). Because LEF1 lacks the C-terminal Clamp domain that TCF7L2 has (33), this domain might contribute to the function of TCF7L2 on the TMEPAI promoter activity. Therefore, we focused on the effect of TCF7L2 on the TGF-β-induced TMEPAI promoter activity in the following experiments.
These results prompted us to test whether Wnt and TGF-β signaling synergistically activates the pGL3ti-850 reporter. Like ALK5ca, which can constitutively activate the TGF-β/Smad pathway in the absence of TGF-β, LiCl, an activator of canonical Wnt signaling, increased the activity of pGL3ti-850. Importantly, the transfection of ALK5ca together with treatment of cells with LiCl revealed synergistic activation of pGL3ti-850 rather than its additional activation (Fig. 2D). Reversely, a dominant negative mutant of TCF7L2 (TCF7L2Δ(1–30)), which cannot interact with β-catenin (34), perturbed TGF-β-induced pGL3ti-850 activity (Fig. 2E). The effect of TCF7L2Δ(1–30) was specific to pGL3ti-850 because TCF7L2Δ(1–30) did not influence TGF-β-induced (CAGA)12-luc activity (22) (supplemental Fig. S6). β-Catenin and TCF7L2 are critical factors for regulating Wnt target genes, whereas Smad3 is one of the intracellular signal transducers of TGF-β signaling. HepG2 cells were therefore transfected with different combinations of β-catenin, TCF7L2, and Smad3, together with pGL3ti-850. After stimulation of the cells with TGF-β, the transcriptional responses were analyzed. The transfection of Smad3 potentiated TGF-β-induced reporter activity. The combination of Smad3 and β-catenin further enhanced the reporter activity induced by TGF-β, whereas cotransfection of Smad3 with TCF7L2 only marginally increased TGF-β-induced pGL3ti-850 activity. When all three components were coexpressed in the cells, the basal reporter activity as well as the TGF-β-induced activity was dramatically augmented (Fig. 2F). It is known that the Smad2 gene has two isoforms (i.e. a long isoform termed Smad2 and its splicing variant termed Smad2Δexon3). Smad2 is also an intracellular signal transducer of TGF-β signaling, although unlike Smad3, it lacks the ability to bind directly to DNA (22, 28). However, Smad2Δexon3 can acquire the ability of direct DNA binding because its inhibitory domain for DNA binding is deleted (23). When Smad2Δexon3 as well as Smad3 was introduced into Smad3KO MEFs, TGF-β-dependent transcriptional activity was restored. On the other hand, Smad2 did not rescue the reporter activity by TGF-β. In addition, TCF7L2-mediated potentiation of pGL3ti-850 activity induced by TGF-β was observed when either Smad2Δexon3 or Smad3 was transfected into Smad3KO MEFs (Fig. 2G). Thus, the ability of DNA binding in Smad proteins might be necessary for Smad proteins to activate pGL3ti-850 reporter.
The Highest Enhancer Activity of the C Region
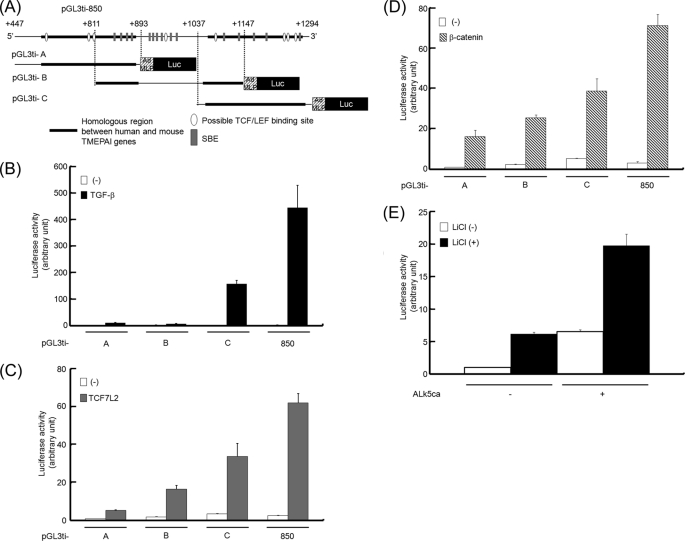
To identify a TGF-β-responsive element(s) within the region spanning from +447 to +1294 in the first intron of the TMEPAI gene, we divided the above 850-nucleotide enhancer region into three parts and conjugated each of them to the luciferase gene (pGL3ti-A, pGL3ti-B, and pGL3ti-C) (Fig. 3A). Subsequently, each reporter was evaluated upon TGF-β stimulation (Fig. 3B), expression of TCF7L2 (Fig. 3C), or expression of β-catenin (Fig. 3D). Of the three parts, the C region possessed the highest enhancer activity when cells were either stimulated with TGF-β or transfected with β-catenin or TCF7L2, although the other two regions showed enhancer activities to a relatively lower extent as well. Thus, we focused on the C region in the following experiments. Because pGL3ti-850 can be synergistically activated by both the TGF-β and Wnt pathways, we asked whether pGL3ti-C was also influenced by both pathways. When cells carrying pGL3ti-C together with or without ALK5ca were stimulated with LiCl, the reporter activity seemed to be enhanced in a synergistic manner rather than in an additive manner (Fig. 3E). Collectively, these results demonstrated that the C region in the first intron of the TMEPAI gene possesses an enhancer of the TGF-β and Wnt signaling pathways.
FIGURE 3.
The highest enhancer activity by TGF-β and Wnt signalings in the C region of the TMEPAI gene. A, schematic presentation of deletion mutants for luciferase (Luc) reporters using the first intron of the TMEPAI gene. B–D, the reporter activity of each deletion mutant upon stimulation of TGF-β, expression of TCF7L2, or expression of β-catenin. HepG2 cells transfected with pGL3ti-A, pGL3ti-B, or pGL3ti-C were stimulated with TGF-β (B), cotransfected with TCF7L2 (C), or cotransfected with β-catenin (D). E, synergistic activation of pGL3ti-C by the combination of ALK5ca with lithium chloride. 293T cells transfected with pGL3ti-850 together with or without ALK5ca were cultured in the presence or absence of 20 mm lithium chloride.
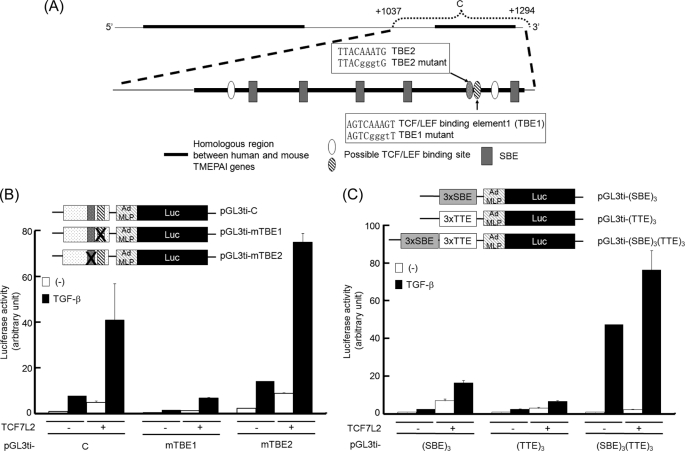
Identification of TTE in the First Intron of the TMEPAI Gene
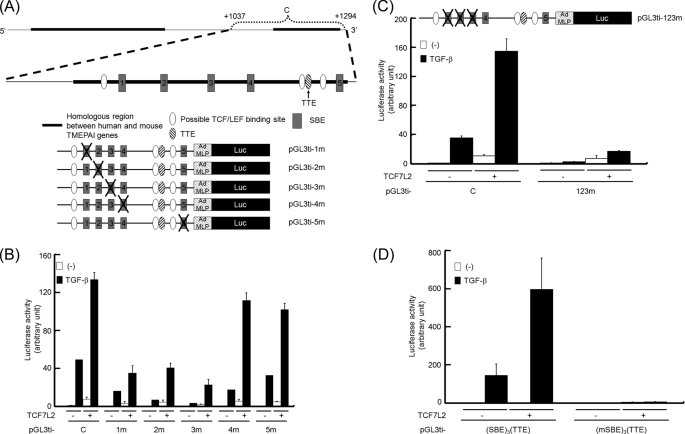
Because TCF7L2 could potentiate TGF-β-induced transcriptional activation of the TMEPAI gene, we examined which TBE in the C region is critical for the activation of the TMEPAI gene upon TGF-β stimulation. When we looked for possible TBEs in the C region, we could find four candidates (Fig. 4A, ovals). We focused on two of those candidates (Fig. 4A, closed and hatched ovals) because both are highly conserved with the consensus TBE (5′-(T/A)(T/A)CAA(T/A)GG-3′) (35). To explore the possibility that these two TBEs contribute to TGF-β-mediated activation of the C region, we introduced mutations for each of the TBEs into pGL3ti-C. Then cells transfected with mutant reporters were stimulated with TGF-β to evaluate their TGF-β responsibility (Fig. 4B). Surprisingly, the activity of pGL3ti-mTBE1 upon stimulation of TGF-β, ectopic expression of TCF7L2, or a combination of both was obviously reduced. In contrast, the mutations in TBE2 did not influence the reporter activity. These results indicate that TBE1 in the C region might play a key role in the TGF-β-induced activation of the TMEPAI gene as an enhancer. Therefore, TBE1 was termed TTE in the following experiments.
FIGURE 4.
Identification of TTE. A, schematic presentation of the C region. The lowercase letters written in squares indicate mutated nucleotides for TBE1 and TBE2. B, requirement of TBE1 for synergistic activation. Upper panel, schematic representation of mutant reporters. Lower panel, HepG2 cells transfected with plasmids described were stimulated with TGF-β. C, effect of concatemers for SBE and/or TTE on TGF-β-mediated responsiveness in the presence of TCF7L2. Upper panel, constructs of artificial reporters. Lower panel, HepG2 cells transfected with plasmids described were stimulated with TGF-β. Luc, luciferase.
Because the mutation of TTE provoked the loss of TGF-β responsiveness in pGL3ti-C, we asked whether TTE alone functions as a TGF-β-responsive element. To test this hypothesis, we inserted three copies of the TTE in front of a minimal promoter (pGL3ti-(TTE)3) and tested this construct in HepG2 cells. However, three copies of the TTE were insufficient for TGF-β-induced luciferase activity (Fig. 4C), although pGL3ti-(mTTE)3 lost the TGF-β-induced luciferase activity even in the presence of TCF7L2 (supplemental Fig. S7). Like pGL3ti-(TTE)3, three copies of the SBE in the pGL3ti vector (pGL3ti-(SBE)3) could not reveal strong inducibility upon TGF-β stimulation, whereas TCF7L2 potentiated TGF-β-dependent activation of pGL3ti-(SBE)3 as well as TGF-β-independent activation. This result was confirmed by the DNA affinity precipitation assay, in which TCF7L2 was able to bind to the SBE upon the activation of ALK5 (see below). On the other hand, pGL3ti-(SBE)3(TTE)3, which includes three copies of the TTE in addition to three copies of the SBE, was highly potent in inducing TGF-β responsiveness. Also, TCF7L2 further potentiated the TGF-β-induced reporter activity of pGL3ti-(SBE)3(TTE)3, although TCF7L2 alone could marginally activate this promoter as much as pGL3ti-(TTE)3 (Fig. 4C).
Next, we were prompted to find which SBEs could act in concert with TTE upon TGF-β stimulation. Because there are five SBEs in the C region, we tried to introduce a mutant to each SBE in the pGL3ti-C reporter (Fig. 5A). Of the five mutants, the activities of pGL3ti-1m, pGL3ti-2m, and pGL3ti-3m upon TGF-β stimulation were drastically decreased. Furthermore, TCF7L2 could not enhance the TGF-β-induced activity of these three mutants as much as pGL3ti-C could (Fig. 5B). When pGL3ti-123m, in which three SBEs are simultaneously mutated, was transfected into HepG2 cells, its inducibility by TGF-β was lost, although the TCF7L2-mediated activity of pGL3ti-123m could remain much weaker than that of pGL3ti-C (Fig. 5C). This evidence encouraged us to confirm that an artificial luciferase reporter consisting of three copies of the SBE and one copy of TTE (pGL3ti-(SBE)3(TTE)) could mimic pGL3ti-C when the cells were stimulated with TGF-β and/or transfected with TCF7L2. As expected, pGL3ti-(SBE)3(TTE) showed a similar response to that of pGL3ti-C, whereas pGL3ti-(mSBE)3(TTE) did not provide any reporter activities upon any combination examined (Fig. 5D). This evidence clearly suggested that TTE can cooperate with SBEs for enhancement of TGF-β-induced transcription by TCF7L2.
FIGURE 5.
Importance of particular SBEs together with TTE in the C region of the TMEPAI gene. A, schematic presentation of point mutants for luciferase (Luc) assay using the C region. Each SBE is numbered from the 5′ upstream of the C region. B, requirement of three distal SBEs for full activation of the pGL3ti-C reporter. HepG2 cells transfected with each reporter and TCF7L2 were stimulated with TGF-β. C and D, loss of synergistic activation by introduction of mutations in three SBEs for pGL3ti-C (C) or pGL3ti-(SBE)3(TTE) (D). HepG2 cells transfected with plasmids described were stimulated with TGF-β.
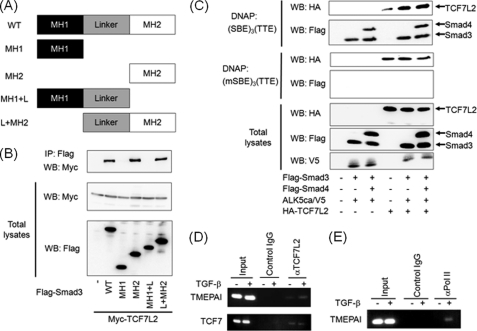
Binding of TCF7L2 to TTE in the TMEPAI Gene
TCF7L2 or its related molecule, LEF1, is known to interact with Smad proteins (17, 27). We also confirmed that Smad3 can interact with TCF7L2 via its MH2 domain (Fig. 6, A and B). We next investigated whether TCF7L2 can bind to TTE when the TGF-β signaling pathway is activated. For this purpose, we performed a DNA affinity precipitation assay using (SBE)3(TTE) as a probe because the TGF-β-induced activity of pGL3ti-(SBE)3(TTE) was drastically enhanced by TCF7L2 (Fig. 5D). TCF7L2 could weakly bind to this probe in the absence of TGF-β signaling, whereas the activation of TGF-β signaling extensively enhanced the affinity of TCF7L2 to the probe in the presence of Smad3. On the other hand, the addition of Smad4 did not affect this interaction (Fig. 6C). When a probe including three copies of the mutant SBE and one TTE (to which neither Smad3 nor Smad4 could bind) was used for the DNA affinity precipitation assay, no TGF-β-dependent binding of TCF7L2 was observed (Fig. 6C). These results support the idea that TCF7L2 is capable of interacting with Smad complex binding to SBE in spite of the fact that Smad complex does not associate with TCF7L2 binding to TTE in the absence of SBE. To further confirm that TCF7L2 lies on TTE of the TMEPAI gene in the chromatin, we employed a ChIP assay using either control rabbit IgG or anti-TCF7L2 rabbit monoclonal antibody. As shown in Fig. 6D (upper panel), TCF7L2 was capable of binding to the sequence around TTE of the TMEPAI gene upon TGF-β stimulation when the sonicated chromatin-protein complex was immunoprecipitated with the anti-TCF7L2 rabbit monoclonal antibody. On the other hand, no interaction was detected when the control IgG antibody was used for immunoprecipitation of the sonicated chromatin-protein complex (Fig. 6D, upper panel). To examine whether TCF7L2 could bind to the typical TCF7L2-binding site in a TGF-β-dependent fashion, we employed the TCF7L2-binding site in the TCF7 gene, which is one of the well known Wnt/β-catenin/TCF7L2 target genes but not a TGF-β target gene. As anticipated, we could not see any TGF-β dependence for TCF7L2-DNA binding (Fig. 6D, lower panel). To further confirm that TTE in the intron 1 contributes to TGF-β-induced transcription of the TMEPAI gene, we also carried out a ChIP assay using anti-RNA polymerase II antibody (36). The results indicated that the region including TTE was covered with RNA polymerase II (Fig. 6E). Thus, TTE might be necessary for RNA polymerase II to contact the promoter region of the TMEPAI gene. Taken together, TCF7L2 is implicated in TGF-β-dependent activation of the TMEPAI gene through its binding to TTE and the adjacent SBEs together with the Smad complex.
FIGURE 6.
TGF-β-dependent interaction of TCF7L2 with TTE. A, depiction of Smad3 mutants. MH, mad homology region. B, interaction of Smad3 with TCF7L2 via its MH2 domain. Immunoprecipitations were carried out using anti-FLAG M5 antibody, and coimmunoprecipitated (IP) TCF7L2 was detected by Western blotting (WB) using anti-Myc 9E10 antibody (top panel). The expression of Myc-TCF7L2 and Smad3 mutants conjugated with FLAG at the N terminus was evaluated using anti-Myc 9E10 (middle panel) and anti-FLAG M5 antibodies (bottom panel), respectively. C, requirement of Smad-SBE complex for enhanced binding of TCF7L2 to TTE. COS7 cell lysates transfected with the plasmids indicated were mixed with either biotinated (SBE)3(TTE) or biotinated (mSBE)3(TTE). TCF7L2-DNA complex and Smads-DNA complex were detected by Western blotting using anti-HA3F10 antibody (top and third panels) and anti-FLAG M5 antibody (second and fourth panels), respectively. The expressions of HA-TCF7L2, FLAG-Smads, and ALK5ca/V5 were evaluated using anti-HA3F10 (fifth panel), anti-FLAG M5 (sixth panel), and anti-V5 antibodies (bottom panel), respectively. D and E, recruitment of TCF7L2 to the C region including TTE upon TGF-β stimulation. Cross-linked chromatin from HepG2 cells were incubated with anti-TCF7L2 (D) and anti-RNA polymerase II antibodies (E). The immunoprecipitated DNA was analyzed by PCR with primers that amplify the fragment including TTE in the TMEPAI gene. As a positive control, DNA sequences including TBE in the TCF7 gene were also amplified (lower panel in D). As a negative control, PCR was performed using DNA immunoprecipitated with mouse control IgG. In parallel, the input DNA was amplified by PCR as described above.
DISCUSSION
The Wnt pathway plays a key role in cell fate determination, self-renewal, and cell differentiation during the process of vertebrate development. Aberrant activation of canonical Wnt pathway leads to neoplasia such as colon cancer and leukemia (37–41). On the other hand, the loss of TGF-β signaling by the mutation of TGF-β type II receptor or Smad4 genes is also known to be associated with tumor progression in colon (2, 5, 42). Thus, it is possible that integration between a loss of TGF-β signaling and constitutively active Wnt signaling might coordinately undergo malignant transformation. Indeed, when compound heterozygotes carrying both mutations of Smad4 and Apc genes in mice were generated, intestinal polyps in these compound mice developed into more malignant tumors than those in mice carrying Apc mutation in one allele (43).
Previous studies have suggested the possibility that the transcript of the TMEPAI gene is regulated by TGF-β and/or Wnt signaling (8, 14, 44). Furthermore, high expression of TMEPAI was observed in several tumors (10, 12, 13). However, it has not been elucidated how the activity of TMEPAI promoter is regulated by TGF-β and/or Wnt signaling. In this study, we showed synergy between TGF-β and Wnt signals in the regulation of the mouse TMEPAI promoter. This cooperative regulation can be mediated by interaction between TCF7L2 and Smad3 on the enhancer within the first intron of the TMEPAI gene. We initially speculated that there was a responsive element(s) needed for integration between Wnt and TGF-β signalings in the promoter upstream of the transcriptional initiation site of the TMEPAI gene because of high similarity within the promoter region between human and mouse TMEPAI genes. However, neither the stimulation of TGF-β nor the expression of TCF7L2 potentiated the activity of −1972TMEPAI-luc and −607TMEPAI-luc (Fig. 1D and data not shown). Although we also found other homologous regions lying just downstream of the first exon between human and mouse TMEPAI genes, the activity of the luciferase reporter including this region (pGL3ti-850) was obviously potent upon TGF-β and Wnt signalings. It is known that nuclear β-catenin interacts with N-terminal region of TCF7L2 to cooperate the transcriptional regulation of Wnt target genes (34). TCF7L2Δ(1–30) lacking the β-catenin-binding region could perturb TGF-β responsiveness of pGL3ti-850. Therefore, the full activation of the TMEPAI gene by TGF-β might require a β-catenin-TCF7L2 complex. However, the family protein of TCF7L2, LEF1, could only marginally activate the promoter of the TMEPAI gene (Fig. 2C). Our data indicate that TCF7L2 might play a more dominant role than LEF1, whereas we are unable to exclude the possibility that the high expression of LEF1 in cells compared with TCF7L2 expression might compensate for its weak transcriptional activity in the TMEPAI promoter. Because the TCF/LEF family consists of four members (TCF7, TCF7L1, TCF7L2, and LEF1) and each member has a number of isoforms, it would be very interesting to investigate in detail which TCF/LEF family member cooperatively functions to activate the TMEPAI promoter together with TGF-β in a context-dependent manner.
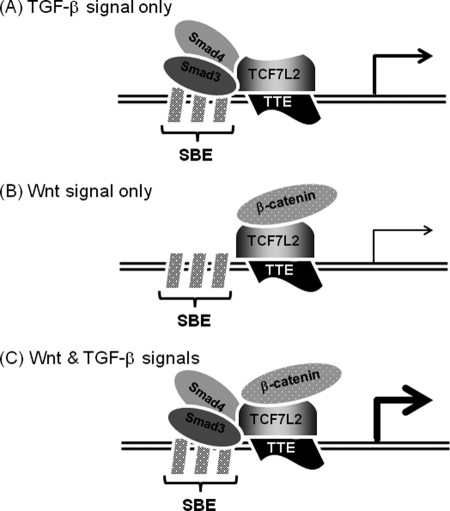
Smad2 lacks the ability to bind to DNA directly because of the presence of extra amino acid sequences proximal to the DNA-binding region, whereas Smad2Δexon3 lacking the extra amino acid sequences can act like Smad3 (23). Thus, we wondered whether Smad2 requires the ability of DNA binding for synergistical activation of the TMEPAI promoter together with TCF7L2. Indeed, Smad2 required its DNA binding ability for its cooperative activation of the TMEPAI promoter together with TCF7L2. When we further narrowed the TGF-β- or TCF7L2-responsive element(s) down within the sequences from +447 to +1294, the C region possessed the highest responsiveness to both TGF-β and Wnt signalings among three regions. It has been reported that the promoters, which are regulated by both TGF-β and Wnt signalings include Smad and TCF/LEF-binding elements very close each other (17–20, 45). As anticipated, there are five SBEs and four possible TBEs in the C region. Surprisingly, the disruption of one possible TBE, termed TTE, drastically reduced the promoter activity of the TMEPAI gene (pGL3ti-mTBE1) upon stimulation of TGF-β, TCF7L2 expression, or the combination. Consequently, TTE was the critical cis-element for TGF-β-mediated activation of the TMEPAI promoter. Like the TMEPAI promoter, the introduction of a mutation into the TCF/LEF-binding sites of the Msx2 and gastrin promoters revealed abrogation of BMP-dependent and TGF-β-dependent activation, respectively (19, 45). Obviously, the nucleotide sequence of TTE present in the TMEPAI gene (5′-AGTCAAAGT-3′) is somehow similar to those of the TCF/LEF-binding sites in the Msx (5′-ACAAAGG-3′) and gastrin genes (5′-AGAGAAATG-3′) (19, 45). Thus, we speculated that synergistic activation of the TMEPAI gene is mediated by a physical association between Smads and TCF7L2. Our current evidence and the reports from other groups seem to indicate that the TCF/LEF family surveils a number of TGF-β-regulated genes by its association with Smad proteins. Also, our artificial pGL3ti-(SBE)3-luc, which possesses only SBE sequences, could be activated by TCF7L2 without TGF-β stimulation (Fig. 4C). This result indicates that TCF7L2 might indirectly be able to bind to SBE via Smad proteins like a coactivator to regulate a TGF-β-target gene(s), whereas the Smad complex did not seem to possess the ability to bind to TTE via TCF7L2 (Fig. 6C). Taken together, these results indicate that synergistic activation of the TMEPAI gene is mediated by a physical association between Smads and TCF7L2. Indeed, we could observe the interaction of Smad3 with TCF7L2 in the coimmunoprecipitation assays despite ligand-independent (or ALK5ca-independent) association (Fig. 6B and supplemental Fig. S8), indicating that C-terminal phosphorylation of Smad3 is not required for their interaction. In contrast to the results of the coimmunoprecipitation assay, TCF7L2 as well as Smad3 could bind to the DNA-containing SBEs and TTE ((SBE)3(TTE)) upon ALK5 activation. However, no ALK5ca dependence was observed when (mSBE)3(TTE) was mixed with TCF7L2. This evidence indicates that the Smad complex might be able to recruit TCF7L2 to TTE in the TMEPAI promoter upon stimulation of TGF-β. Actually, the CHIP assay further supported our concept because TCF7L2 could bind to the endogenous TTE in the TMEPAI gene upon TGF-β stimulation. In addition to the cooperative activity between Smads and TCF7L2, TGF-β and Wnt pathways might also independently regulate a TMEPAI promoter (Fig. 7). Because we showed that TCF7L2 lacking the β-catenin-binding domain can perturb the activation of a TMEPAI promoter by TGF-β, the canonical Wnt pathway through stabilization of β-catenin seems to be required for the full activation of a TMEPAI promoter.
FIGURE 7.
A model for activation of a TMEPAI gene by TGF-β and Wnt pathways. The TMEPAI promoter with SBEs adjacent to TTE can be activated upon TGF-β stimulation in the absence of Wnt signal (A), upon Wnt stimulation without TGF-β signal (B), or cooperatively in the presence of both TGF-β and Wnt signals (C).
The transcript of the TMEPAI gene has been reported to be induced by testosterone, its derivatives, or mutated p53 and to be implicated in tumorigenesis (8–11). Thus, it is suspected that the cross-talk among androgen, p53, TGF-β, and Wnt signaling with respect to TMEPAI expression might be involved in malignant transformation. In fact, when intestinal polyps from the ApcMin/+ mouse were stained with anti-phosphorylated Smad2 (index for TGF-β signaling), anti-β-catenin (index for Wnt signaling), and anti-TMEPAI antibodies, the cells stained with both anti-phosphorylated Smad2 and anti-β-catenin antibodies express TMEPAI at a higher level than those stained with anti-phosphorylated Smad2 alone (supplemental Fig. S3).
In conclusion, the TMEPAI gene has been shown to require both a SBE and a TTE for synergistic activation in cells. Similar to the gastrin and Msx2 promoters, the introduction of a mutation in TTE showed a more inhibitory effect on the TMEPAI expression than did disruption of SBEs, supporting the notion that TCF7L2 plays a key role in TGF-β-mediated activation of the TMEPAI gene.
Supplementary Material
Acknowledgments
We thank F. Miyamasu for excellent English proofreading, Dr. K. Watanabe for Myc-TCF7L2, Dr. A. B. Roberts for Smad3KO mice, Dr. K. Iwata for TGF-β3, and Dr. K. Sampath for BMP-6.
This work was supported by Grants-in-aid for Scientific Research 17390073, 20012007, and 21590328 (to M. K. and S. I.), the Takeda Science Foundation (to S. I.), a Genome Network Project grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M. K.), a grant-in-aid for Japan Society for the Promotion of Science Fellows (to Y. W. and F. I.), and a long range research initiative grant from the Japan Chemical Industry Association (to M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- TCF
- T cell factor
- LEF
- lymphoid enhancer-binding factor
- TTE
- TGF-β-responsive TCF7L2-binding element
- SBE
- Smad-binding element
- MEF
- mouse embryonic fibroblast
- TBE
- TCF/LEF-binding element
- TMEPAI
- transmembrane prostate androgen-induced RNA
- TGF-β
- transforming growth factor
- BHP
- bone morphogenic protein
- Apc
- adenomatous polyposis coli.
REFERENCES
- 1.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 2.Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh S., ten Dijke P. (2007) Curr. Opin. Cell Biol. 19, 176–184 [DOI] [PubMed] [Google Scholar]
- 4.ten Dijke P., Arthur H. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 857–869 [DOI] [PubMed] [Google Scholar]
- 5.Gordon K. J., Blobe G. C. (2008) Biochim. Biophys. Acta 1782, 197–228 [DOI] [PubMed] [Google Scholar]
- 6.Margadant C., Sonnenberg A. (2010) EMBO Rep. 11, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosimann C., Hausmann G., Basler K. (2009) Nat. Rev. Mol. Biol. 10, 276–286 [DOI] [PubMed] [Google Scholar]
- 8.Watanabe Y., Itoh S., Goto T., Ohnishi E., Inamitsu M., Itoh F., Satoh K., Wiercinska E., Yang W., Shi L., Tanaka A., Nakano N., Mommaas A. M., Shibuya H., Ten Dijke P., Kato M. (2010) Mol. Cell 37, 123–134 [DOI] [PubMed] [Google Scholar]
- 9.Li H., Xu L. L., Masuda K., Raymundo E., McLeod D. G., Dobi A., Srivastava S. (2008) J. Biol. Chem. 283, 28988–28995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L. L., Shi Y., Petrovics G., Sun C., Makarem M., Zhang W., Sesterhenn I. A., McLeod D. G., Sun L., Moul J. W., Srivastava S. (2003) Cancer Res. 63, 4299–4304 [PubMed] [Google Scholar]
- 11.Anazawa Y., Arakawa H., Nakagawa H., Nakamura Y. (2004) Oncogene 23, 7621–7627 [DOI] [PubMed] [Google Scholar]
- 12.Xu L. L., Shanmugam N., Segawa T., Sesterhenn I. A., McLeod D. G., Moul J. W., Srivastava S. (2000) Genomics 66, 257–263 [DOI] [PubMed] [Google Scholar]
- 13.Giannini G., Ambrosini M. I., Di Marcotullio L., Cerignoli F., Zani M., MacKay A. R., Screpanti I., Frati L., Gulino A. (2003) Mol. Carcinog. 38, 188–200 [DOI] [PubMed] [Google Scholar]
- 14.Reichling T., Goss K. H., Carson D. J., Holdcraft R. W., Ley-Ebert C., Witte D., Aronow B. J., Groden J. (2005) Cancer Res. 65, 166–176 [PubMed] [Google Scholar]
- 15.Rae F. K., Hooper J. D., Nicol D. L., Clements J. A. (2001) Mol. Carcinog. 32, 44–53 [DOI] [PubMed] [Google Scholar]
- 16.Brunschwig E. B., Wilson K., Mack D., Dawson D., Lawrence E., Willson J. K., Lu S., Nosrati A., Rerko R. M., Swinler S., Beard L., Lutterbaugh J. D., Willis J., Platzer P., Markowitz S. (2003) Cancer Res. 63, 1568–1575 [PubMed] [Google Scholar]
- 17.Labbé E., Letamendia A., Attisano L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8358–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishita M., Hashimoto M. K., Ogata S., Laurent M. N., Ueno N., Shibuya H., Cho K. W. (2000) Nature 403, 781–785 [DOI] [PubMed] [Google Scholar]
- 19.Hussein S. M., Duff E. K., Sirard C. (2003) J. Biol. Chem. 278, 48805–48814 [DOI] [PubMed] [Google Scholar]
- 20.Szeto D. P., Kimelman D. (2004) Development 131, 3751–3760 [DOI] [PubMed] [Google Scholar]
- 21.Nakashima A., Katagiri T., Tamura M. (2005) J. Biol. Chem. 280, 37660–37668 [DOI] [PubMed] [Google Scholar]
- 22.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi K., Goto D., Hamamoto T., Takenoshita S., Kato M., Miyazono K. (1999) J. Biol. Chem. 274, 703–709 [DOI] [PubMed] [Google Scholar]
- 24.Sasaki T., Suzuki H., Yagi K., Furuhashi M., Yao R., Susa S., Noda T., Arai Y., Miyazono K., Kato M. (2003) Cancer Res. 63, 801–806 [PubMed] [Google Scholar]
- 25.Itoh S., Thorikay M., Kowanetz M., Moustakas A., Itoh F., Heldin C. H., ten Dijke P. (2003) J. Biol. Chem. 278, 3751–3761 [DOI] [PubMed] [Google Scholar]
- 26.Noda D., Itoh S., Watanabe Y., Inamitsu M., Dennler S., Itoh F., Koike S., Danielpour D., ten Dijke P., Kato M. (2006) Oncogene 25, 5591–5600 [DOI] [PubMed] [Google Scholar]
- 27.Hirota M., Watanabe K., Hamada S., Sun Y., Strizzi L., Mancino M., Nagaoka T., Gonzales M., Seno M., Bianco C., Salomon D. S. (2008) Cell Signal. 20, 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonk L. J., Itoh S., Heldin C. H., ten Dijke P., Kruijer W. (1998) J. Biol. Chem. 273, 21145–21152 [DOI] [PubMed] [Google Scholar]
- 29.Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., Deng C. (1999) EMBO J. 18, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang B., Vu M., Booker T., Santner S. J., Miller F. R., Anver M. R., Wakefield L. M. (2003) J. Clin. Invest. 112, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerji J., Rusconi S., Schaffner W. (1981) Cell 27, 299–308 [DOI] [PubMed] [Google Scholar]
- 32.Benoist C., Chambon P. (1981) Nature 290, 304–310 [DOI] [PubMed] [Google Scholar]
- 33.Weise A., Bruser K., Elfert S., Wallmen B., Wittel Y., Wöhrle S., Hecht A. (2010) Nucleic Acids Res. 38, 1964–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 35.Brannon M., Gomperts M., Sumoy L., Moon R. T., Kimelman D. A. (1997) Genes Dev. 11, 2359–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuman N. A., Ma S., Schnitzler G. R., Zhu Y., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 13202–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weerkamp F., Pike-Overzet K., Staal F. J. (2006) Trends Immunol. 27, 125–131 [DOI] [PubMed] [Google Scholar]
- 38.Moon R. T., Bowerman B., Boutros M., Perrimon N. (2002) Science 296, 1644–1646 [DOI] [PubMed] [Google Scholar]
- 39.Reya T., Clevers H. (2005) Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 40.Nusse R. (2005) Cell. Res. 15, 28–32 [DOI] [PubMed] [Google Scholar]
- 41.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 42.Massagué J., Blain S. W., Lo R. S. (2000) Cell 103, 295–309 [DOI] [PubMed] [Google Scholar]
- 43.Takaku K., Oshima M., Miyoshi H., Matsui M., Seldin M. F., Taketo M. M. (1998) Cell 92, 645–656 [DOI] [PubMed] [Google Scholar]
- 44.Levy L., Hill C. S. (2005) Mol. Cell. Biol. 25, 8108–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei S., Dubeykovskiy A., Chakladar A., Wojtukiewicz L., Wang T. C. (2004) J. Biol. Chem. 279, 42492–42502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.