Abstract
The calnexin cycle is a process by which glycosylated proteins are subjected to folding cycles in the endoplasmic reticulum lumen via binding to the membrane protein calnexin (CNX) or to its soluble homolog calreticulin (CRT). CNX and CRT specifically recognize monoglucosylated Glc1Man9GlcNAc2 glycans, but the structural determinants underlying this specificity are unknown. Here, we report a 1.95-Å crystal structure of the CRT lectin domain in complex with the tetrasaccharide α-Glc-(1→3)-α-Man-(1→2)-α-Man-(1→2)-Man. The tetrasaccharide binds to a long channel on CRT formed by a concave β-sheet. All four sugar moieties are engaged in the protein binding via an extensive network of hydrogen bonds and hydrophobic contacts. The structure explains the requirement for glucose at the nonreducing end of the carbohydrate; the oxygen O2 of glucose perfectly fits to a pocket formed by CRT side chains while forming direct hydrogen bonds with the carbonyl of Gly124 and the side chain of Lys111. The structure also explains a requirement for the Cys105–Cys137 disulfide bond in CRT/CNX for efficient carbohydrate binding. The Cys105–Cys137 disulfide bond is involved in intimate contacts with the third and fourth sugar moieties of the Glc1Man3 tetrasaccharide. Finally, the structure rationalizes previous mutagenesis of CRT and lays a structural groundwork for future studies of the role of CNX/CRT in diverse biological pathways.
Keywords: Calcium Binding Proteins, Carbohydrate Binding Protein, Chaperone Chaperonin, Endoplasmic Reticulum (ER), Lectin, X-ray Crystallography
Introduction
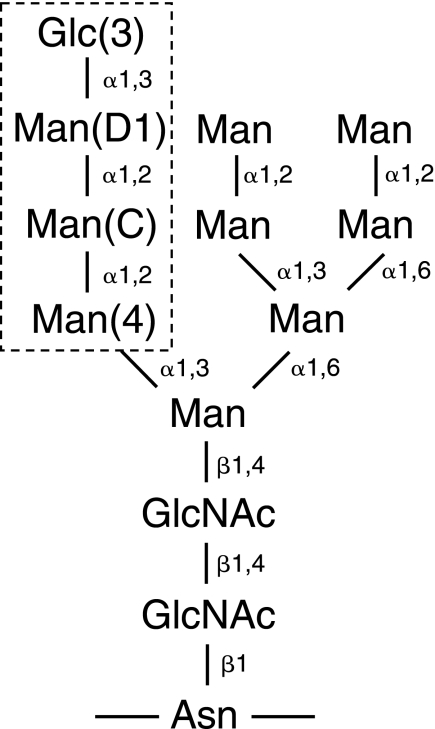
Protein glycosylation plays an important role in maturation and quality control of proteins in the endoplasmic reticulum (ER)3 (1–3). Nascent polypeptide chains of the secretory pathway are translocated into the lumen of the ER where they immediately become N-glycosylated with Glc3Man9GlcNAc2 on the side chain of asparagine residues in Asn-X-Ser/Thr motifs (see Scheme 1). The processing of the glycan starts with removal of the outermost glucose by the action of glucosidase I. Glucosidase II then trims the next glucose, upon which the monoglucosylated proteins selectively bind to the membrane ER protein calnexin (CNX) or its soluble homolog calreticulin (CRT) to enter the calnexin cycle. In the calnexin cycle, the nascent protein bound to CNX or CRT interacts with chaperones to promote its folding. The glucose and adjacent three mannose residues are critically important for carbohydrate recognition by CNX and CRT (Scheme 1) (4–6). Removal of the terminal glucose from correctly folded protein substrates releases them from binding to CNX or CRT and allows their exit from the ER to their final destination. Non-native proteins, which lack the terminal glucose residue, can be reglucosylated by the enzyme UDP-glucose-glycoprotein glucosyltransferase to re-enter the calnexin cycle. Multiple rounds of deglucosylation-reglucosylation cycles can occur until a glycoprotein reaches its native fold or, if terminally misfolded, it is targeted for degradation via the ER-associated degradation pathway (7).
SCHEME 1.
Structure of monoglucosylated N-linked glycan. The labels 3, D1, C, and 4 indicate the sugar positions. The tetrasaccharide that binds CRT is boxed.
The previous crystal structure of CNX revealed two main structural components, a globular lectin domain and an extended arm-like domain called the P-domain (8). The lectin domain shows a fold similar to leguminous lectins and largely consists of a β-sandwich formed by two curved β-sheets. It also contains a single high affinity calcium-binding site that plays an important role in stabilizing the protein (9) but does not participate in carbohydrate recognition (8). The proline-rich P-domain interrupts the lectin domain between residues Pro270 and Phe415. The P-domain consists of four copies of a repeat motif (termed type 1) and four copies of second repeat motif (type 2) in a “11112222” configuration to form a long hook-like arm that interacts with the thiol oxidoreductase ERp57 (10, 11). The CRT P-domain shows a similar modular arrangement but consists of only three repeats of each type (10, 12, 13). CRT also possesses a highly negative C terminus, which binds calcium ions with millimolar affinity, but the C terminus does not contribute to glycan binding (14, 15).
A portion of the carbohydrate-binding site was identified in CNX by soaking the crystals with glucose, but the study did not address the question of specificity toward monoglucosylated substrates and how remaining mannose moieties of the glycan recognize CNX (8). Subsequent mutagenesis studies confirmed the importance of CNX residues for carbohydrate binding (16). Parallel studies of CRT identified the residues that are essential for oligosaccharide interactions but also hinted at possible differences between carbohydrate recognition by CNX and CRT (17–19). The CNX structure did not explain the earlier observation that treatment with dithiothreitol impairs carbohydrate binding by CRT (6).
Here, we determined the high resolution crystal structure of a fragment of CRT corresponding to lectin domain in complex with a tetrasaccharide fragment from the glucosylated arm of the Glc1Man9GlcNAc2 glycan. The structure explains CRT specificity for monoglucosylated protein substrates and rationalizes mutagenesis studies of the protein family.
MATERIALS AND METHODS
Protein Expression, Preparation, and Purification
The C163S mutant of the mouse CRT lectin domain (residues 18–206 and 301–368 linked by Gly-Ser-Gly-Ser-Gly) was cloned into pET29a (Amersham Biosciences-Pharmacia) and expressed in Escherichia coli BL21(DE3) in rich (LB) medium as a fusion protein with both N-terminal and C-terminal His-tags. Residues are numbered according to the unprocessed, immature protein sequence. For labeling for NMR experiments, cells were grown in M9 minimal medium with 15N-NH4Cl and [U-13C]glucose. For production of a selenomethionine-labeled protein, the expression plasmid was transformed into the E. coli methionine auxotroph strain DL41(DE3), and the protein was produced using LeMaster medium (20). Cells were harvested and broken in TSC buffer (50 mm Tris, 300 mm NaCl, 3 mm CaCl2, pH 8.0). The fusion protein was purified by affinity chromatography on Ni2+-charged Sepharose resin, and the N-terminal tag was removed by cleavage with thrombin, leaving the Gly-Ser-Met N-terminal extension and Leu-Glu-His6 C-terminal extension. The cleaved protein was additionally purified using size-exclusion chromatography using HPLC buffer (20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5). Selenomethionine-labeled protein was purified in a similar manner. The tetrasaccharide, α-d-glucopyranose-(1→3)-α-d-mannopyranose-(1→2)-α-d-mannopyranose-(1→2)-d-mannose (Glc1Man3), was purchased from the Alberta Research Council (Edmonton, Canada) and used without further purification.
Crystallization
Initial crystallization conditions were identified utilizing hanging drop vapor diffusion using Classics II screen (Qiagen). The best crystals for unliganded CRT were obtained by equilibrating a 1.0-μl drop of a protein (6 mg/ml) in buffer (20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5) and then mixed with 1.0 μl of reservoir solution containing 29% (w/v) PEG monomethyl ether 2000, 0.2 m KSCN, 10 mm taurine, and 0.1 m Tris (pH 8.5) and suspended over 1 ml of reservoir solution. Crystals grew in 3–7 days at 22 °C. For cryoprotection, the reservoir composition with the addition of 7% glycerol was used. For data collection, crystals were picked up in a nylon loop and flash cooled in an N2 cold stream (Oxford Cryosystem). The crystals contain one molecule in the asymmetric unit (Z = 4), corresponding to Vm = 2.08 Å3 Da−1 and a solvent content of 40.8% (21).
The best crystals for tetrasaccharide-bound CRT were obtained by equilibrating a 1.0-μl drop of a protein (6 mg/ml)/tetrasaccharide mixture at 1:2 molar ratio in buffer (20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5), mixed with 1.0 μl of reservoir solution containing 25.5% (w/v) PEG monomethyl ether 2000, 0.15 m KSCN, and 0.1 m Tris (pH 8.0) and suspended over 1 ml of reservoir solution. Crystals grew in 3–7 days at 22 °C. For cryoprotection, the reservoir composition with the addition of 7% glycerol was used. For data collection, crystals were picked up in a nylon loop and flash cooled in a N2 cold stream (Oxford Cryosystem). The crystals contain one molecule in the asymmetric unit (Z = 4), corresponding to Vm = 1.96 Å3 Da−1 and a solvent content of 37.4% (21).
Structure Solution and Refinement
The single-wavelength anomalous diffraction ion data sets from selenomethionine-labeled crystals of tetrasaccharide-bound CRT and a native data set from a crystal of unliganded CRT were collected at a wavelength of 0.9769 Å on an Area Detector Systems Corp. Quantum-210 CCD detector at beamline A1 at the Cornell High Energy Synchrotron Source (Table 1). Data processing and scaling were performed with HKL2000 (22). The starting phases for the orthorhombic crystal of tetrasaccharide-bound CRT were obtained using molecular replacement with CNX structure (Protein Data Bank code 1JHN) followed by direct refinement against experimentally derived selenium sites using PHASER (23). The resulting map was subjected to density modification with the program ARP/wARP (24) that allowed for automated model building of ∼90% of the residues.
TABLE 1.
Data collection and refinement statistics
| CRT | CRT/Glc1Man3 | CRT/Glc1Man3 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 43.11, 75.32, 79.59 | 42.73, 43.77, 133.34 | 74.62, 43.23, 84.63 |
| α, β, γ | 90.00°, 90.00°, 90.00° | 90.00°, 90.00°, 90.00° | 90.00°, 96.05°, 90.00° |
| Resolution (Å)a | 50-2.30 (2.34-2.30) | 50-1.95 (1.98-1.95) | 50-2.00 (2.03-2.00) |
| Rsym | 0.071 (0.280) | 0.062 (0.142) | 0.069 (0.248) |
| I/σI | 25.9 (6.3) | 23.2 (5.9) | 19.8 (5.4) |
| Completeness (%) | 99.8 (100.0) | 96.5 (80.2) | 100.0 (99.9) |
| Redundancy | 8.1 (7.2) | 4.5 (2.3) | 4.5 (3.8) |
| Refinement | |||
| Resolution (Å) | 54.7-2.30 | 66.7-1.95 | 84.2-2.01 |
| No. of reflections | 11,377 | 17,355 | 34,462 |
| Rwork/Rfree | 0.214/0.274 | 0.197/0.251 | 0.191/0.240 |
| No. of atoms | |||
| Protein | 2018 | 1994 | 4033 |
| Glc1Man3 | 0 | 45 | 90 |
| Calcium ions | 1 | 1 | 2 |
| Water | 56 | 177 | 374 |
| B-factors | |||
| Protein | 25.9 | 22.2 | 22.7 |
| Glc1Man3 | 24.5 | 27.1 | |
| Calcium ions | 27.5 | 16.4 | 21.0 |
| Water | 29.2 | 28.0 | 35.8 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.009 |
| Bond angles | 0.94° | 1.27° | 1.22° |
| Ramachandran statistics (%) | |||
| Most favored regions | 87.8 | 90.9 | 88.8 |
| Additional allowed regions | 12.2 | 9.1 | 11.2 |
a Highest resolution shell is shown in parentheses.
The partial model obtained from ARP/wARP was extended manually with the help of the program Coot (25) and was improved by several cycles of refinement, using the program REFMAC (26). Of 273 residues of the construct, the final model does not include GSM of the cloning linker at the N terminus, P204PK206, GSGSG of the linker replacing the P-domain and E363EQRLK368LEHHHHHH C-terminal residues. In addition, one Glc1Man3 molecule, one calcium ion and 177 water molecules were included in the model. The final model had good stereochemistry with no outliers in the Ramachandran plot computed using PROCHECK (27).
For the unliganded CRT and monoclinic crystals of tetrasaccharide-bound CRT, the structures were obtained by molecular replacement with the orthorhombic tetrasaccharide-bound CRT structure using PHASER (23) and improved by several cycles of refinement, using the program REFMAC (26) and model refitting followed by the translation-libration-screw (TLS) refinement (28). For the unliganded CRT, the final model does not include L203PPK206, GSGSG of the linker and E364QRLK368LEHHHHHH C-terminal residues. One calcium ion and 56 water molecules were included in the model. The final model has good stereochemistry with no outliers in the Ramachandran plot computed using PROCHECK (27).
Isothermal Titration Calorimetry
Experiments were carried out on a MicroCal iTC200 titration calorimeter (GE Healthcare) using the VPViewer software for instrument control and data acquisition. The buffer used for isothermal titration calorimetry experiments contained 20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5. During a titration experiment, a sample of the CRT lectin domain was kept at 293 K in a stirred (1000 rpm) reaction cell of 0.2 ml. Nineteen injections, each of 2-μl volume and 4-s duration with a 150-s interval between injections, were carried out using a 39.4-μl syringe filled with Glc1Man3 solution. Titration experiments were performed with 30 μm protein solution in the cell and 300 μm carbohydrate solution in the syringe. The calorimetric data were processed using the software package ORIGIN (version 7) to determine the Gibbs free energy of binding, molar binding stoichiometry (N), molar binding entropy (ΔS), and molar binding enthalpy (ΔH).
RESULTS
Crystallization and Structure Determination of CRT Lectin Domain
Previous attempts to crystallize CRT were likely hindered by the intrinsic mobility of the arm-like P-domain and unstructured C terminus (supplemental Fig. 1). To overcome this, we deleted the P-domain (residues 207–300) of mouse CRT and replaced it with a short linker. Additionally, Cys163 was mutated to serine to avoid intermolecular disulfide bond formation. Although we were able to obtain purified protein, extensive screening did not yield any crystals. We hypothesized that the presence of an unstructured C-terminal tail additionally hindered crystallization. To define the boundaries of the folded domain, we subjected full-length and P-domain-deleted mouse CRT to limited proteolysis using trypsin, chymotrypsin, proteinase K, and V8 protease. Characterization by mass spectrometry of fragments from trypsin digestion suggested the C terminus could be removed by cleavage at Lys368 to produce a stable fragment and NMR spectroscopy confirmed that it was soluble and well folded (supplemental Figs. 2 and 3). Based on these results, we recloned the lectin domain of mouse CRT to include residues 18–206 and 301–368. Crystallization trials produced long needle-like crystals that could be improved using additives including a Glc1Man3 tetrasaccharide. Large, well diffracting crystals were obtained in the primitive orthorhombic (space group P212121) and monoclinic (P21) forms with the best diffraction extending to beyond 2.0 Å.
We obtained unbiased, experimental phases for the orthorhombic crystals using selenomethionine-labeled protein and single wavelength anomalous diffraction (Table 1). Subsequently, this structure was used for molecular replacement to solve the structure of the monoclinic crystal form. The asymmetric units of orthorhombic and monoclinic crystal structures contain one and two copies of the CRT-Glc1Man3 complex, respectively. Despite different crystal contacts in both crystal forms, all three copies are nearly identical suggesting that the structures are not influenced by crystal packing.
Structure of CRT Lectin Domain
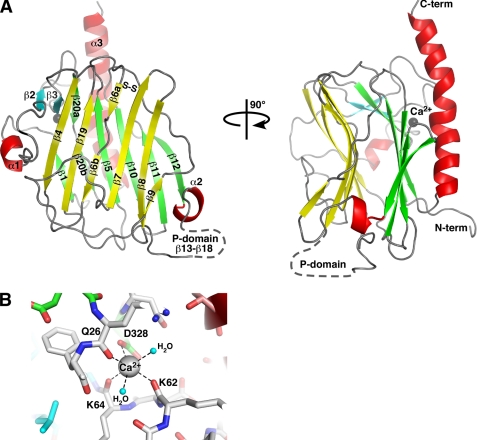
The structure gives an accurate definition of the CRT lectin domain boundaries running from Asp18 to Phe202 and from Pro301 to Glu363. No density was observed for Leu203–Lys207 and the following linker GSGSG due to disorder in the crystal. Also, the C-terminal sequence Glu364–Lys368 with the C-terminal His-tag were disordered. The structure of CRT lectin domain displays a jelly roll fold largely formed by a sandwich of two large β-sheets (Fig. 1A). The hydrophobic interactions between the seven-stranded concave β-sheet and six-stranded convex β-sheet are crucial for structural integrity of the domain. There is also a small β-sheet covering the surface where the concave and convex sheets create space between them. The structure also shows two short α-helices (Ala32–Arg36 and Leu196–Asp199) and a long α-helix (Glu336–Asp362) that runs along and beyond the convex β-sheet. There are also several protruding loops in the domain. Thus, a flap-like β6–β7 loop is covering a part of the concave β-sheet while packing against the α1-β2 loop that rises against the β4 strand. Another long loop between strands β2 and β3 is stabilizing the C-terminal half of the long helix α3. The Cys105–Cys137 disulfide bridges the beginning of strand β6 with the end of strand β7.
FIGURE 1.
Structure of the CRT lectin domain. A, schematic representation of the structure. Helices are shown in red, β-strands in the concave β-sheet are yellow, the convex β-sheet is green, and the two additional strands β2 and β3 are cyan. The bound calcium ion is shown as a gray sphere. The position of the P-domain that contains strands β13–β20 is indicated by a dashed line. Cys105 and Cys137 in the concave β-sheet form a disulfide bond (S–S). B, enlarged view of the calcium-binding site. Residues and hydrogen bonds (dashed lines) coordinating the calcium ion are shown along with two coordinating water molecules (cyan spheres). Residue color coding is the same as in A. N-term, N terminus; C-term, C terminus.
The structure clearly defines a calcium-binding site in the CRT lectin domain. The calcium ion is coordinated by the side chain of Asp328, backbone carbonyls of Gln26, Lys62, and Lys64, and by two water molecules (Fig. 1B).
Structural Basis of Carbohydrate Recognition
The tetrasaccharide Glc1Man3 corresponds to the branch of the Glc1Man9GlcNAc2 glycan that binds to the lectin domain of CRT/CNX. By isothermal titration calorimetry, we confirmed that the isolated, recombinant domain retained all of the lectin functions. The affinity of 0.7 μm for Glc1Man3 is very close to the reported value for intact CRT (supplemental Fig. 4) (29).
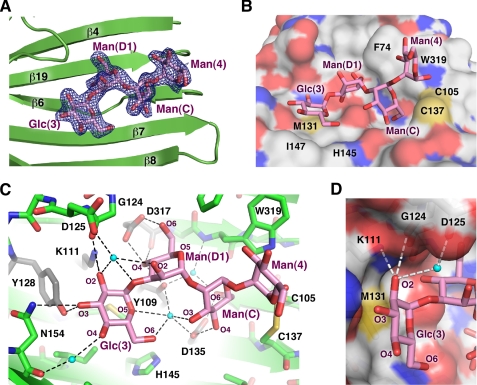
The electron density map showed clear, easily interpretable density for all four sugar moieties (Fig. 2A) that we refer to according to their numbering in the natural glycan, Glc(3)-Man(D1)-Man(C)-Man(4). The tetrasaccharide runs along the long channel formed by the curved β-sheet with all sugar moieties engaged in protein binding (Fig. 2B). Among them, the last mannose, Man(4), has somewhat looser contacts with CRT and higher B-factors. The first three moieties Glc(3), Man(D1), and Man(C) interact tightly with CRT and have B-factors similar to those of the protein residues.
FIGURE 2.
Structural basis of Glc1Man3 recognition by CRT. A, omit map calculated in the absence of tetrasaccharide shows well defined electron density (blue) for all four sugar moieties. The tetrasaccharide binds in a cavity on the concave β-sheet. B, surface representation of CRT shows the side chains of Phe74, Met131, His145, Ile147, Trp319, and the Cys105–Cys137 disulfide bridge form the walls of the cavity in contact with the glycan (magenta). C, oxygens in the tetrasaccharide form a network of hydrogen bonds (dotted lines) with ordered water molecules (cyan spheres) and CRT. Residues that disrupt CRT binding when mutated are shown in gray (17–19). D, the equatorial oxygen (O2) of glucose makes hydrogen bonds with the side chain of Lys111 and backbone carbonyl of Gly124. Mannose has an axial O2, which clashes sterically with the underlying side chain of Met131 to prevent binding in that position.
At the nonreducing end of the tetrasaccharide, the glucose moiety lies flat in the shallow cavity, the base of which is formed by side chains of Met131 and Ile147 (Fig. 2B). In addition to these hydrophobic contacts, Glc(3) forms a number of hydrogen bonds with protein. Thus, O2 of Glc(3) forms hydrogen bonds with side chain of Lys111 and backbone carbonyl of Gly124, whereas O3 hydrogen bonds with the side chains of Tyr128 and Asn154 (Fig. 2C). Additionally, several protein-carbohydrate hydrogen bonds are mediated through ordered water molecules. These include hydrogen bonds between side chain of Asp125 and O1 and O2 of Glc(3), the backbone carbonyl of Asn154 and Glc(3) O4, and Glc(3) O6 and side chains of Tyr109 and Asp135. In summary, every oxygen of the glucose Glc(3) is in involved in direct or indirect hydrogen bonds with CRT.
Importantly, the structure explains the requirement for glucose at the nonreducing end of the carbohydrate. Glucose and mannose are C2 epimers of each other. Our structure shows that equatorial O2 of glucose perfectly fits to the groove formed by CRT side chains (Fig. 2D). Mannose in this position would cause a steric clash with the sulfur atom of Met131 and the loss of hydrogen bonds with carbonyl of Gly124 and the side chain of Lys111. Previous mutagenesis studies showed that even the single K111A mutation impairs CRT-carbohydrate interactions (17, 18).
The structure also explains the specificity for monoglucosylated glycans (Glc1Man9GlcNAc2) over the precursor with two glucose residues (Glc2Man9GlcNAc2). Although the carbohydrate-binding site can accommodate a glucose residue in the second position, the sugar linkages are different. Binding of the tetrasaccharide α-Glc-(1→3)-α-Glc-(1→3)-α-Man-(1→2)-Man would result in the loss of hydrogen bonds and unfavorable interactions as the last mannose residue intersects the protein surface.
In contrast to the extensive protein contacts by the first sugar, Man(D1) and Man(C) mainly use their O4–O6 edges for interactions with CRT (Fig. 2C). In particular, O4 of Man(D1) occupies a crucial position engaging in three direct hydrogen bonds with side chain of Tyr109 and both the side chain and backbone carbonyl of Asp317. The side chain of Asp317 also makes a direct hydrogen bond with O6 of Man(D1). Although O2 of Man(D1) is directed away from the β-sheet, it forms a water-mediated hydrogen bond with the side chain of Asp125 located in the carbohydrate-interacting loop. Asp135 of CRT is crucial for Man(C) binding, as its side chain forms direct hydrogen bonds with O4 and O6 of Man(C). Two water molecules assist the other Man(C)-CRT hydrogen bonds: between O3 and the side chains of Tyr109 and Asp135, and between O6 and the side chain of Tyr109 and backbone amide of Trp319.
Previous studies showed that treatment with dithiothreitol abrogates carbohydrate binding by CRT (6). We similarly see no binding in the presence of the non-thiol-reducing agent TCEP (tris(2-carboxyethyl)phosphine) (supplemental Fig. 4B). These cysteines are also essential to the chaperone function of CRT (30). The CRT/Glc1Man3 structure provides a basis to explain these observations as the Cys105–Cys137 disulfide bond is involved in contacts with the Man(C) and Man(4) moieties of the Glc1Man3 tetrasaccharide. Namely, the C5–C6 bond of Man(C) and C1–O1 bond of Man(4) partially wrap around the CRT disulfide bond (Fig. 2C, right). Reduction of this disulfide bond would clearly disrupt binding of the last two mannose moieties. Both Man(C) and Man(4) are also engaged in hydrophobic interactions with the side chain of Trp319.
The C1–O1 bond of Man(4) is directed away from the protein. Therefore, it is likely that the following mannose residue Man(3) of Glc1Man9GlcNAc2 glycan does not make any significant interactions with CRT, and the full essence of CRT-carbohydrate recognition is captured in our structure. In agreement with this conclusion, Glc1Man3 was shown to compete effectively with Glc1Man9 for binding to CNX (6).
Identical tetrasaccharide conformations were observed in the two crystal forms. This clearly demonstrates that the conformation is not affected by crystal packing. There is a single crystal contact between the C6–O6 bond of Man(4) at the far end of tetrasaccharide and the side chain of Glu345 of another CRT molecule in the P212121 crystal form, whereas the carbohydrate does not contact any symmetry-related molecules in the P21 crystal form. Additionally, we note that the bound calcium ion is positioned far from the carbohydrate-binding site and is not involved in glycan recognition.
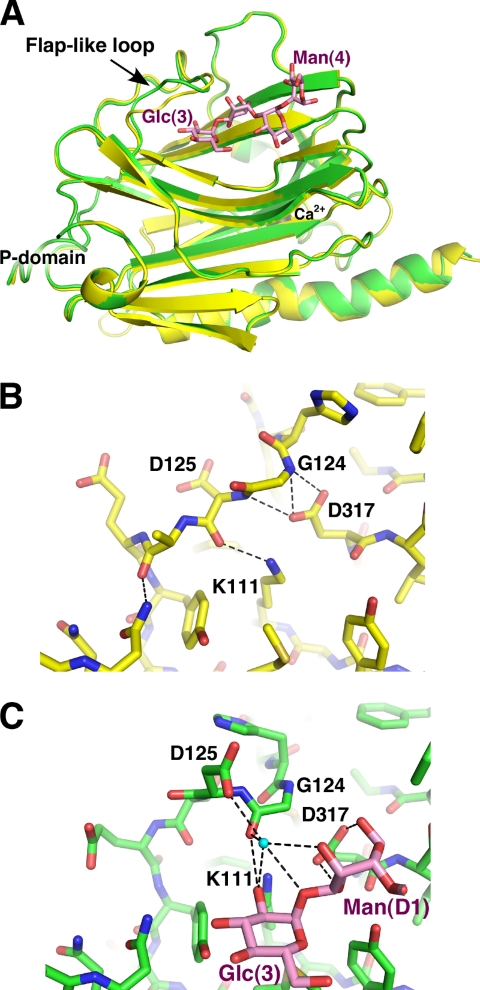
We overlaid the tetrasaccharide-bound and unliganded CRT lectin domain structures to assess the conformational changes in CRT upon carbohydrate binding, The overlay results in an root mean square deviation of 0.2 Å over 229 Cα atoms demonstrating that the two structures are nearly identical (Fig. 3A). The only significant change occurs in the flap-like loop containing Gly124 and Asp125, residues involved in glucose binding. The loop conformation in the unliganded state is stabilized by side chains of Lys111 and Asp317 that make hydrogen bonds with the backbone carbonyl of Asp125 and amides of Gly124 and Asp125, respectively (Fig. 3B). When oligosaccharide binds CRT, side chains of Lys111 and Asp317 engage in direct hydrogen bonding with sugar moieties. The released Gly124 and Asp125 rotate allowing carbonyl of Gly124 to form a hydrogen bond with Glc(3), whereas the side chain of Asp125 interacts with carbohydrates via an ordered water molecule (Fig. 3C).
FIGURE 3.
CRT undergoes limited conformational changes upon carbohydrate binding. A, overlay of unliganded (yellow) and Glc1Man3-bound (green) CRT lectin domain structures shows differences in the large loop between strands β6 and β7. B, the conformation of the loop in unliganded CRT is stabilized by hydrogen bonds between the side chain of Asp317 and amides of Gly124 and Asp125 and between the side chain of Lys111 and carbonyl of Asp125. C, in the complex, sugar residues Glc(3) and Man(D1) engage loop residues Gly124 and Asp125 through a 60 ° rotation in the ψ backbone angle of Gly124. This rotation enables the side chain of Asp125 to participate in carbohydrate binding via an ordered water molecule (cyan sphere). The interactions of Asp317 with Gly124 are replaced by hydrogen bonds of Asp317 with Man(D1).
Structure Correlates well with Mutagenesis Results
Due to the interest in recognition of glycosylated proteins by CRT/CNX, vast mutagenesis data have been obtained with CRT to characterize its binding to oligosaccharides. Mutagenesis of CRT by different groups showed that Tyr109, Lys111, Tyr128, Asp135, and Asp317 are critical for carbohydrate binding (17–19). The crystal structure shows that the side chains of all of these residues form direct hydrogen bonds with carbohydrate moieties (Fig. 2C). The structure is also in perfect agreement with the results showing reduced carbohydrate binding for the W319I and W319A CRT mutants (19). The bulky side chain of this residue is involved in hydrophobic contacts with the reducing end mannose residue in our structure (Fig. 2B). Similarly, substitutions of Met131 reduced but did not prevent binding (17, 18). On the other hand, mutations of Asp125 did not affect oligosaccharide binding (17), as the contacts of this residue with sugar are mediated through a water molecule. The structure further explains the observation that D160G and D160A mutations do not affect carbohydrate binding (18, 19), as this residue is 13 Å away from the bound ligand. The structure also reveals that the 75% decrease in carbohydrate affinity in the R73L CRT mutant (17) results from mid-range conformational changes, as this residue is >10 Å away from the bound tetrasaccharide and is unable to participate directly in binding.
Comparison with CNX and Other ER Lectins
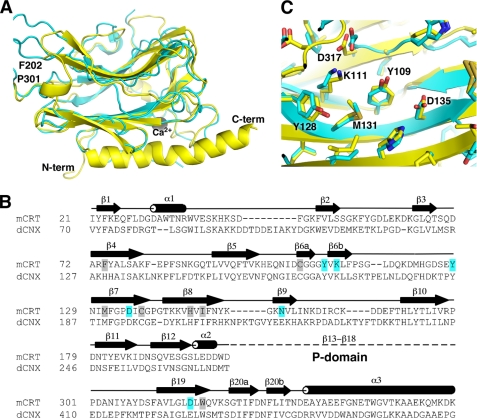
Sequence similarity between homologous regions of CRT and CNX led to assumptions that the structures of these proteins are very similar and resulted in use of homology models of CRT based on the CNX structure. Consequently, these models were used to interpret results from single point and deletion mutagenesis that proved to be inaccurate in some cases. As an example, the Glu217 of CNX, which participates in ligand binding, was incorrectly assigned as homologous to Asp160 of CRT, which is far from the carbohydrate-binding site (17, 18). On the other hand, while the CRT construct used for glycan-independent substrate binding studies was missing several residues of the long C-terminal helix (31), the thermal denaturation curve for that construct is similar to that of the lectin domain studied here (supplemental Fig. 5).
The CRT structure allows for an accurate structural alignment of CRT and CNX. The structural overlay of unliganded CRT and CNX shows 33% sequence identity between their lectin domains and a root mean square deviation of 1.7 Å over 195 CA atoms. The structural similarities are strongest in the β-sheet regions, whereas the loops often adopt divergent conformations (Fig. 4A). Conspicuous differences between the two structures occur in the helical regions. Thus, the short α1 and α2 CRT helices are absent in the CNX structure. Similarly, the C-terminal helix is much shorter (10 versus 25 residues) in the CNX structure. The reason for this divergence is unclear, as this region is well conserved between the two proteins (Fig. 4B). Although this may reflect a genuine difference between CNX and CRT, it is possible that the native CNX C-terminal helix is longer than observed previously.
FIGURE 4.
Structural comparison of the CRT and CNX lectin domains. A, overlay of the CRT (yellow) and CNX (Protein Data Bank code 1JHN; cyan) structures. The termini and boundaries of the P-domain of CRT are indicated. B, structure-based sequence alignment of mouse CRT and dog CNX lectin domains. Secondary structure of CRT is shown above the alignment with β-strands and α-helices labeled. Residues that make direct hydrogen bonds with Glc1Man3 are highlighted in cyan, and those making van der Waals contacts are highlighted in gray. The position of the internal P-domain comprising strands β13 to β18 of CRT is indicated. C, enlarged view of the carbohydrate-binding site shows nearly identical positioning of key residues in CRT and CNX. Residue numbers refer to CRT. N-term, N terminus; C-term, C terminus.
Despite differences elsewhere, the oligosaccharide-binding surface is nearly identical in both proteins (Fig. 4C). The residues that are critical for carbohydrate binding are very well conserved and adopt very similar conformations in both CRT and CNX. Glucose soaking of CNX crystals showed that glucose contacts Met189 of CNX (equivalent to Met131 of mouse CRT) and makes hydrogen bonds with Tyr165 (Tyr109), Lys167 (Lys111), Tyr186 (Tyr128), Glu217 (Asn154), and possibly Glu426 (Asp317) (8). This is slightly shifted from the position observed in our crystal structure and involves residues (Tyr109 and Asp317 of CRT) that form hydrogen bonds with the following mannose in the CRT-tetrasaccharide complex. The reason for this discrepancy could simply result from low binding affinity of glucose to CNX coupled with the low resolution of crystallographic data set leading to some positional shift of the glucose moiety in the CNX structure. Or, the isolated glucose moiety could bind at a slightly different location when it is not part of an oligosaccharide chain. Nonetheless, the superposition of the carbohydrate binding residues is striking and suggests that CNX and CRT bind carbohydrates in an identical fashion.
The calcium ion is coordinated in the CNX structure by side chains of Asp437 (Asp328 of CRT) and Asp118 (Asp63), carbonyl of Ser75 (Gln26) and, possibly, by carbonyl of Lys119 (Lys64). The missing coordinating groups were not observed possibly due to insufficient resolution. In CRT, an equivalent calcium ion is coordinated by both oxygen atoms of Asp328 side chain, carbonyls of Gln26, Lys62, and Lys64 and by two water molecules.
A structural similarity search using the Dali database (32) showed that the CRT lectin domain is most similar to CNX (Z-score, 27.5) as expected. In addition, the CRT structure is similar to Emp47p (Z-score, 15.5), ERGIC-53 (Z-score, 14.3), and VIP36 (Z-score, 13.7). VIP36 and ERGIC-53 are transport lectin proteins that are involved in trafficking of glycosylated proteins out of the ER, whereas Emp46/47p is a yeast homolog of ERGIC-53. Interestingly, these proteins have specificity toward the deglucosylated D1 arm of high mannose glycans (33–35), the same arm recognized by CRT/CNX in its monoglucosylated state. The crystal structures of VIP36 in complex with α-Man-(1→2)-Man and α-Man-(1→2)-α-Man-(1→3)-β-Man-(1→4)-GlcNAc have been determined (36). Despite a significant overlap in oligosaccharide specificity and the use of a similar structural scaffold between CRT/CNX and VIP36, they use differing surfaces to bind carbohydrates (supplemental Fig. 6). This is an example of how a similar fold is adapted for binding somewhat differing ligands.
DISCUSSION
The high resolution crystal structure of the CRT lectin domain in complex with the Glc1Man3 tetrasaccharide illuminates the molecular basis of monoglucosylated Glc1Man9GlcNAc2 glycan function in the calnexin cycle. The structure explains the requirement for a single glucose at the nonreducing end of the carbohydrate and allows for an accurate structural alignment between CRT and CNX. The striking similarity in the sugar-binding sites suggests that CNX and CRT interact with monoglucosylated substrates in identical fashion.
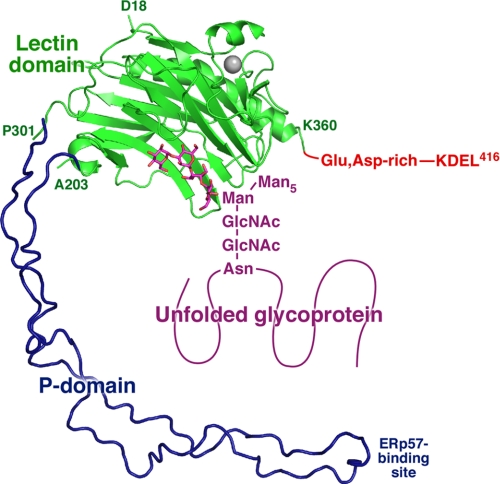
To gain insight into the structure of full-length CRT with a bound glycoprotein, we overlaid the CRT lectin domain with a model of the P-domain derived from the structure of CNX (Fig. 5). As the CRT P-domain is shorter and has only three repeat modules, we removed the third module of the CNX P-domain. The CNX P-domain was chosen as a model because the crystal structure includes information about the relative orientation of the P-and lectin domains that is absent from the NMR structure of the isolated CRT P-domain (12, 13). The tip of the CRT P-domain contains a binding site for ERp57, an oxidoreductase involved in disulfide bond formation in glycoproteins. The model gives an idea of how the bound glycoprotein might be positioned relative to the lectin domain, the P-domain and ERp57. CRT also contains an ∼55-residue-long C-terminal extension that is rich (>60%) in glutamate and aspartate residues. Because of its abundant negative charges, this C-terminal domain is unlikely to be structured in solution, but it becomes more ordered upon binding calcium (37).
FIGURE 5.
Structural model of full-length CRT. The lectin domain is shown in green, and the bound carbohydrate is shown in magenta. The approximate orientation of the P-domain (dark blue) is shown based on the CNX structure with one repeat unit removed to match the shorter CRT P-domain. The bound carbohydrate is shown as part of the N-linked glycan linked to an asparagine residue of an unfolded protein. The C terminus of CRT contains a Glu-, Asp-rich sequence, which binds calcium, and a KDEL ER retention signal. The residues that define the boundaries of crystallized CRT fragment and the portion of the P-domain that binds the thiol oxidoreductase, ERp57, are labeled.
To exit the CNX/CRT cycle, monoglucosylated glycoproteins are processed by glucosidase II to remove the terminal glucose residue. The accessibility of the glycan for processing while bound to CNX/CRT has been the subject of some debate (38, 39). Our structure shows that the bond between Glc(3) and Man(D1) targeted by glucosidase II is not easily accessible. It seems likely that the glycosylated substrate has to dissociate from CRT for deglucosylation to occur.
Earlier reports also suggested that CRT undergoes conformational changes upon carbohydrate binding (40). Comparison of the tetrasaccharide-bound and unliganded CRT lectin domain structures demonstrates that the only significant change occurs in the flap-like loop that is involved in glucose binding. On the other hand, loss of the bound calcium ion is likely to strongly destabilize the CRT lectin domain. Numerous studies have shown calcium-dependent conformational changes in CRT (41–43). Although our carbohydrate-bound CRT structure confirms that the calcium-binding site is too far to affect interactions with glycans, the tightly bound calcium ion is important for the structural integrity of the lectin domain. With a Kd of ∼2 μm (14), loss of this high affinity calcium ion is unlikely to occur within the ER, but it may occur for CRT outside of the ER (44).
CNX and CRT exhibit overlapping but distinct patterns of interaction with folding glycoproteins in vivo (45–47). Given that the lectin sites of these chaperones are essentially identical, the basis for observed differences in glycoprotein binding specificity must reside in other properties. Previous studies have shown that that the distinct luminal versus membrane-bound topologies of CRT and CNX contribute to selection of client glycoproteins (45, 48, 49). It is also likely that the reported ability of CRT and CNX to bind directly to peptides (31, 50, 51) or to polypeptide segments of non-native protein conformers (9, 40) contributes to substrate selection. Consequently, the identification of such peptide binding sites on these chaperones is of considerable interest. However, despite the fact that the lectin domains of both CNX (50) and CRT4 are capable of binding hydrophobic peptides with micromolar Kd, our efforts to obtain co-crystals of the CRT lectin domain with such peptides have so far been unsuccessful. Likewise, we have been unable to co-crystallize the lectin domain of CRT with ATP. Both CNX and CRT have been reported to bind ATP (41, 42), and the presence of this nucleotide has been shown to enhance their abilities to bind non-native polypeptides and to suppress their aggregation in vitro (9, 40). Given the conformational changes that have been reported to accompany nucleotide or peptide binding (9, 41, 42, 50), it may be that such conformations are less amenable to crystallization.
CRT has also been reported to bind zinc, and four histidines within the lectin domain have been implicated in binding (40, 43, 52–56). Examination of these histidines reveals that some are buried but that His42 is exposed and adjacent to other residues, Asp118, Asp121, His123, and Asp125, that could potentially bind zinc.
In conclusion, we have determined the structure of the CRT lectin domain in complex with its physiological ligand. The structure provides the framework for the design and interpretation of mutants that affect the multiple physiological functions of CRT.
Supplementary Material
Acknowledgments
Data acquisition at the Macromolecular Diffraction (MacCHESS) facility at the Cornell High Energy Synchrotron Source was supported by National Science Foundation Award DMR 0225180 and National Institutes of Health Award RR-01646.
This work was supported by Canadian Institutes of Health Research Grants MOP-81277 (to K. G.) and MOP-53310 (to D. B. W.).
The atomic coordinates and structure factors (codes 3O0V, 3O0W, and 3O0X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
C. L. Pocanschi, unpublished observations.
- ER
- endoplasmic reticulum
- CNX
- calnexin
- CRT
- calreticulin.
REFERENCES
- 1.Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 2.Caramelo J. J., Parodi A. J. (2008) J. Biol. Chem. 283, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lederkremer G. Z. (2009) Curr. Opin. Struct. Biol. 19, 515–523 [DOI] [PubMed] [Google Scholar]
- 4.Ware F. E., Vassilakos A., Peterson P. A., Jackson M. R., Lehrman M. A., Williams D. B. (1995) J. Biol. Chem. 270, 4697–4704 [DOI] [PubMed] [Google Scholar]
- 5.Spiro R. G., Zhu Q., Bhoyroo V., Söling H. D. (1996) J. Biol. Chem. 271, 11588–11594 [DOI] [PubMed] [Google Scholar]
- 6.Vassilakos A., Michalak M., Lehrman M. A., Williams D. B. (1998) Biochemistry 37, 3480–3490 [DOI] [PubMed] [Google Scholar]
- 7.Soldà T., Galli C., Kaufman R. J., Molinari M. (2007) Mol. Cell. 27, 238–249 [DOI] [PubMed] [Google Scholar]
- 8.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 9.Brockmeier A., Williams D. B. (2006) Biochemistry 45, 12906–12916 [DOI] [PubMed] [Google Scholar]
- 10.Frickel E. M., Riek R., Jelesarov I., Helenius A., Wuthrich K., Ellgaard L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozlov G., Maattanen P., Schrag J. D., Pollock S., Cygler M., Nagar B., Thomas D. Y., Gehring K. (2006) Structure 14, 1331–1339 [DOI] [PubMed] [Google Scholar]
- 12.Ellgaard L., Riek R., Herrmann T., Güntert P., Braun D., Helenius A., Wüthrich K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3133–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellgaard L., Bettendorff P., Braun D., Herrmann T., Fiorito F., Jelesarov I., Güntert P., Helenius A., Wüthrich K. (2002) J. Mol. Biol. 322, 773–784 [DOI] [PubMed] [Google Scholar]
- 14.Baksh S., Michalak M. (1991) J. Biol. Chem. 266, 21458–21465 [PubMed] [Google Scholar]
- 15.Peterson J. R., Helenius A. (1999) J. Cell Sci. 112, 2775–2784 [DOI] [PubMed] [Google Scholar]
- 16.Leach M. R., Williams D. B. (2004) J. Biol. Chem. 279, 9072–9079 [DOI] [PubMed] [Google Scholar]
- 17.Kapoor M., Ellgaard L., Gopalakrishnapai J., Schirra C., Gemma E., Oscarson S., Helenius A., Surolia A. (2004) Biochemistry 43, 97–106 [DOI] [PubMed] [Google Scholar]
- 18.Thomson S. P., Williams D. B. (2005) Cell Stress Chaperones 10, 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gopalakrishnapai J., Gupta G., Karthikeyan T., Sinha S., Kandiah E., Gemma E., Oscarson S., Surolia A. (2006) Biochem. Biophys. Res. Commun. 351, 14–20 [DOI] [PubMed] [Google Scholar]
- 20.Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Sweet R. M. eds) pp. 307–326, Vol. 276, Part A, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 23.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 28.Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 29.Kapoor M., Srinivas H., Kandiah E., Gemma E., Ellgaard L., Oscarson S., Helenius A., Surolia A. (2003) J. Biol. Chem. 278, 6194–6200 [DOI] [PubMed] [Google Scholar]
- 30.Martin V., Groenendyk J., Steiner S. S., Guo L., Dabrowska M., Parker J. M., Müller-Esterl W., Opas M., Michalak M. (2006) J. Biol. Chem. 281, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 31.Rizvi S. M., Mancino L., Thammavongsa V., Cantley R. L., Raghavan M. (2004) Mol. Cell 15, 913–923 [DOI] [PubMed] [Google Scholar]
- 32.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 33.Hara-Kuge S., Ohkura T., Seko A., Yamashita K. (1999) Glycobiology 9, 833–839 [DOI] [PubMed] [Google Scholar]
- 34.Kamiya Y., Yamaguchi Y., Takahashi N., Arata Y., Kasai K., Ihara Y., Matsuo I., Ito Y., Yamamoto K., Kato K. (2005) J. Biol. Chem. 280, 37178–37182 [DOI] [PubMed] [Google Scholar]
- 35.Kamiya Y., Kamiya D., Yamamoto K., Nyfeler B., Hauri H. P., Kato K. (2008) J. Biol. Chem. 283, 1857–1861 [DOI] [PubMed] [Google Scholar]
- 36.Satoh T., Cowieson N. P., Hakamata W., Ideo H., Fukushima K., Kurihara M., Kato R., Yamashita K., Wakatsuki S. (2007) J. Biol. Chem. 282, 28246–28255 [DOI] [PubMed] [Google Scholar]
- 37.Villamil Giraldo A. M., Lopez Medus M., Gonzalez Lebrero M., Pagano R. S., Labriola C. A., Landolfo L., Delfino J. M., Parodi A. J., Caramelo J. J. (2010) J. Biol. Chem. 285, 4544–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodan A. R., Simons J. F., Trombetta E. S., Helenius A. (1996) EMBO J. 15, 6921–6930 [PMC free article] [PubMed] [Google Scholar]
- 39.Zapun A., Petrescu S. M., Rudd P. M., Dwek R. A., Thomas D. Y., Bergeron J. J. (1997) Cell 88, 29–38 [DOI] [PubMed] [Google Scholar]
- 40.Saito Y., Ihara Y., Leach M. R., Cohen-Doyle M. F., Williams D. B. (1999) EMBO J. 18, 6718–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou W. J., Bergeron J. J., Li Y., Kang C. Y., Thomas D. Y. (1995) J. Biol. Chem. 270, 18051–18059 [DOI] [PubMed] [Google Scholar]
- 42.Corbett E. F., Michalak K. M., Oikawa K., Johnson S., Campbell I. D., Eggleton P., Kay C., Michalak M. (2000) J. Biol. Chem. 275, 27177–27185 [DOI] [PubMed] [Google Scholar]
- 43.Li Z., Stafford W. F., Bouvier M. (2001) Biochemistry 40, 11193–11201 [DOI] [PubMed] [Google Scholar]
- 44.Gold L. I., Eggleton P., Sweetwyne M. T., Van Duyn L. B., Greives M. R., Naylor S. M., Michalak M., Murphy-Ullrich J. E. (2010) FASEB J. 24, 665–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danilczyk U. G., Cohen-Doyle M. F., Williams D. B. (2000) J. Biol. Chem. 275, 13089–13097 [DOI] [PubMed] [Google Scholar]
- 46.Peterson J. R., Ora A., Van P. N., Helenius A. (1995) Mol. Biol. Cell 6, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pieren M., Galli C., Denzel A., Molinari M. (2005) J. Biol. Chem. 280, 28265–28271 [DOI] [PubMed] [Google Scholar]
- 48.Wada I., Imai S., Kai M., Sakane F., Kanoh H. (1995) J. Biol. Chem. 270, 20298–20304 [DOI] [PubMed] [Google Scholar]
- 49.Hebert D. N., Zhang J. X., Chen W., Foellmer B., Helenius A. (1997) J. Cell Biol. 139, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brockmeier A., Brockmeier U., Williams D. B. (2009) J. Biol. Chem. 284, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 51.Sandhu N., Duus K., Jørgensen C. S., Hansen P. R., Bruun S. W., Pedersen L. Ø., Højrup P., Houen G. (2007) Biochim. Biophys. Acta 1774, 701–713 [DOI] [PubMed] [Google Scholar]
- 52.Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 53.Baksh S., Spamer C., Heilmann C., Michalak M. (1995) FEBS Lett. 376, 53–57 [DOI] [PubMed] [Google Scholar]
- 54.Guo L., Groenendyk J., Papp S., Dabrowska M., Knoblach B., Kay C., Parker J. M., Opas M., Michalak M. (2003) J. Biol. Chem. 278, 50645–50653 [DOI] [PubMed] [Google Scholar]
- 55.Khanna N. C., Tokuda M., Waisman D. M. (1986) J. Biol. Chem. 261, 8883–8887 [PubMed] [Google Scholar]
- 56.Tan Y., Chen M., Li Z., Mabuchi K., Bouvier M. (2006) Biochim. Biophys. Acta 1760, 745–753 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.