Summary
The e (P4) phosphatase from Haemophilus influenzae functions in a vestigial NAD+ utilization pathway by dephosphorylating NMN to nicotinamide riboside. P4 is also the prototype of class C acid phosphatases, which are nonspecific 5′-, 3′-nucleotidases localized to the bacterial outer membrane. To understand substrate recognition by P4 and other class C phosphatases, we have determined the crystal structures of a substrate-trapping mutant P4 enzyme complexed with NMN, 5′-AMP, 3′-AMP, and 2′-AMP. The structures reveal an anchor-shaped substrate-binding cavity comprising a conserved hydrophobic box that clamps the nucleotide base, a buried phosphoryl binding site, and three solvent-filled pockets that contact the ribose and hydrogen-bonding edge of the base. The span between the hydrophobic box and phosphoryl site is optimal for recognizing nucleoside monophosphates, which explains the general preference for this class of substrate. The base makes no hydrogen bonds with the enzyme, which is consistent with observed lack of base specificity. Two solvent-filled pockets flanking the ribose are key to the dual recognition of 5′- and 3′-nucleotides. These pockets minimize the enzyme’s direct interactions with the ribose and provide sufficient space to accommodate 5′ substrates in an anti conformation and 3′ substrates in a syn conformation. Finally, the structures suggest that class B and C acid phosphatases share a common strategy for nucleotide recognition.
Keywords: X-ray crystallography, phosphatase, nucleotidase, substrate recognition, steady-state kinetics
Introduction
The lipoprotein e (P4)1,2 is a 28-kDa outer membrane acid phosphatase from Haemophilus influenzae, a common commensal inhabitant of the human nasopharynx and the etiologic agent of local and invasive infections in humans, particularly children.3,4 The enzyme is a major component of the outer membrane and is highly conserved among H. influenzae strains. The high conservation and outer membrane location of e (P4) has motivated investigations of the enzyme as a potential vaccine component. Studies have shown that recombinant P4 (rP4) and rP4 mutant enzymes are highly immunogenic, anti-rP4 antibodies exhibit bactericidal activity, and immunization of mice with rP4 reduces nasal colonization of nontypeable H. influenzae strains.5–7
The main biological role of e (P4) is to catalyze the conversion of nicotinamide mononucleotide (NMN) to nicotinamide riboside (NR) as part of vestigial NAD+ utilization pathway.8,9 H. influenzae lacks the full repertoire of enzymes needed for the de novo biosynthesis of NAD+, therefore, it must obtain this essential cofactor from the host. The NAD+ utilization pathway includes an uptake system that imports NAD+, NMN, and NR into the periplasm. Within the periplasm, the NAD+ nucleotidase NadN catalyzes the hydrolysis of NAD+ to generate NMN and AMP. NMN produced by NadN, or imported by the uptake system, is dephosphorylated to NR by e (P4). NadN also has NMN 5′-nucleotidase activity, but e (P4) has higher efficiency for NMN and is thus thought to be the major catalyst for the production of NR for the pathway.8 NR is then transported across the inner membrane into the cytosol by the NR-specific permease PnuC, where it is converted into NAD+ by the bifunctional NR kinase/NMN adenylyltransferase NadR10. Although the biological function of e (P4) in NAD+ utilization is well established, the structural basis for the recognition of NMN by P4 is unknown.
The larger context for the research described here is that e (P4) is the prototype of class C acid phosphatases (CCAPs). First recognized as a family of related bacterial enzymes by Thaller et al. in 1998,11 CCAPs belong to the DDDD superfamily of phosphohydrolases and are defined at the primary structure level by the conserved bipartite sequence motif of [IV]-[VAL]-D-[IL]-D-E-T-[VM]-L-X-[NT]-X(2)-Y and [IV]-[LM]-X(2)-G-D-[NT]-L-X-D-F (Asp residues of the DDDD motif in bold).
In addition to e (P4),1,2,12–14 several CCAPs have been characterized to various degrees, including those from Elizabethkingia meningosepticum (OlpA15), Streptococcus equisimilis (LppC16), Helicobacter pylori (HppA17), and Clostridium perfringens.18,19 CCAPs are dimeric enzymes that exhibit phosphomonoesterase activity for commonly used aryl phosphate substrates such as p-nitrophenyl phosphate (pNPP) and 4-methylumbelliferyl phosphate. Among the physiologically relevant molecules that have been tested, the highest catalytic efficiencies have been achieved with nucleoside 5′-monophosphates. CCAPs do not exhibit a strong base preference among this class of substrate. OlpA, LppC,20 rP4 (vide infra) and the C. perfringens enzyme18 also exhibit activity with nucleoside 3′-monophosphates, but with lower efficiency than nucleoside 5′-monophosphate substrates. Thus, the available in vitro data suggest that CCAPs function primarily as nonspecific 5′-, 3′-nucleotidases.
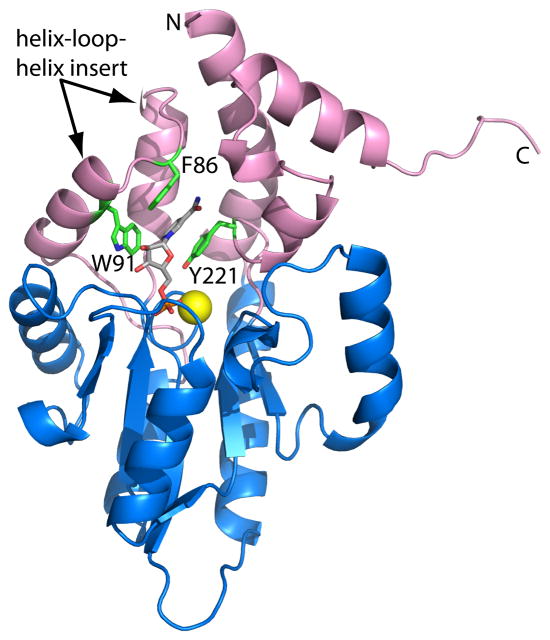
The structure of one CCAP - rP4 - has been determined.14 rP4 has a two-domain fold consisting of a core α/β domain (Fig. 1, blue) and an α-helical cap domain (Fig. 1, pink). The four Asp residues of the DDDD motif are clustered around a Mg2+ ion at the base of active site (Fig. 1, yellow sphere). The core domain fold indicates that rP4 belongs to the haloacid dehalogenase (HAD) structural superfamily.21
Fig. 1.
Ribbon representation of D66N complexed with NMN. The core and cap domains are colored blue and pink, respectively. NMN is colored gray. The yellow sphere represents Mg2+. Residues of the aromatic box are colored green. This figure and others were created with PyMOL.39
The structure of rP4 complexed with the inhibitor tungstate provided insight into the identities of the nucleophile that attacks the substrate phosphoryl group (Asp64), the residue that protonates the leaving group (Asp66), and side chains that stabilize the substrate phosphoryl group (Lys161, Thr124). However, the residues that interact with the non-phosphoryl groups of substrates have not been identified. Thus, the structural elements that enforce the preference for nucleoside monophosphates are unknown. Furthermore, the question of how CCAPs achieve the dual recognition of nucleoside 5′- and 3′-monophosphates remains unanswered.
Within this context we initiated a structure-based study of substrate recognition having the goals of understanding how rP4 binds its known biological substrate, NMN, and more generally, elucidating the structural features of CCAPs that are responsible for recognizing nucleoside monophosphate substrates. To this end, we have determined high resolution crystal structures of a substrate-trapping mutant of rP4 complexed with NMN, 5′-AMP, 3′-AMP, and 2′-AMP, as well as a structure of rP4 complexed with the product inorganic phosphate (Pi) (Table 1). The structures and the accompanying kinetic data provide insight into the basis of the nucleotidase activity of P4 and other CCAPs.
Table 1.
Data collection and refinement statistics
| NMN | 5′-AMP | 3′-AMP | 2′-AMP | phosphate | |
|---|---|---|---|---|---|
| Beamline | 24-ID-C | 4.2.2 | 4.2.2 | 4.2.2 | 24-ID-C |
| Wavelength | 0.9792 | 1.00 | 1.00 | 1.00 | 0.9792 |
| Data collection resolution (Å) | 50 – 1.35 (1.40 – 1.35) | 50 – 1.55 (1.61–1.55) | 50 – 1.85 (1.92 – 1.85) | 50 – 1.90 (1.93 – 1.90) | 50 – 1.40 (1.45 – 1.40) |
| No. of observations | 622380 | 944628 | 536096 | 499757 | 340873 |
| No. of unique reflections | 66003 | 44418 | 26166 | 24575 | 57486 |
| Rmerge(I) | 0.074 (0.603) | 0.088 (0.427) | 0.114 (0.439) | 0.120 (0.534) | 0.064 (0.557) |
| Average I/σ | 39.6 (4.3) | 47.0 (5.9) | 27.6 (4.35) | 30.2 (3.47) | 25.4 (2.8) |
| Completeness (%) | 99.7 (100) | 100 (100) | 99.9 (99.4) | 99.9 (98.3) | 95.9 (98.3) |
| Redundancy | 9.4 (9.4) | 21.3 (19.0) | 20.5 (14.3) | 20.3 (13.0) | 5.9 (5.4) |
| Refinement resolution (Å) | 50 – 1.35 (1.37 – 1.35) | 50 – 1.55 (1.58–1.55) | 50 – 1.85 (1.92 – 1.85) | 50 – 1.90 (1.98 – 1.90) | 50 – 1.40 (1.42 – 1.40) |
| Rcryst | 0.135 (0.211) | 0.141 (0.159) | 0.164 (0.177) | 0.178 (0.196) | 0.139 (0.207) |
| Rfreea | 0.154 (0.245) | 0.171 (0.219) | 0.189 (0.202) | 0.217 (0.245) | 0.168 (0.277) |
| No. of protein residues | 247 | 247 | 247 | 247 | 247 |
| No. of protein atoms | 1933 | 1938 | 1931 | 1912 | 1929 |
| No. of water molecules | 274 | 280 | 231 | 173 | 256 |
| Average B-factor (Å2) | |||||
| Protein | 14.1 | 15.8 | 16.8 | 26.8 | 14.2 |
| Water | 25.0 | 27.5 | 24.4 | 30.4 | 24.9 |
| Ligand | 12.2 | 15.7 | 22.1 | 23.3 | 11.7 |
| Mg+2 | 7.7 | 9.7 | 11.8 | 15.8 | 7.6 |
| rmsdb | |||||
| Bonds (Å) | 0.006 | 0.005 | 0.006 | 0.006 | 0.005 |
| Angles (deg) | 1.17 | 1.04 | 1.00 | 1.02 | 1.03 |
| Ramachandran plotc | |||||
| Favored (%) | 98.0 | 98.0 | 98.0 | 97.5 | 98.0 |
| Allowed (%) | 2.0 | 2.0 | 2.0 | 2.5 | 2.0 |
| Coordinate error (Å)d | 0.13 | 0.17 | 0.18 | 0.21 | 0.14 |
| PDB code | 3OCU | 3OCV | 3OCW | 3OCX | 3OCY |
Values for the outer resolution shell of data are given in parenthesis.
A common set of test reflections (5 %) was used for refinement of all structures.
Compared to the parameters of Engh and Huber.37
The Ramachandran plot was generated with RAMPAGE.38
Maximum likelihood-based coordinate error estimate.
Results
Structures D66N complexed NMN and 5′-AMP
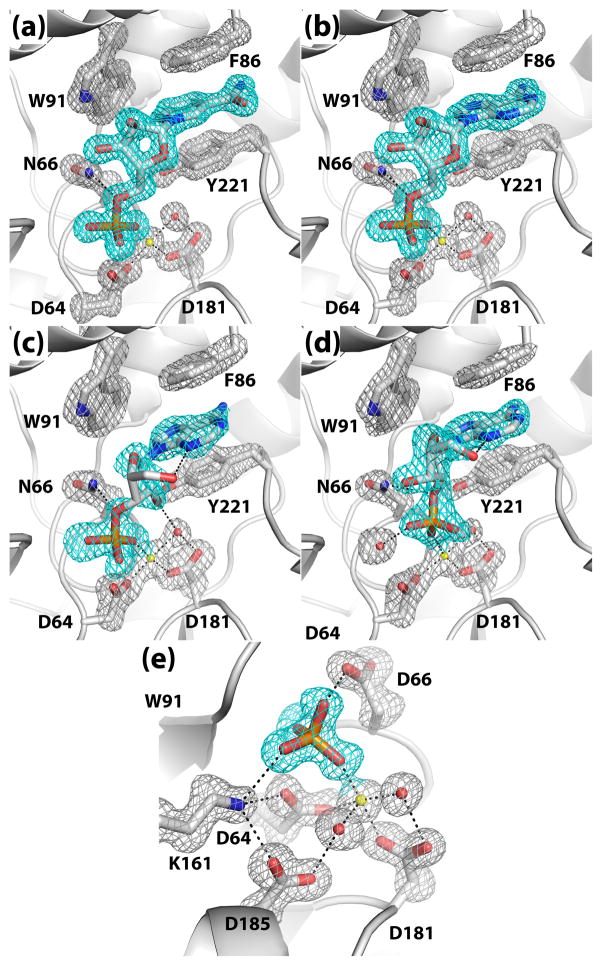
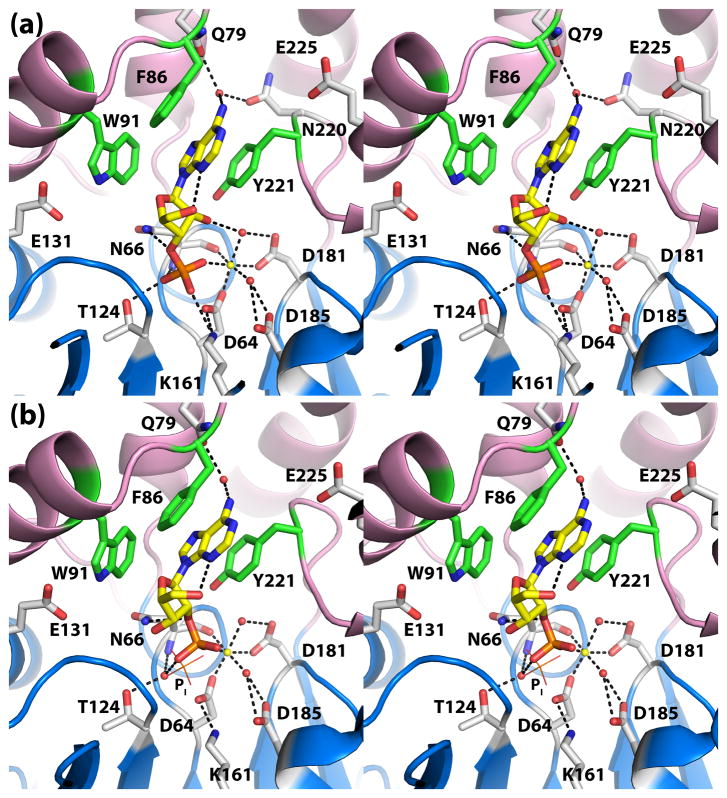
Crystal structures of rP4 complexed with NMN and 5′-AMP were determined to understand how the enzyme recognizes nucleoside 5′-monophosphate substrates. A mutant of rP4 in which the residue that protonates the leaving group, Asp66, has been changed to Asn (D66N) was used for structure determination of enzyme-substrate complexes. Structures of D66N complexed with NMN (Fig. 1) and 5′-AMP were determined at high resolution limits of 1.35 Å and 1.55 Å, respectively (Table 1). The electron density maps at these resolutions allowed unambiguous determination of the conformations of the bound substrates, enumeration of enzyme-substrate interactions, and identification of water molecules involved in substrate binding (Fig. 2a, 2b).
Fig. 2.
Conformations of ligands bound to rP4: (a) NMN, (b) 5′-AMP, (c) 3′-AMP, (d) 2′-AMP, and (e) Pi. In each panel, the cage (cyan for the ligand, silver for protein side chains) represents a simulated annealing σA-weighted Fo - Fc omit map contoured at 3.0 σ. Prior to map calculation, the ligand and surrounding residues and water molecules were removed, and simulated annealing refinement was performed using PHENIX.
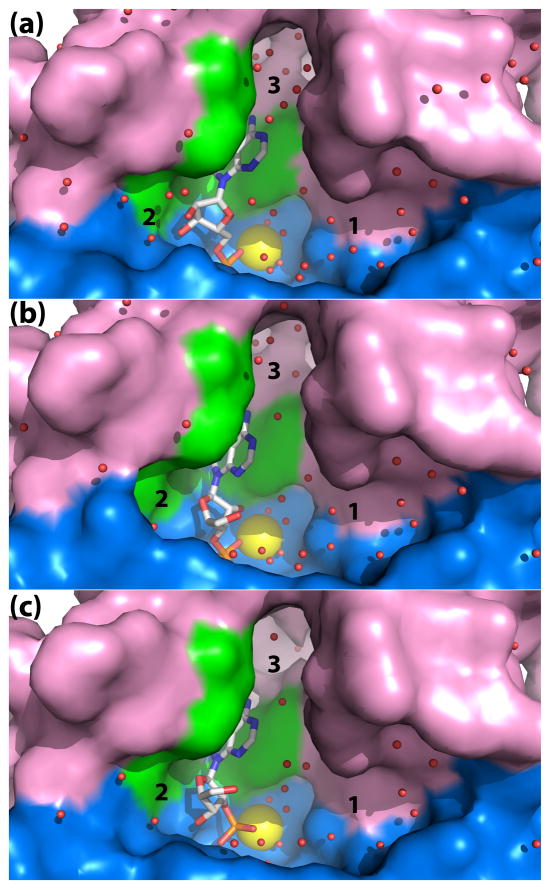
NMN and 5′-AMP bind in an anchor-shaped cavity located in the junction of the α/β core domain and the cap domain, as shown for 5′-AMP in Fig. 3a. NMN and 5′-AMP adopt identical conformations when bound to the enzyme (Fig. 4). Using the nomenclature described by Sanger,22 the ribose in each case displays an unsymmetrical twist with major C3′-endo and minor C2′-exo pucker (3T2). The bases adopt anti orientations about the glycosyl bond.
Fig. 3.
Close-up view of the rP4 active site emphasizing the shape and solvent content. The panels correspond to D66N complexed with (a) 5′-AMP, (b) 3′-AMP, and (c) 2′-AMP. Three solvent-filled pockets are labeled 1, 2, and 3. The core and cap domains are colored blue and pink, respectively, and residues of the aromatic box are colored green. The yellow sphere represents Mg2+.
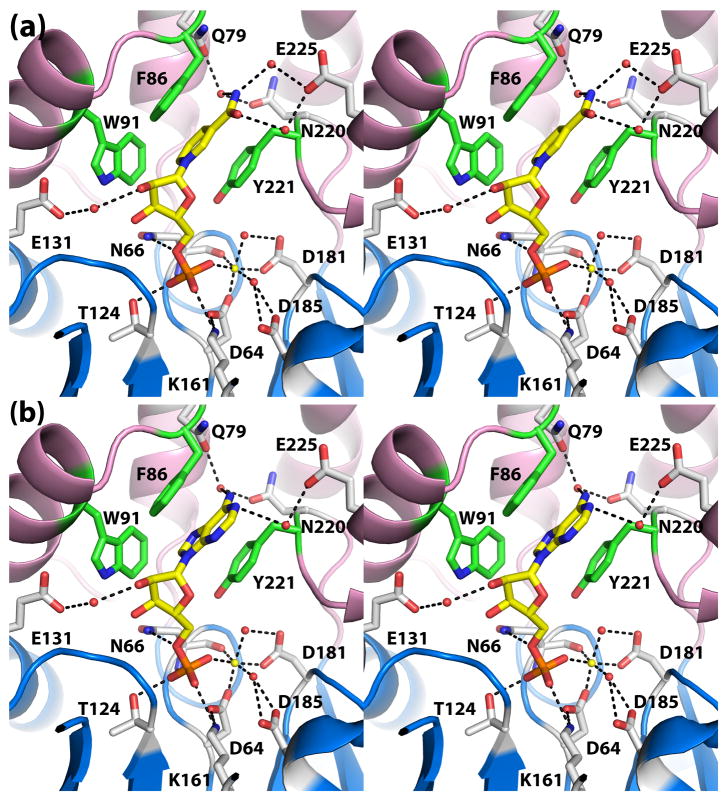
Fig. 4.
Recognition of the nucleoside 5′-monophosphate substrates (a) NMN and (b) 5′-AMP (stereographic views). In both panels, the substrate is represented in yellow sticks, and Mg2+ is depicted as a yellow sphere. Secondary structural elements of the core and cap domains are colored blue and pink, respectively, and residues of the aromatic box are colored green.
The substrate phosphoryl group occupies the well-known phosphoryl binding site of HAD superfamily enzymes (Fig. 4). In both structures, the phosphoryl group interacts with the active site Mg2+, conserved residues Thr124 and Lys161, and the backbone N-H group of Asn66. These interactions are identical to those observed in the structure of rP4 complexed with Pi (Fig. 2e). Furthermore, the substrate phosphoryl sits above the nucleophilic O atom of Asp64 in an orientation suggestive of backside nucleophilic attack. In particular, the nucleophilic O atom is poised 3.0 Å from the P atom, and the angle formed by the nucleophile, P, and O5′ is 179°. Finally, the O atom of the scissile bond forms a hydrogen bond with Asn66 (2.8 Å). This interaction is consistent with Asp66 functioning as the acid that protonates the leaving group.
The ribose moieties of NMN and 5′-AMP form identical interactions with rP4 (Fig. 4). In each structure, the ribose is oriented such that the C2′-C3′ locus contacts the pyrrole ring of Trp91, while the opposite edge of the ring (i.e., O4′) points towards a large solvent-filled pocket on the right hand side of the active site (pocket 1 in Fig. 3A). The hydroxyl groups of the ribose occupy a second, smaller pocket on the left side (pocket 2 in Fig. 3A). Within pocket 2, the 2′-hydroxyl group forms a water-mediated hydrogen bond with Glu131 (Fig. 4).
Finally, the bases of NMN and 5′-AMP bind in an aromatic box formed by Phe86, Trp91, and Tyr221 (Fig. 4). The first two residues are part of the helix-loop-helix substructure of the cap domain, while Tyr221 is located on the loop that follows the last strand of the core domain (Fig. 1). Phe86 and Tyr221 form the sides of the box, while Trp91 forms the floor. In both complexes, the base stacks in parallel between Phe86 and Tyr221 forming an aromatic sandwich. Trp91 contacts the C5 – C6 locus of the NMN nicotinamide and the pyrrole ring of adenine. The hydrogen bonding groups of the bases are directed toward a solvent-filled cavity at the top of the active site (pocket 3 in Fig. 3A). As a result, neither base forms direct hydrogen bonds with the enzyme, but there are water-mediated hydrogen bonds with Gln79, Asn220, and Glu225.
Structures D66N complexed with 3′-AMP and 2′-AMP
Structures of D66N complexed with 3′-AMP and 2′-AMP were determined to understand how rP4 and other CCAPs accommodate nucleoside monophosphate substrates differing in the position of the phosphoryl group on the ribose. Both electron density maps exhibited a strong feature representing the bound substrates. The quality of the 2′-AMP map (Fig. 2d) rivaled those of NMN and 5′AMP. Although the 3′-AMP map exhibited lower quality, the locations of the adenine ring, phosphoryl group, 2′ hydroxyl, and O4′ were unambiguous (Fig. 2c). Density for the C5′-O5′ bond of the ribose was weaker and suggested the same conformation as in 2′-AMP. The weaker density for 3′-AMP also suggested that the occupancy of the ligand is less than 1.0; refinement of the occupancy resulted in a value of 0.81.
The conformations of 2′- and 3′-AMP differ substantially from that of 5′-AMP (Fig. 5). In both 2′-AMP and 3′-AMP, the ribose adopts the C2′-endo pucker (2E), and the base is in a syn orientation. We note that the C2′-endo pucker is favored for syn nucleosides.22 The syn conformation is stabilized by an intramolecular hydrogen bond between N3 and O5′ (2.9 Å in 3′-AMP, 2.8 Å in 2′-AMP). We note that this type of interaction is commonly found in syn nucleotides.22 As observed with 5′-AMP, the bases of 2′-AMP and 3′-AMP bind in the aromatic box with the N6-N7 edge directed toward pocket 3 (Fig. 3b, 3c). However, the ribose rings of 2′-AMP and 3′-AMP are oriented with the O4′ pointing towards the left into pocket 2, which is the reverse of the orientation of the bound 5′ substrates (compare Figs. 3b and c to Fig. 3a).
Fig. 5.
The active sites of D66N complexed with (a) 3′-AMP (b) 2′-AMP (stereographic views). In both panels, the substrate is represented in yellow sticks, and Mg2+ is depicted as a yellow sphere. Secondary structural elements of the core and cap domains are colored blue and pink, respectively, and residues of the aromatic box are colored green. In panel (b), the location of normal phosphoryl binding site is indicated by the phosphate ion shown in lines and labeled Pi.
Finally, there is a difference in the position of the phosphoryl group of 2′-AMP compared to the other substrates (Fig. 5b). The phosphoryl of 2′-AMP is shifted by 1.8 Å from the expected site such that the P atom is 4.3 Å from the nucleophilic O atom of Asp64, which is obviously not optimal for catalysis. As a result of the shift, the phosphoryl group does not form the expected interactions with the conserved residues Thr124 and Lys161.
Kinetic characterization of rP4
The catalytic efficiencies (kcat/Km) of rP4 for certain nucleoside monophosphate substrates were estimated using steady-state kinetic assays. NMN, 5′-AMP, 3′-AMP, and 2′-AMP were used as substrates; the results are summarized in Table 2. Among the substrates tested, the enzyme has the highest efficiency for NMN, followed by 5′-AMP, 3′-AMP, and 2′-AMP. The efficiency for NMN is only 2.5 times that for 5′-AMP, suggesting that rP4 does not exhibit a strong preference for NMN over other nucleoside 5′-monophosphatases. Comparing the data for the adenosine monophosphate substrates, rP4 exhibits only a 2-fold preference for 5′-AMP over 3′-AMP. 2′-AMP is the poorest substrate tested, having a 20-fold lower efficiency than 5′-AMP. Thus, like some other CCAPs, it is reasonable to classify rP4 as a dual 5′-, 3′-nucleotidase.
Table 2.
Kinetic parameters for rP4
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (%)* |
|---|---|---|---|
| NMN | 0.7 ± 0.2 | 0.52 ± 0.05 | 100 |
| 5′-AMP | 0.23 ± 0.04 | 0.070 ± 0.004 | 40 |
| 3′-AMP | 0.8 ± 0.2 | 0.10 ± 0.01 | 20 |
| 2′-AMP | 6 ± 2 | 0.11 ± 0.01 | 2 |
relative to NMN
Discussion
Structural basis of substrate recognition
The main result of our work is to provide structure-based insight into substrate preference and promiscuity of rP4 and CCAPs closely related to rP4. CCAPs are somewhat selective in the sense that the in vivo substrates are thought to be nucleoside monophosphates. For example, genetic and molecular studies suggest that NMN is a biological substrate for e (P4)9. Although the in vivo substrates of other CCAPs have not been similarly identified, the available in vitro kinetic data suggest nucleoside monophosphates as biologically relevant substrates. On the other hand, CCAPs are generally promiscuous with regard to the identity of the base and whether the phosphoryl is attached at the 5′ or 3′ position of the sugar. For example, we have shown here that the catalytic efficiency of rP4 for 5′-AMP is approximately half that of NMN and only twice that of 3′-AMP. Similar results have been reported for other CCAPs.15,17,18
The basis for the preference for nucleoside monophosphates is evident from the D66N complexes. The aromatic box is well suited for binding the aromatic ring systems of nucleotides. In particular the box provides two aromatic residues that stack in parallel with the base forming a sandwich. Furthermore, the span between the aromatic box and phosphoryl binding pocket is optimal for nucleoside monophosphates.
The structures also provide insight into substrate promiscuity with respect to the base. The base is aligned in the aromatic box such that the hydrogen bonding groups point into a solvent-filled pocked. As a result, there are no direct hydrogen bonds with the enzyme implying low base selectivity. Indeed we have shown here that the catalytic efficiency of rP4 for 5′-AMP rivals that with NMN. These results imply that P4 is not tuned to exclusively recognize NMN, and that the enzyme may have other biological functions beyond NAD+ utilization such as acquiring inorganic phosphate from nucleoside monophosphates found in the bacterium’s environment.
The D66N structures also shed light on how rP4 achieves the dual recognition of 5′- and 3′-nucleoside monophosphates, a characteristic of some CCAPs. The 5′ substrates bind with the base in an anti conformation, whereas 3′-AMP adopts a syn conformation. The somewhat higher catalytic efficiency of 5′ substrates likely reflects the lower conformational energy of the anti conformation. The two solvent-filled pockets flanking the ribose appear to be important for the enzyme’s ability to bind both types of substrates. These pockets minimize the enzyme’s direct interactions with the ribose and provide sufficient space to accommodate the different ribose orientations. Indeed, the ribose occupies the widest part of the active site (Fig. 3). Thus, the open active site of rP4 appears to underlie both the weak base specificity and the dual recognition of 5′- and 3′-nucleoside monophosphates.
The structure of D66N complexed with 2′-AMP further demonstrates the ability of the active site to bind different nucleoside monophosphates. The structure is unusual in that the phosphoryl is shifted away from the catalytic Asp into a solvent-filled pocket and is thus not aligned optimally for catalysis. This conformation perhaps represents a non-productive complex. As discussed by Cornish-Bowden, non-productive binding of the substrate is a form of competitive inhibition, and the measured values of Vmax and Km are lower than expected by an unknown and typically immeasurable amount.23 Nevertheless, Vmax/Km does provide a correct measure of the catalytic properties of the enzyme in such cases.23 We found that the apparent catalytic efficiency of rP4 for 2′-AMP is substantially lower (10 – 50 times) than those for the other substrates tested, indicating that it is a poor substrate. Regardless of whether the observed conformation represents non-productive substrate binding, the rP4/2′-AMP structure provides another illustration of how the open active site plays a role in binding different nucleoside monophosphate ligands.
Connections with other CCAPs and class B acid phosphatases
Sequence conservation suggests that the rP4 complexes reported here are representative of other CCAPs. The residues of the phosphate binding pocket (Asp64 Asp66, Lys161, and Thr124) are highly conserved in the HAD superfamily, and as expected, these residues are highly conserved among CCAPs. In fact, Asp64, Asp66, and Lys161 are invariant among the CCAPs, and Thr124 appears as Ser, a conservative substitution, in some CCAPs. Residues of the aromatic box are also highly conserved. Tyr221 appears to be universally conserved among CCAPs. An aromatic ring in the form of Phe, Tyr, Trp, or His is always present at the residue corresponding to rP4 Phe86. Thus, all CCAPs appear to have residues capable of forming an aromatic sandwich with the nucleotide base. Finally, the floor of the box, Trp91, is almost invariant; substitution with Phe, a conservative change, is observed in some CCAP sequences. Thus, we suggest that the structures reported here provide a model for understanding substrate recognition in other CCAPs.
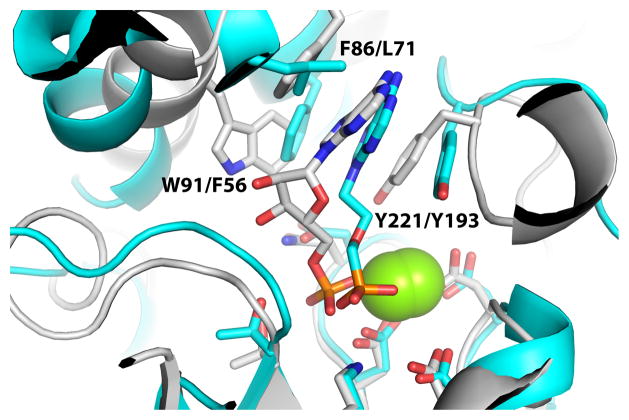
The rP4 structures also reveal a new relationship between CCAPs and the related phosphatases known as class B acid phosphatases (CBAPs). The D66N/5′-AMP complex is reminiscent of the structure of the CBAP AphA complexed with the 5′-AMP analog 9-[(R)-2-(phosphono-methoxy)ethyl]adenine (PMEA, PDB code 2G1A24). CBAPs also belong to the DDDD superfamily, and AphA is regarded as the prototype of the family. CBAPs have a bipartite sequence motif that is similar to that of CCAPs, and like CCAPs, class B enzymes also show a preference for nucleoside 5′- and 3′-monophosphate substrates. Although rP4 and AphA share a common HAD superfamily core domain, their sequences have negligible similarity outside of the bipartite sequence motif (13 % overall identity), and their cap domains have different folds. The latter difference is related to the different quaternary structures of the two enzymes (dimer for CCAPs, tetramer for CBAPs).
Despite the different cap domain structures, AphA and P4 exhibit commonalities in substrate recognition. In particular, the adenine ring of PMEA packs into a hydrophobic pocket that is similar to the aromatic box of rP4 (Fig. 6). The hydrophobic pocket of AphA consists of Leu71, Tyr193, and Phe56, which are analogous to the rP4 aromatic box residues Phe86, Tyr221, and Trp91, respectively. Note also that the adenine rings have nearly identical orientations in the two structures. We thus suggest that class B and C acid phosphatases share a common strategy for nucleotide recognition.
Fig. 6.
Comparison of the active sites of D66N/5′-AMP (white) and AphA/PMEA (cyan). Residue numbers for the aromatic boxes are listed as rP4/AphA. The yellow spheres represent Mg2+.
Materials and Methods
Subcloning and mutagenesis
Previous structural studies of rP4 used a recombinant enzyme lacking a polyhistidine affinity tag, but the purification of that enzyme was inefficient,13 and crystallization was not highly reproducible. Therefore, for this work, an rP4 construct encoding the enzyme fused to a C-terminal hexahistidine tag was created to aid purification. The hel gene was subcloned into pET20b using Nco1 and Xho1 sites such that the N-terminal signal sequence was replaced with the pelB leader sequence from Erwinia chrysanthemi and the N-terminal Cys of the mature protein was replaced with Met. As a result, the rP4 protein used here contains a C-terminal hexahistidine tag, is free of lipid modification, and targeted to the Escherichia coli periplasm.
A site-directed mutant of rP4 in which the residue that protonates the leaving group, Asp66, is changed to Asn (D66N) was created for the purpose of determining crystal structures of enzyme/substrate complexes. We note that an analogous strategy has been used to trap substrate complexes of other phosphatases.25,26 The mutation was introduced into the aforementioned plasmid using the QuickChange kit (Stratagene) and confirmed by DNA sequencing.
Expression and purification of rP4
The rP4-pET20b plasmid was transformed into E. coli BL21AI cells and plated on LB medium containing ampicillin (50 μg/mL). A single colony of the transformant was picked and used to inoculate 1 L of culture. The protein was expressed using auto-induction27 at 37 °C with constant shaking at 300 rpm. The cells were harvested by centrifugation at 3500 rpm for 30 min at 4°C and resuspended in 20 mM phosphate, 20 mM imidazole, and 500 mM NaCl at pH 7.0. The cell pellet was quick frozen in liquid nitrogen and stored at −80°C.
Frozen cells were thawed at 4 °C and ruptured using a French press at 1000 psi. Unbroken cells and cellular debris were removed by centrifugation for 60 min. at 17500 rpm and 4 °C. The supernatant was collected and subjected to a second centrifugation step (30 min, 17500 rpm, 4 °C). The resulting supernatant was used for further purification by immobilized metal ion affinity chromatography (Ni2+-charged HiTRAP, GE Healthcare) followed by cation exchange chromatography (HiTRAP SP, GE Healthcare). The purified enzyme was dialyzed into 50 mM sodium acetate, 50 mM NaCl, 2.5 mM MgCl2 at pH 6.0. The sample was concentrated using a centrifugal ultrafiltration device (10 kDa cutoff) to 2 – 5 mg/mL (based on the bicinchoninic acid assay, Pierce).
Expression and purification of D66N
The D66N mutant plasmid was transformed into E. coli BL21(DE3) cells and plated on LB medium containing ampicillin (50 μg/mL). A single colony of the transformant was picked and used to inoculate a 10 mL starter culture containing ampicillin (50 μg/mL). After overnight growth at 37 °C with 250 rpm shaking, the starter culture was used to inoculate 1 L of LB medium supplemented with ampicillin (50 μg/mL). The culture was then grown at 37 °C with 250 rpm shaking until the optical density at 600 nm reached 0.6. Protein expression was induced by adding isopropyl β-D-thiogalactoside (0.5 mM), and the culture was incubated for 8 hours at 25 °C with 200 rpm shaking. The expressed protein was purified as described above for rP4. After purification, the sample was dialyzed overnight into 50 mM sodium acetate, 50 mM NaCl at pH 6.0 and concentrated to 10 mg/ml.
Crystallization and preparation of enzyme-ligand complexes
Crystallization trials of rP4 and D66N were performed at 20 °C using the sitting drop method with drops formed of 2 μL each of the enzyme and reservoir solutions. Initial crystallization conditions were identified using commercially available screens (Hampton Research). Promising results were obtained with reservoirs containing ammonium citrate and PEG 3350. After optimization, diffraction quality crystals having hexagonal external morphology were grown over reservoirs containing 0.05 – 0.2 M ammonium citrate, 0.05–0.15 mM MgCl2, and 18 – 28 % (w/v) PEG 3350 in the pH range of 6.8 – 7.2. The best rP4 crystals typically grew in 18 – 23 % (w/v) PEG 3350, whereas a higher concentration of 23 – 28 % (w/v) PEG 3350 was used for crystallization of D66N. Typical protein concentrations used for optimal crystal growth were 1 – 3 mg/mL for rP4 and 8 – 10 mg/mL for D66N.
Crystals of the D66N/substrate complexes were obtained by soaking as follows. Stock solutions of the substrates were prepared in water, and the pH was adjusted to 6.0. D66N crystals were cryoprotected in 28 – 30 % PEG 3350, 0.1 M ammonium citrate buffer pH 7.0, and 20% PEG 200. The cryoprotected crystals were transferred to a solution of the cryobuffer supplemented with 5 – 20 mM of a substrate and 100 – 200 mM MgCl2. The soaking time was in the range of 5 – 45 minutes.
Crystals of rP4 complexed with Pi were also obtained by soaking. A stock solution of potassium phosphate (100 mM KH2PO4 at pH 6.0) was first prepared. Next, crystals of rP4 were cryoprotected at room temperature in 23–28 % (w/v) PEG 3350, 0.1 M ammonium citrate buffer pH 7.0, and 20% PEG 200. The cryoprotected crystals were transferred to a solution of the cryobuffer supplemented with 25 mM Pi and 200 mM MgCl2. After 30 minutes the crystals were picked up with Hampton loops and plunged into liquid nitrogen.
Structure determination
X-ray diffraction data sets were collected at the Advanced Light Source beamline 4.2.2 and Advanced Photon Source beamline 24-ID-C (Table 1). The data sets were processed with HKL2000.28 The crystals have space group P6522 with unit cell lengths of a = 98 Å, c = 107 Å, one molecule in the asymmetric unit, 54 % solvent, and Vm of 2.65 Å3/Da.29,30 We note that this form is different from the tetragonal one used in our earlier work.14 Initial phases were estimated using molecular replacement as implemented in PHASER31 with the search model derived from a previously determined rP4 structure (PDB code 3ET414). COOT32 was used for model building, and PHENIX33 was used for refinement. A common set of test reflections (5 %) was used for the refinement calculations. For each structure, the B-factor model using during the initial rounds of refinement consisted of an isotropic B-factor for each non-hydrogen atom and TLS refinement with one TLS group corresponding to the protein chain. Anisotropic B-factors were used during the final few rounds of refinement of the NMN and Pi complexes. The introduction of anisotropic B-factors decreased Rfree by 0.08 for the NMN complex and 0.06 for the Pi complex.
Kinetic characterization
Steady-state enzymatic activity was assessed at 25 °C using a discontinuous assay that measures the production of inorganic phosphate.34,35 The assay buffer consisted of 100 mM sodium acetate, 100 mM NaCl, 1 mM MgCl2 at pH 5.5. For each substrate concentration, the reaction was stopped using the malachite green reagent after reaction times of 15 s, 75 s, 135 s and 195 s, and the citrate color development reagent (34 % (w/v) sodium citrate) was added 60 s after stopping each reaction. After 30 minutes, the inorganic phosphate concentrations were determined spectrophotometrically at 625 nm by reference to a standard curve constructed from solutions of known Pi concentration. The initial rate was estimated by fitting data from the four time points to a line. Apparent values of Km and Vmax were estimated by fitting the initial rate data to the Michaelis-Menten equation using Origin 8 software.
Accession codes
Atomic coordinates and structure factor amplitudes have been deposited in the PDB36 with accession codes 3OCU (NMN), 3OCV (5′-AMP), 3OCW (3′-AMP), 3OCX (2′-AMP), and 3OCY (Pi).
Acknowledgments
We thank Dr. Jay Nix of ALS beamline 4.2.2 for help with data collection. This research was supported by the NIH grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research (MRCE) and the University of Missouri Research Board. H.S. was supported by a pre-doctoral fellowship from National Institutes of Health grant DK071510 and a Chancellor’s Dissertation Completion Fellowship from the University of Missouri-Columbia. Part of this research was performed at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Part of this work is based upon research conducted at the Northeastern Collaborative Access Team beam lines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38.
Abbreviations used
- rP4
recombinant Haemophilus influenzae e (P4) class C acid phosphatase
- CCAP
class C acid phosphatase
- CBAP
class B acid phosphatase
- PDB
Protein Data Bank
- NMN
nicotinamide mononucleotide
- NR
nicotinamide riboside
- 5′-AMP
adenosine 5′-monophosphate
- 3′-AMP
adenosine 3′-monophosphate
- 2′-AMP
adenosine 2′-monophosphate
- Pi
inorganic phosphate
- D66N
mutant of rP4 in which Asp66 is replaced by Asn
- OlpA
class C acid phosphatase from Elizabethkingia meningoseptica
- LppC
class C acid phosphatase from Streptococcus equisimilis
- HppA
class C acid phosphatase from Helicobacter pylori
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reilly TJ, Chance DL, Smith AL. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J Bacteriol. 1999;181:6797–6805. doi: 10.1128/jb.181.21.6797-6805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reilly TJ, Smith AL. Purification and characterization of a recombinant Haemophilus influenzae outer membrane phosphomonoesterase e (P4) Protein Expr Purif. 1999;17:401–409. doi: 10.1006/prep.1999.1157. [DOI] [PubMed] [Google Scholar]
- 3.Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J. 2009;28:43–8. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- 5.Green BA, Baranyi E, Reilly TJ, Smith AL, Zlotnick GW. Certain site-directed, nonenzymatically active mutants of the Haemophilus influenzae P4 lipoprotein are able to elicit bactericidal antibodies. Infect Immun. 2005;73:4454–4457. doi: 10.1128/IAI.73.7.4454-4457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason KW, Zhu D, Scheuer CA, McMichael JC, Zlotnick GW, Green BA. Reduction of nasal colonization of nontypeable Haemophilus influenzae following intranasal immunization with rLP4/rLP6/UspA2 proteins combined with aqueous formulation of RC529. Vaccine. 2004;22:3449–56. doi: 10.1016/j.vaccine.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 7.Hotomi M, Ikeda Y, Suzumoto M, Yamauchi K, Green BA, Zlotnick G, Billal DS, Shimada J, Fujihara K, Yamanaka N. A recombinant P4 protein of Haemophilus influenzae induces specific immune responses biologically active against nasopharyngeal colonization in mice after intranasal immunization. Vaccine. 2005;23:1294–300. doi: 10.1016/j.vaccine.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 8.Kemmer G, Reilly TJ, Schmidt-Brauns J, Zlotnik GW, Green BA, Fiske MJ, Herbert M, Kraiss A, Schlor S, Smith A, Reidl J. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J Bacteriol. 2001;183:3974–3981. doi: 10.1128/JB.183.13.3974-3981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlach G, Reidl J. NAD+ utilization in Pasteurellaceae: simplification of a complex pathway. J Bacteriol. 2006;188:6719–27. doi: 10.1128/JB.00432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Kurnasov OV, Chen B, Robinson H, Grishin NV, Osterman AL, Zhang H. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J Biol Chem. 2002;277:33291–9. doi: 10.1074/jbc.M204368200. [DOI] [PubMed] [Google Scholar]
- 11.Thaller MC, Schippa S, Rossolini GM. Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 1998;7:1647–52. doi: 10.1002/pro.5560070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reilly TJ, Green BA, Zlotnick GW, Smith AL. Contribution of the DDDD motif of H. influenzae e (P4) to phosphomonoesterase activity and heme transport. FEBS Lett. 2001;494:19–23. doi: 10.1016/s0014-5793(01)02294-3. [DOI] [PubMed] [Google Scholar]
- 13.Ou Z, Felts RL, Reilly TJ, Nix JC, Tanner JJ. Crystallization of recombinant Haemophilus influenzae e (P4) acid phosphatase. Acta Cryst. 2006;F62:464–6. doi: 10.1107/S1744309106012358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felts RL, Ou Z, Reilly TJ, Tanner JJ. Structure of Recombinant Haemophilus Influenzae e (P4) Acid Phosphatase Reveals a New Member of the Haloacid Dehalogenase Superfamily. Biochemistry. 2007;46:11110–9. doi: 10.1021/bi701016m. [DOI] [PubMed] [Google Scholar]
- 15.Passariello C, Schippa S, Iori P, Berlutti F, Thaller MC, Rossolini GM. The molecular class C acid phosphatase of Chryseobacterium meningosepticum (OlpA) is a broad-spectrum nucleotidase with preferential activity on 5′-nucleotides. Biochim Biophys Acta. 2003;1648:203–9. doi: 10.1016/s1570-9639(03)00147-x. [DOI] [PubMed] [Google Scholar]
- 16.Malke H. Cytoplasmic membrane lipoprotein LppC of Streptococcus equisimilis functions as an acid phosphatase. Appl Environ Microbiol. 1998;64:2439–42. doi: 10.1128/aem.64.7.2439-2442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reilly TJ, Calcutt MJ. The class C acid phosphatase of Helicobacter pylori is a 5′ nucleotidase. Protein Expr Purif. 2004;33:48–56. doi: 10.1016/j.pep.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Reilly TJ, Chance DL, Calcutt MJ, Tanner JJ, Felts RL, Waller SC, Henzl MT, Mawhinney TP, Ganjam IK, Fales WH. Characterization of a unique class C acid phosphatase from Clostridium perfringens. Appl Environ Microbiol. 2009;75:3745–54. doi: 10.1128/AEM.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Ohtani K, Wang Y, Yuan Y, Hassan S, Shimizu T. Genetic and biochemical analysis of a class C non-specific acid phosphatase (NSAP) of Clostridium perfringens. Microbiology. 2010;156:167–73. doi: 10.1099/mic.0.030395-0. [DOI] [PubMed] [Google Scholar]
- 20.Malke H, Steiner K. XIV Lancefield International Symposium on Streptococci and Streptococcal Diseases. Aukland New Zealand: 1999. [Google Scholar]
- 21.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Saenger W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 23.Cornish-Bowden A. Fundamentals of enzyme kinetics. Butterworths; London: 1979. [Google Scholar]
- 24.Leone R, Cappelletti E, Benvenuti M, Lentini G, Thaller MC, Mangani S. Structural insights into the catalytic mechanism of the bacterial class B phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases. J Mol Biol. 2008;384:478–88. doi: 10.1016/j.jmb.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 25.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–5. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh H, Felts RL, Schuermann JP, Reilly TJ, Tanner JJ. Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition. J Mol Biol. 2009;394:893–904. doi: 10.1016/j.jmb.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 30.Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–71. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 35.Carter SG, Karl DW. Inorganic phosphate assay with malachite green: an improvement and evaluation. J Biochem Biophys Methods. 1982;7:7–13. doi: 10.1016/0165-022x(82)90031-8. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engh RA, Huber R. Accurate bond and angle parameters for x-ray protein structure refinement. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 38.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]