Abstract
Background
Maternally inherited endosymbionts like Wolbachia pipientis are in linkage disequilibrium with the mtDNA of their hosts. Therefore, they can induce selective sweeps, decreasing genetic diversity over many generations. This sex ratio distorter, that is involved in the origin of parthenogenesis and other reproductive alterations, infects the parthenogenetic weevil Naupactus cervinus, a serious pest of ornamental and fruit plants.
Results
Molecular evolution analyses of mitochondrial (COI) and nuclear (ITS1) sequences from 309 individuals of Naupactus cervinus sampled over a broad range of its geographical distribution were carried out. Our results demonstrate lack of recombination in the nuclear fragment, non-random association between nuclear and mitochondrial genomes and the consequent coevolution of both genomes, being an indirect evidence of apomixis. This weevil is infected by a single Wolbachia strain, which could have caused a moderate bottleneck in the invaded population which survived the initial infection.
Conclusions
Clonal reproduction and Wolbachia infection induce the coevolution of bacterial, mitochondrial and nuclear genomes. The time elapsed since the Wolbachia invasion would have erased the traces of the demographic crash in the mtDNA, being the nuclear genome the only one that retained the signal of the bottleneck. The amount of genetic change accumulated in the mtDNA and the high prevalence of Wolbachia in all populations of N. cervinus agree with the hypothesis of an ancient infection. Wolbachia probably had great influence in shaping the genetic diversity of N. cervinus. However, it would have not caused the extinction of males, since sexual and asexual infected lineages coexisted until recent times.
Background
Parthenogenetic reproduction is fairly common in Curculionidae [1,2]. Many cases have been reported in three different subfamilies: Scolytinae (bark beetles), Listroderinae and Entiminae (broad-nose weevils), especially in species from the Old World (e.g. [1,3-6]). Although less studied, weevils from South America also concentrate a huge number of asexual species. For instance, Lanteri and Normark [7] provided a list of more than 30 parthenogenetic or presumptive parthenogenetic species within the tribe Naupactini (subfamily Entiminae). There is no direct evidence of parthenogenesis for the majority of these species. However, Floyd F. Smith and colleagues from the United States Department of Agriculture (USDA) proved this reproductive mode through rearing experiments for the species Naupactus cervinus and N. leucoloma respectively (see [8]), and Marvaldi [9] did the same with N. ruizi.
Parthenogenesis in Curculionidae is thelytokous and apomictic (i.e. ameiotic) [2]. Apomixis was confirmed for several Old World broad-nosed weevils [3,10], although nothing is known for South American species. As a consequence of this kind of reproduction, progeny would result in a group of females genetically identical to their mothers [11]. Nevertheless, the absence of recombination would originate heterozygote genotypes, as rare mutations accumulate [12,13]. The lack of meiosis could be detrimental in the short term [14-16], although heterozygosity could explain the higher dispersion and adaptation ability of some parthenogenetic species over the sexual ones in the long term [1].
Traditionally, hybridization and polyploidy were the main traits invoked to explain the origin of asexuality in weevils [2,17]. However, the report of the parthenogenesis inductor bacterium Wolbachia pipientis (hereafter "Wolbachia") in several species of the tribe Naupactini [18,19] and also in the genera Cathormiocerus (Entiminae, Trachyphloeini) [20] and Otiorhynchus (Entiminae, Otiorhynchini) [21], suggested another possible explanation for the origin of this reproductive mode in broad-nosed weevils.
Wolbachia infection can produce drastic consequences on the evolution of its host species, such as extinction or sex role reversal [22,23]. However, it can also affect the population genetics and molecular evolution of the vertical transmitted genomes (mitochondrial, and for apomictic species also nuclear DNA), because of the linkage disequilibrium among these cotransmitted molecules [24-26].
When an arthropod population is invaded by a Wolbachia strain that rapidly spreads, the host mtDNA (and perhaps the nDNA) associated to the initial infection (i.e. in linkage disequilibrium with the bacterial genome) will hitchhike through the population to fixation [26,27]. Consequently, this selective sweep ("indirect selection on the mtDNA") will lead to a loss of mtDNA/nDNA diversity in part of the host species distribution. Since the expected coalescent time of mtDNA in an uninfected species is twice the effective population size of females (Nf), a decrease in mtDNA/nDNA diversity will be evident for ca. 2Nf generations after the Wolbachia invasion [25]. Then, current levels of genetic diversity in infected arthropod populations should be analyzed in the context of the infection age.
The spread and prevalence of the infection can also be related to other important factors. For instance, the success of the infected females in producing more daughters than uninfected females [28]. If infected females have a higher fitness than unifected ones, a perfect transmission of the endosymbiont will lead to a high prevalence or to its fixation [23].
From the considerations stated before, we formulate the following hypotheses:
i) If parthenogenesis is apomictic, the lack of meiosis precludes the occurrence of recombination;
ii) Then, asexuality can generate linkage disequilibrium among mitochondrial and nuclear genomes;
iii) Vertical transmission of Wolbachia can generate linkage disequilibrium among mitochondrial, nuclear and bacterial genomes;
iv) Infection with Wolbachia can sweep the genetic variation on both mitochondrial and nuclear DNAs of the hosts;
v) Infection age correlates with recovery of genetic diversity of host genomes and also with the high prevalence of Wolbachia in its populations.
To test these hypotheses we carried out a study on the genetic diversity of the species Naupactus cervinus, as part of a broader research project on the evolution of parthenogenesis in South American weevils of the tribe Naupactini. This pest insect, commonly known as the Fuller's rose weevil, is very attractive for studying the evolution of asexuality, because it is worldwide distributed and it is very abundant in nature. Although probably native to Northeastern Argentina, Southern Brazil and Uruguay, in currently occurs in Australia, Azores Islands, Canary Islands, Chile, France, Italy, Japan, Mexico, Morocco, New Zealand, Spain, USA, etc. as a consequence of commercial trade of several crops, especially ornamental and fruit plants [29-31].
Naupactus cervinus reproduces by parthenogenesis, although some sexual lineages have been recorded from Northeastern Argentina and Southern Brazil in the 1940's (see [32]). Rodriguero et al. [19] and Rodriguero [33] reported that all asexual lineages of this species are infected with the Wolbachia strain wNau5, which belongs to the supergroup B.
The main goal of this contribution is to understand the conquences of Wolbachia infection and asexuality on the genomes of N. cervinus through a mito-nuclear genetic comparison. To test the hypotheses previously formulated, particularly the occurrence of apomixis, we will quantify the minimum number of recombination events and the linkage disequilibrium between both genomes. Additionally, the estimation of the number of Wolbachia strains, their prevalence and the infection age, will bring insights on the influence of this endosymbiont on both host genomes, and will contribute to explain the distribution of their genetic variation.
Results
Genetic variation estimates
Three hundred and nine individuals from 38 different locations were screened for genetic variation in a 748 bp fragment of the COI gene. All of them were females. Seventeen mitochondrial haplotypes (arbitrarily named A-R [GenBank: GQ406827 - GQ406843]) and 25 segregating sites were identified, of which only three were singletons. Some haplotypes were found in multiple locations (e.g. haplotype B, Table 1), while others occurred at a single site (e.g. haplotypes A, D, E, G, H, J, L, N, P, Table 1).
Table 1.
Geographic distribution and genetic diversity of Naupactus cervinus samples
| Sampling location | Acronym | Lat/Long | N | Infection Status | mtDNA haplotypes | nDNA haplotypes | Multilocus genotypes |
|---|---|---|---|---|---|---|---|
| Alegrete (BR) | Al | 29° 46'S, 55° 47'W | 5 | √ | F | VI VIII * | 1? *, 1 ? VIII, 3 F VI |
| Brazo Largo(AR) | BL | 33° 54'S, 58° 53'W | 11 | √ | C M | VII * | 4 C *, 7 M VII |
| Bozzano (BR) | Bo | 28° 35'S, 53° 59'W | 4 | √ | C Q | II | 1 C II, 3 Q II |
| Buenos Aires (AR) | BA | 34° 36' S, 58° 26' W | 5 | √ | B F H | VI | 1 B ?, 3 F VI, 1 H VI, |
| Cardales (AR) | Ca | 34° 18'S, 58° 57' W | 5 | √ | B G | VI VII | 3 B VII, 2 G VI |
| Cerro Azul (AR) | CA | 27° 38' S, 55° 30' W | 10 | √ | Q | IV | 10 Q IV |
| Chajarí (AR) | Chj | 30° 47' S, 57° 59' W | 6 | √ | F | VI VII | 5 F VI, 1 F VII |
| El Palmar (AR) | EP | 31° 50' S, 58° 17'W | 14 | √ | F | VI | 14 F VI |
| Easter Island (CH) | IP | 27° 08' S, 109° 26' W | 7 | √ | I | --- | 7 I ? |
| French Polynesia | PF | 23° 08' S, 134° 58' W | 1 | √ | B | VII | 1 B VII |
| Godoy Cruz (AR) | GC | 32° 56' S, 68° 50' W | 3 | √ | A | VII | 3 A VII |
| Gualeguaychú (AR) | Gu | 33° 01' S, 58° 31' W | 18 | √ | F M | VI VII | 13 F VI, 5 M VII |
| Ijui (BR) | Ij | 28° 23' S, 53° 54' W | 3 | √ | C R | * | 2 C *, 1 R * |
| Itaára (BR) | It | 29° 36' S, 53° 45' W | 4 | √ | C | VIII * | 1 C VIII, 3 C * |
| Jari (BR) | Ja | 29° 17' S, 54° 13' W | 3 | √ | C | * | 3 C * |
| La Falda (AR) | LF | 31° 05' S, 64° 29' W | 13 | √ | B | VII | 13 B VII |
| Laranjeiras do Sul (BR) | LS | 25° 24' S, 52° 24' W | 2 | √ | R | * | 2 R * |
| Libertad (UR) | Li | 34° 37' S, 56° 37' W | 2 | √ | B | VI | 2 B VI |
| Mendoza (AR) | Me | 33° 30' S, 69° 00' W | 3 | √ | B | VII | 3 B VII |
| Oberá (AR) | Ob | 27° 29' S, 55° 08' W | 2 | √ | Q R | III | 1 Q III, 1 R ? |
| Otamendi Res. (AR) | RO | 34° 14' S, 58° 52' W | 21 | √ | N | VII | 21 N VII |
| P. P. Pereyra Iraola (AR) | PI | 34° 50' S, 58° 8' W | 13 | √ | B D F | VI VII | 1 B VII, 9 D VII, 3 F VI |
| Pergamino (AR) | Pe | 33° 54' S, 60° 35' W | 2 | √ | B | VII | 2 B VII |
| Ponta Grossa (BR) | PG | 25° 05' S, 50° 09' W | 4 | √ | C F R | I VI * | 1 C *, 2 F VI, 1 R I |
| Río Cuarto (AR) | RC | 33° 08' S, 64° 21' W | 5 | √ | A B | VII | 3 A VII, 2 B VII |
| Salto Grande (AR) | SG | 31° 23' S, 58° 01' W | 12 | √ | F | VI | 12 F VI |
| Santa Maria (BR) | SM | 29° 40' S, 53° 47' W | 10 | √ | Q | IV | 10 Q IV |
| Santiago de Chile (CH) | SC | 33°26' S, 70°29' W | 8 | √ | B | VII | 8 B VII |
| Sao Sepé (BR) | SS | 30° 10' S, 53° 34' W | 11 | √ | P | IV | 11 P IV |
| Tahiti | Th | 17° 52'S, 149° 56'W | 3 | √ | B | VII | 3 B VII |
| Talavera Island (AR) | IT | 34° 10' S, 58° 30' W | 19 | √ | B F K M | VI VII | 2 B VII, 6 F VI, 1 K VII, 10 M VII |
| Tandil(AR) | Ta | 37° 19' S, 59° 08' W | 18 | √ | B F L | V VI | 5 B V, 10 F VI, 3 L ? |
| Tenerife (SP) | Te | 27° 27' N, 16° 14' W | 5 | √ | B | VII | 5 B VII |
| Toledo (BR) | To | 24° 42' S, 53° 44' W | 5 | √ | C | * | 5 C * |
| Tres Lomas (AR) | TL | 36° 28' S, 62° 52' W | 8 | √ | B | VII | 8 B VII |
| Valencia (SP) | Val | 39° 29' N, 00° 23' W | 12 | √ | B | VII | 12 B VII |
| Vallenar (CH) | Var | 28° 57' S, 71° 15' W | 11 | √ | I J | VII | 4 I VII, 7 J VII |
| Yapeyú (AR) | Ya | 29° 28' S, 56° 50' W | 4 | √ | C E | * | 3 C *, 1 E * |
| Zárate (AR) | Za | 34° 06' S, 59° 01' W | 20 | √ | B K | VII | 1 B VII, 19 K VII |
Sampling sites of Naupactus cervinus. Acronyms, latitude, longitude, sampling size, Wolbachia infection status, mitochondrial and nuclear haplotypes and multilocus genotypes are specified for each location. The asterisk indicates ITS1 "double string" sequences. Whenever one of the haplotypes could not be obtained, it was indicated through a question mark.
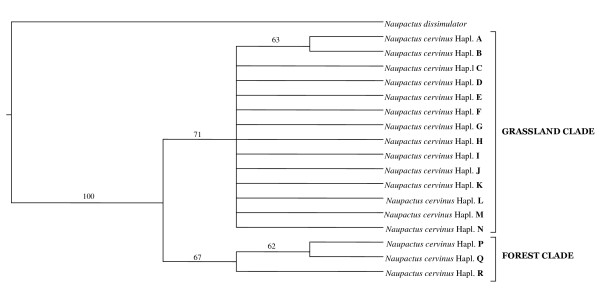
Alignment of the translated COI sequences showed a distribution of the genetic variation similar to that reported by Lunt et al. [34]. The absence of stop codons and mutations that alter the reading frame excluded numt amplifications. The total proportions of nucleotides were 31.8% A, 17.0% C, 15.7% G and 35.6% T, with a strong A + T bias (67.4%), which is higher in the third codon position. Estimates of genetic variation for the total sample were θπ = 0.007 ± 0.001, θW = 0.007 ± 0.002 and Hd = 0.844 ± 0.011. Maximum parsimony search yielded 13 most parsimonious trees 113 steps long. A strict consensus tree is shown, where two main clades have been recovered (Figure 1). One of these clades includes mitochondrial haplotypes from forests ("forest clade"), and the other one mitochondrial haplotypes from open vegetation areas ("grassland clade"), except "F", "G" and "H" that come from a transition zone along the Uruguay River (Figure 1, Table 1). The most parsimonious trees differentiate in minor changes in the relationships among haplotypes within the "grassland clade", yielding a highly unresolved consensus tree with several branches of zero length (i.e. a large polytomy).
Figure 1.
Mitochondrial diversity. Strict consensus of 13 most parsimoniuos trees of COI haplotypes. Numbers above the branches are 50% or higher bootstrap values.
Sequencing of ITS1 in 282 individuals yielded a fragment of ca. 1,100 bp, 155 bp of it belonging to the highly conserved 18 S rDNA region, and the remaining to the ITS1 region. Most specimens analyzed were a subsample of those sequenced for COI. Twenty three of these individuals showed chromatograms with double peaks ("double string sequences") (Table 1). This finding was indicative of at least two simultaneously amplified sequences from the same individual.
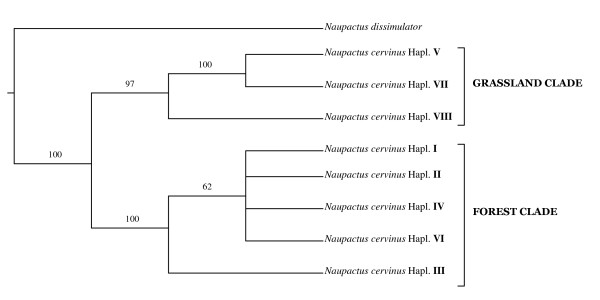
Forty-four segregating sites were identified for ITS1 (only four singletons) yielding eight different nuclear haplotypes (arbitrarily named I-VIII [GenBank: GQ406818 - GQ406825]). Unlike COI, insertion/deletion events were frequent in this dataset. However, based on a gap insertion:substitution cost ratio 10:1, primary homologies could be unambiguously established. Nuclear haplotype distribution depicted a pattern similar to COI (e.g. haplotypes VI and VII are widely distributed, and haplotypes I, II and III are restricted to a single site, Table 1). The following values of genetic variability were obtained: θπ = 0.026 ± 0.001, θW = 0.013 ± 0.002 and Hd = 0.574 ± 0.028. The Hd value was remarkably lower than that obtained for COI, in agreement with the minor number of ITS1 haplotypes. Maximum parsimony search yielded one most parsimonious tree 153 steps long (Figure 2). In agreement with the mitochondrial dataset, two divergent clades were recovered: one from open vegetation areas ("grassland clade") and the other from forests ("forest clade"), although the later includes a single haplotype (VI) from the transition zone previously mentioned.
Figure 2.
Nuclear diversity. Cladogram of ITS1 haplotypes. Numbers above the branches are 50% or higher bootstrap values.
Linkage Disequilibrium Analysis
Direct analysis of multilocus genotypes (COI and ITS1) in 282 individuals shows that in most cases, nuclear and mitochondrial genotypes cosegregate (Table 1). In fact, ten out of 17 mitochondrial haplotypes (A, D, G, H, I, J, K, M, N and P) from 104 individuals are exclusively associated with a single nuclear haplotype. For instance, 20 individuals bearing the mitochondrial haplotype "K" always carry the nuclear haplotype "VII", 11 individuals with "P" always carry "IV", 22 individuals with "M" always carry "VII", and so on (see Table 1). Additionally, the most frequent haplotypes "B" and "F" are linked with nuclear haplotypes "VII" and "VI" respectively, in 118 out of 126 individuals (Table 1).
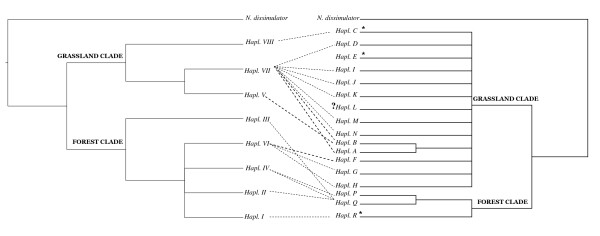
Moreover, the analysis of congruence between COI and ITS1 gene trees reveals signatures of coevolution between both genomes. For instance, eight mitochondrial haplotypes belonging to the "grassland clade" ("A", "B", "D", "I", "J", "K", "M" and "N") share the grassland nuclear haplotype "VII" (Figure 3). The remaining haplotypes from this same clade "F", "G" and "H, which occur in a transition zone between forests and grasslands (Table 1), are linked to the nuclear haplotype "VI". More interestingly, within the mitochondrial "forest clade" the sister haplotypes "P" and "Q" share the forest nuclear haplotype "IV", and the "R" haplotype bears the nuclear genotype "I", phylogenetically related to "IV" (Figure 3). The inverse association was also observed, since some related nuclear haplotypes are linked to the same mitochondrial haplotype. For example "II", "III" and "IV" are linked with "Q", whereas "V" and "VII" are associated with "B" (Figure 3).
Figure 3.
Phylogenetic congruence. Congruent phylogenetic relationships between mitochondrial and nuclear haplotypes of N. cervinus.
Only four individuals show traces of recombination. They are those carrying the haplotype combinations "B-VI" (N = 2), "C-II" (N = 1) and "F-VII" (N = 1). Moroever, ITS1 double strings sequences were found in three mitotypes ("C", "E" and "R"), a fact that could be the consequence of hybridization events.
The hypothesis of mito-nuclear coevolution is also supported by the result of the statistical assessment of the genetic linkage between both genomes. In fact, the 1,403 comparisons of nuclear vs. mitochondrial sites yielded 772 significant values of linkage disequilibrium (see Additional File 1, Table S1). After Bonferroni's correction, the number of significant associations decreased to 606. This led to the rejection of the null hypothesis of linkage equilibrium between the nuclear and mitochondrial fragments herein sequenced.
Analysis of recombination in nuclear sequences
The estimation of the recombination parameter according to RM indicates lack of recombination in the nuclear sequence under study (RM = 0, p = 0). Although ITS1 is a small fragment in comparison with the whole nuclear genome, this result may be regarded as an indirect evidence of apomixis. Then, the null hypothesis of apomixis cannot be rejected.
Selective sweeps
The MK test, based on polymorphism and divergence, showed a significant deficit of fixed non-synonymous differences in COI (G = 8.199; p < 0.01). Then, purifying or negative selection penalizes the non-synonymous substitutions between N. cervinus and N. dissimulator. The NI and the α parameter support this conclusion (NI = 9.474; α = -8.474). Tajima's DT and Fu and Li's F were nonsignificant (DT = 0.183, p > 0.05 and F = 1.151, p > 0.10), suggesting that genetic variation of COI is under selective neutrality or in drift-mutation equilibrium.
The models selected by MrModelTest were GTR + G [35,36] for the mitochondrial dataset, and HKY85 + I [37,38] for the nuclear dataset. Bayesian phylogenetic analyses converged after 100,000 generations, based on the inspection of the burn-in plot of log-likelihood scores, tree lengths, all model parameters, and the analysis of cumulative posterior probabilities and the standard deviation of split frequencies. Therefore, the first 250 samples from each analysis were discarded, resulting in two posterior distributions containing 750 samples each. A plot of the posterior probabilities of all splits from the two separate MCMCMC runs demonstrated a linear relationship, suggesting that these analyses were not restricted to local optima. The ITS1 sites were optimized onto this phylogram. The likelihood ratio tests obtained were LRT1 = 1.001 (p = 0.606) for test 1, and LRT2 = 2.001 (p = 0.317) for test 2. Therefore, the null hypothesis of no positive selection could not be rejected for the nuclear dataset. In fact, out of the 838 nuclear sites examined, 837 were under negative selection (0.99 >p > 0.50) and only one was under positive selection (p > 0.99) (data not shown). On the other hand, Tajima's DT and Fu and Li's F were significantly positive for ITS1 (DT = 2.979, p < 0.01 and F = 2.193, p < 0.02), not accounting for a selective sweep.
Demographic analysis
ModelTest selected the GTR model for ITS1 [35]. The values of the parameters and confidence intervals (α = 0.05) were: g = -1.748 [(-4.950)-(-0.289)] and θ = 0.004 [0.002-0.008]. The sign of g indicates that N. cervinus most likely passed through a bottleneck (i.e. a decline in population size). In turn, the equation of g implies that θ was higher in the past.
Prevalence and diversity of Wolbachia in Naupactus cervinus
A total of 247 individuals of N. cervinus from different locations were screened by PCR assay using Wolbachia 16 S rDNA gene-specific primers. All of them were positive for Wolbachia infection, as it was demonstrated by the amplification of a product of approximately 800 bp. This fragment size was similar to that of the positive control. No PCR band was obtained from the negative control.
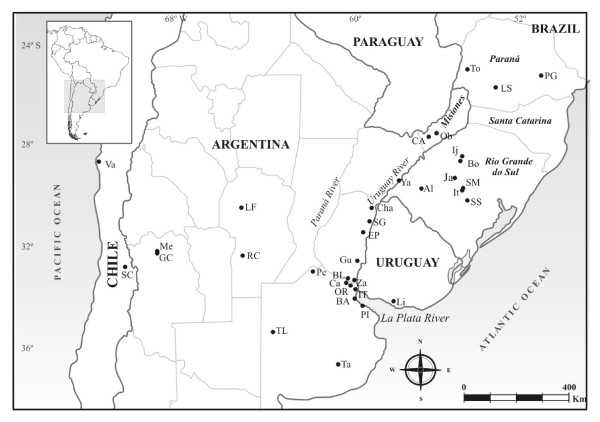
To investigate multiple infections within N. cervinus, we focused on 16 sampling sites, including weevils from both the "forest" and the "grassland" clades (Brazo Largo, Cerro Azul, Chajarí, El Palmar, Isla Talavera, Oberá, Pergamino, Río Cuarto, Salto Grande, Tandil and Yapeyú from Argentina, and Alegrete, Laranjeiras do Sul, Santa Maria, São Sepé and Toledo from Brazil, Figure 4). Sixteen randomly chosen gene fragments wsp, coxA and fbpA were amplified (one from every sampling site) and sequenced. All the individuals assayed yielded identical gene sequences (wsp: ca. 580 bp in length [GenBank: GQ402145]; coxA: ca. 400 bp in length [GenBank: GU079631.2]; fbpA ca. 430 bp in length [GenBank: GU079632.1]). Based on these results, we concluded that only one Wolbachia strain infects this weevil species. Therefore, although multiple infections could have occurred, only one would have succeded.
Figure 4.
Geographic distribution of Naupactus cervinus. Sampling locations of Naupactus cervinus. Countries included in this study are indicated in capital letter.
Divergence time estimation
The age of the demographic event (i.e. moderate bottleneck) was calculated through a molecular clock, using both the nuclear and the mitochondrial datasets.
Table 2 shows a matrix of uncorrected genetic distances among all the nuclear haplotypes (I-VIII). The relative rate tests indicate that all these nDNA lineages may have split at a constant substitution rate (p > 0.10 for all comparisons). The average pairwise divergence among them was D = 0.03198, so the divergence time for this dataset would be ca. 1.9 Myrs.
Table 2.
Nuclear divergence
| I | II | III | IV | V | VI | VII | VII | |
|---|---|---|---|---|---|---|---|---|
| I | --- | |||||||
| II | 0.05170 | --- | ||||||
| III | 0.05296 | 0.00883 | --- | |||||
| IV | 0.04918 | 0.00504 | 0.00378 | --- | ||||
| V | 0.05044 | 0.00631 | 0.00504 | 0.00126 | --- | |||
| VI | 0.05044 | 0.00631 | 0.00504 | 0.00126 | 0.00252 | --- | ||
| VII | 0.03657 | 0.05296 | 0.05422 | 0.05044 | 0.05170 | 0.05170 | --- | |
| VIII | 0.03657 | 0.05296 | 0.05422 | 0.05044 | 0.05170 | 0.05170 | 0.00000 | --- |
Uncorrected genetic distances among nuclear haplotypes of N. cervinus. The values with constant substitution rate, as verified by the relative-rate test, are in bold (p > 0.10).
Table 3 shows a matrix of uncorrected genetic distances among all the mitochondrial haplotypes (A-R). As in the former case, the relative rate test indicate that all these mtDNA lineages may have splitted at a constant substitution rate (p > 0.10 for all comparisons). Divergence time among all mitochondrial haplotypes is more recent than that obtained for the nuclear dataset. Based on an average distance D = 0.00962, it was 400,000 years. The difference between both markers will be discussed later.
Table 3.
Mitochondrial divergence
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | P | Q | R | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | --- | ||||||||||||||||
| B | 0.00000 | --- | |||||||||||||||
| C | 0.00174 | 0.00174 | --- | ||||||||||||||
| D | 0.00348 | 0.00348 | 0.00174 | --- | |||||||||||||
| E | 0.00348 | 0.00348 | 0.00174 | 0.00348 | --- | ||||||||||||
| F | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00348 | --- | |||||||||||
| G | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00522 | 0.00174 | --- | ||||||||||
| H | 0.00696 | 0.00522 | 0.00522 | 0.00696 | 0.00696 | 0.00348 | 0.00174 | --- | |||||||||
| I | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00522 | 0.00522 | 0.00348 | 0.00522 | --- | ||||||||
| J | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00348 | --- | |||||||
| K | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00174 | 0.00174 | --- | ||||||
| L | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00348 | 0.00348 | 0.00174 | --- | |||||
| M | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00174 | 0.00174 | 0.00000 | 0.00174 | --- | ||||
| N | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00522 | 0.00522 | 0.00348 | 0.00522 | 0.00348 | 0.00348 | 0.00174 | 0.00348 | 0.00174 | --- | |||
| P | 0.02261 | 0.02261 | 0.02087 | 0.02261 | 0.02261 | 0.02261 | 0.02087 | 0.02261 | 0.02087 | 0.02087 | 0.01913 | 0.02087 | 0.01913 | 0.02087 | --- | ||
| Q | 0.02435 | 0.02435 | 0.02261 | 0.02435 | 0.02435 | 0.02435 | 0.02261 | 0.02435 | 0.02261 | 0.02261 | 0.02087 | 0.02261 | 0.02087 | 0.02261 | 0.00174 | --- | |
| R | 0.02435 | 0.02435 | 0.02261 | 0.02435 | 0.02435 | 0.02435 | 0.02261 | 0.02435 | 0.02261 | 0.02261 | 0.02087 | 0.02261 | 0.02087 | 0.02261 | 0.00870 | 0.01043 | --- |
Uncorrected genetic distances among mitochondrial haplotypes of N. cervinus. The values with constant substitution rate, as verified by the relative-rate test, are in bold (p > 0.10).
The wsp is the most rapid evolving gene in Wolbachia [39], but strickingly, it remains invariant in all the individuals assayed. Assuming 0.002 changes per site per Myrs, the wsp sequence should have accumulated 0.0008-0.0036 changes per site in 0.4-1.9 Myrs. In a sequence ca. 580 bp in lenght, we should have found 0.46-2 changes. Therefore, invariancy is not an unexpected result.
Discussion
The estimates of mtDNA genetic variation of N. cervinus are similar to those reported for other species of Curculionidae having similar life history traits (e.g. [40-42]). There is a remarkable bias in the nucleotide proportions of mtDNA toward A/T, mainly in the third positions, which is very common in insects [43,44], including weevils (e.g. [41,45-49]).
The comparison of the haplotype diversity between the two markers herein investigated suggests that the substitution rate of the nuclear genome would be slower than that of the mitochondrial genome, as it has been reported for several animal species (see [50] for a review), including N. cervinus [51].
Apomixis implies clonal transmission of all genomes and lack of recombination. As a consequence, this kind of asexual reproduction will induce cosegregation of mitochondrial and nuclear genotypes and the consequent linkage disequilibrium among all genetic markers [52,53]. The cosegregation of nuclear and mitochondrial haplotypes seen in most individuals herein analyzed provides evidence of the clonal transmission of both genomes in N. cervinus. Furthermore, the congruence between ITS1 and COI phylogenetic trees indicates that coevolution between these genomes have occurred, since many phylogenetically related mitotypes share the same nuclear genotype or viceversa. This phylogenetic congruence suggests that an ancestral mitochondrial haplotype and all its derived lineages evolved along with the same nuclear haplotype across evolutionary times, possibly as a consequence of the higher substitution rate for the mitochondrial gene. Therefore, the hypothesis of ancient linkage disequilibrium and long history of mito-nuclear genomic association is strongly supported.
Only a few number of individuals (1.4%) showed signals of recombination. Besides, traces of hybridization were inferred from individuals carrying at least two nuclear haplotypes or "double strings" ITS1 sequences (8.2%) (a more detailed study of this exciting finding is currently being undertaken). These individuals could be either remnants of historical gene flow, or the consequence of occasional crosses between parthenogenetic females of N. cervinus and males of the same or other related species (as it was reported by Saura et al. [17] and Stenberg et al. [54] for other weevils with similar reproductive behavior). However, these sporadic sexual events would not invalidate the hypothesis of apomixis for this weevil, since there is clonal transmission of nuclear and mitochondrial genomes, lack of recombination (RM = 0) and non-random association for several mitochondrial and nuclear sites.
The cytoplasmic location of the Wolbachia endosymbiont and the mitochondrial organelle will naturally induce linkage disequilibrium among all their genetic markers. Furthermore, the non-random association among mitochondrial and nuclear markers would indicate that the nuclear genome is also linked with the other two. Then, the selective sweep on mtDNA induced by Wolbachia should have affected also the nuclear genome.
If the initial Wolbachia infection have sweeped the mitochondrial diversity, a significant and negative Tajima's DT, and a significant and negative NI should be expected. However, the first statistics suggests selective neutrality and the second, negative selection. The neutral result from Tajima's DT test could be explained by the antiquity of the infection. Most probably, the time elapsed since this event had been long enough to recover the genetic variation to levels prior to the initial selective sweep, increasing Hd and θW values. A similar result was reported by Shoemaker et al. [55], Keller et al. [56] and Marshall [57], who proposed ancient infections for Solenopsis invicta (Hymenoptera, Formicidae), Chelymorpha alternans (Coleoptera, Chrysomelidae) and Allonemobius fasciatus-socius species complex (Orthoptera, Gryllidae). On the other hand, the apparent contrasting result of the MK test (i.e. negative selection) might be interpreted under the Nearly Neutral Theory [27,58-62]. In fact, the value for this statistic would indicate an excess of deleterous mutations, which according to the mentioned theory, would be fixed by chance only if the effective population size is small. In other words, the selective pressure coefficients that operate on the COI gene would be so small (i.e. nearly neutral), that elimination cannot occur.
Low nucleotide diversity in the mitochondrial genome is expected in recently infected species [25,26]. However, a comparison with the related sexual species Naupactus xanthographus, which is unifected [19], would indicate that both species exhibit comparable levels of mitochondrial genetic diversity [63]. This result is interpreted as another evidence of the ancient Wolbachia infection in N. cervinus.
Regarding the nuclear gene, even though ITS1 is a non-functional region, it does not evolve freely in insects [64]. In sexual species, ca. 40% of nucleotide sites are implied in complex secondary structures, and hence under natural selection [65]. Among the 838 sites analyzed herein, 837 showed evidence of negative selection, which is overwhelming in comparison with the result of Schlötterer et al. [65]. This bias in the sites under negative selection could be explained by the genetic linkage among the three genomes, in agreement with Schön et al. [52]. Then, the negative selection operating on bacterial, mitochondrial or nuclear genes could also drive other linked nuclear sites to a similar fate.
The positive Tajima's DT obtained for the nuclear gene may suggest a balancing selection, a spatial population structure of the genetic variation or a moderate bottleneck (after Depaulis et al. [66]). The fact that directional negative selection was detected for ITS1 rules out the first possibility and points to a demographic phenomenon as the most likely explanation for the DT result. Indeed, the LAMARC analysis supported this conclusion, suggesting that N. cervinus could have suffered a bottleneck, probably as a consequence of the Wolbachia invasion [67]. The moderate perturbation in the population size of the initial uninfected population could have allowed some lineages to survive this invasion.
Why this demographic phenomenon was not detected for the mitochondrial gene? The mitochondrial genome usually undergoes faster evolution than the nuclear genome (e.g. [68,69]). Thus, the signatures of the bottleneck could have been erased in N. cervinus mtDNA. Then, the significant Tajima's DT suggests that the bottleneck was probably ancient, since the traces of this demographic event are still recovered from the nuclear dataset, but not from the mitochondrial one.
The fact that all the individuals tested were infected with the same Wolbachia strain (identified by three genes of rapid evolution) favors the hypothesis that the infection of N. cervinus occurred once, and that all extant haplotypes of this weevil species are descended from a single ancestral infected female. However, the positive Tajima's DT for nDNA suggests a different scenario, according to which at least two genetic lineages had been infected with the same strain.
The divergence time estimated from mitochondrial and nuclear datasets allowed us to infer that the bottleneck in N. cervinus population occurred between 400,000-1,900,000 years ago, i.e. during the Plio-Pleistocene, a geological period of enormous cyclical changes in the South American forests and grasslands [70,71]. The older age obtained for nDNA can be attributed to those lineages that survived the demographic event, which could produce an overestimation of the calculated age. On the other side, the younger age inferred for the mitochondrial dataset can be attributed to the faster accumulation of genetic variation, which yielded younger lineages that decreases the mean distance, leading to an underestimation of the age. Therefore, nDNA and mtDNA estimates can be considered as the upper and the lower age limits of the demographic crash.
The amount of evolutionary change accumulated in mtDNA is very high in contrast to the lack of change in wsp, which is the most rapidly evolving gene known in Wolbachia [39]. However, according to the age estimated for the infection in N. cervinus, wsp might have accumulated ca. 0.46-2 substitutions in 0.4-1.9 Myrs. So, lack of variation is not an unexpected result. A similar wsp invariant pattern was found in other organisms like sandflies, leaf beetles and fruitflies (e.g. [25,56,72]. An alternative explanation could be that substitution rates for mitochondrial and nuclear genomes are different from those assumed herein, due to asexual reproduction. In fact, acceleration [73] and deceleration [52] of substitution rates have been reported for species with clonal reproduction. The first alternative implies a more recent infection, in agreement with the lack of variation of wsp Wolbachia gene. However, a decelerated substitution rate would indicate an age of infection much older than that herein estimated.
Although our present data do not allow an accurate inference of the infection age, the high prevalence of Wolbachia in the whole species distribution contradict the hypothesis of a recent invasion. In fact, although prevalence of Wolbachia is usually related to different phenotypes [74], it can also be associated to the infection age [23]. As Wolbachia was detected in 100% of the individuals of N. cervinus assayed, its high prevalence is compatible with the hypothesis of an ancient infection.
Our results suggest that Wolbachia "lived together" with its host N. cervinus at least for a third of its lifespan [see [33]]. If the origin of the apomictic lineages were related to this infection, could Wolbachia be responsible for the extinction of all the bisexual populations of N. cervinus? Given that some males were collected in 1945 and 1947 in the forests of Northern Argentina and Southern Brazil [32], it is unlikely that this bacterium caused their extinction. Rather, the intense deforestation of the Paranaense forest that occurred during the last 40 years [75] is the most important factor to explain the extinction of the bisexual populations of this weevil.
Conclusion
We provide first genetic evidence of apomixis for the weevil Naupactus cervinus. This kind of parthenogenesis, in addition to Wolbachia infection, induces linkage disequilibrium among three genomes: the nuclear and mitochondrial weevil genomes and the bacterial genome. Hence, the mito-nuclear genetic variation of the host would have been shaped by apomictic reproduction, the moderate bottleneck probably caused by the initial Wolbachia infection, the long time passed since then, and the high prevalence of the unique Wolbachia strain infecting this weevil.
Even if Wolbachia accounts for the origin of parthenogenesis in N. cervinus, this infection would have not caused the extinction of the bisexual population. The current absence of males and the lack of sexual reproduction during the last 50 years would be a consequence of the destruction of the native forest where these populations occurred.
Methods
Sampling and specimens examined
Adults of N. cervinus (Entiminae: Naupactini) were collected during the summers of 2004-2007, on wild and cultivated plants from several geographic locations of Argentina, Southern Brazil and Uruguay. Samples from the countries where the species has been introduced were also included (see Figure 4 and Table 1) (N = 309). Specimens were collected using a beating sheet (55 cm × 55 cm) and stored at -80°C or in 100% ethanol at 4°C for molecular analyses.
PCR assay and sequencing
Total genomic DNA was extracted following the protocol of Reiss et al. [76]. The negative controls were samples lacking DNA template.
A segment of ca. 700 bp of the Cytochrome Oxidase I (COI) mitochondrial gene of N. cervinus was amplified using the specific primers S1718 (5'-GGA GGA TTT GGA AAT TGA TTA GTT CC-3') and A2442 (5'-GCT AAT CAT CTA AAA ATT TTA ATT CCT GTT GG-3') [77] and a nuclear region of ca. 1100 bp using the primers rDNA2 (5'-TTG ATT ACG TCC CTG CCC TTT-3') [78] and rDNA 1.58 S (5'-ACG AGC CGA GTG ATC CAC CG-3') [79], which are suitable for amplifying the region 3' of the 18 S rDNA gene, plus the complete ITS1 region (Internal Transcribed Spacer 1) and the 5' region of the 5.8 S rDNA gene.
Search of multiple Wolbachia lineages within N. cervinus was accomplished through amplification and sequencing of the wsp gene fragment using the primers brought by Braig et al. [80]: wsp 81F (5'-TGG TCC AAT AAG TGA TGA AGA AAC-3') and wsp 691R (5'-AAA AAT TAA ACG CTA CTC CA -3'), and the most variables MLST genes for the B supergroup coxA and fbpA, using the primers designed by Baldo et al. [81]: coxA F1 (5'-TTG GRG CRA TYA ACT TTA TAG-3'), coxA R1(5'-CTA AAG ACT TTK ACR CCA GT-3'), fbpA F1 (5'-GCT GCT CCR CTT GGY WTG AT-3') and fbpA R1 (5'-CCR CCA GAR AAA AYY ACT ATT C-3').
Prevalence of Wolbachia infection was studied through amplification of 16 S rDNA gene in ten weevils (whenever it was possible) from every sampling site, using the primers designed by O'Neill et al. [82]: forward (5'-TTG TAG CCT GCT ATG GTA TAA CT-3') and reverse (5'-GAA TAG GTA TGA TTT TCA TGT-3').
Total genomic DNA from Drosophila melanogaster naturally infected with Wolbachia was used as a positive control. Negative controls consisted of samples lacking DNA template from insects and D. melanogaster treated with tetracyline. D. melanogaster DNA was kindly provided by Dr. Scott O'Neill (Queensland University, Australia). All experiments were repeated at least twice.
Amplification was carried out in a 50 μl volume reaction with 50-100 ng of DNA used as template, 0.5 μM of each primer, 0.1 mM of each dNTP, 3.0 mM MgCl2, 0.05 units of Taq polymerase and reaction buffer 1× (Invitrogen). The reactions were performed in a GeneAmp® PCR System 2700 thermal cycler (Applied Biosystems) under the conditions described by Scataglini et al. [48] for COI and Szalanski and Owens [83] for ITS1 fragments.
Double-stranded PCR products were separated by electrophoresis on a 1% agarose gel with TAE buffer containing 0.5 mg⁄ml of ethidium bromide. The bands were excised from the gel and the DNA was purified with a QIAquick Gel Extraction Kit (Qiagen Inc.). DNA was sequenced using a 3130-XL Automatic Sequencer (Applied Biosystems).
Sequence analysis
Standard chromatographic curves of forward and reverse sequences were edited using the Bioedit program [84]. Sequences were translated with the program MEGA v. 4.0.1 [85] to check for the presence of stop codons or frame shifts which might indicate the amplification of pseudogenes [86-89]. Aminoacid sequences were inferred according to the invertebrate mitochondrial code [34]. Alignment was done using CLUSTAL W [90] and adjusted by eye.
Quantifying genetic variability
Nucleotide diversity for each gene region was estimated using Watterson's (θW) [91] and Tajima's (θπ) [92] estimators. Haplotype diversity (Hd) was also estimated according to Nei [93]. All the calculations were performed with DnaSP v. 5.10.00 [94].
Phylogenetic analyses of mitochondrial and nuclear haplotypes were performed by maximum parsimony using haplotypes as "terminal taxa" with the program NONA v. 2.0 [95], executed through the interface WinClada v. 1.00.08 [96]. The implicit enumeration search option was used to get the most parsimonious trees (number of taxa < 25). All characters were regarded as unordered and unweighted. For the nuclear dataset, gaps were treated as fifth state. Clade stability was assessed by 10,000 parsimony bootstrap replications [97]. Naupactus dissimulator was sequenced to use it as outgroup [GenBank: GQ406844 for COI and GQ406826 for ITS1].
Detecting Linkage disequilibrium
Significant associations between mtDNA and nDNA sites were tested using linkage disequilibrium analysis based on the D parameter [98]. This parameter was obtained with DnaSP v. 5.10.00 [94]. Bonferroni's correction for multiple comparisons [99,100] was used for avoiding spurious rejections of the null hypothesis (i.e. linkage equilibrium).
Detecting Recombination
The population recombination parameter was estimated with the method of Hudson and Kaplan [101] based on the minimum number of recombination events in a sample (RM) using DnaSP v. 5.10.00 [94]. The significance of the test was calculated by performing 10,000 coalescent simulations based on a Monte Carlo process with no recombination [102].
Investigating selective sweeps
Tajima's DT [103] and Fu and Li's F [104] test statistics were applied to evaluate whether the frequency spectrum of segregating sites departs from neutral expectations [105] for both datasets. In addition, the MK test [106] was applied to evaluate a possible correlation between polymorphism and divergence for the synonymous to non-synonymous variation ratio predicted by the Neutral Theory [105] only for the mtDNA dataset (coding sequences). The neutrality index (NI) [107] and the α parameter [108] were used as indicators of the degree and direction of departures from neutrality; NI = 1 indicates strict neutrality and NI < 1 suggests an excess of non-synonymous fixation (i.e. adaptive evolution). The α parameter reflects the proportion of fixed mutations by positive selection. Unlike DT, MK is highly recommended when demographic factors can obscure the results of neutrality tests [27,109,110]. These estimations were performed with DnaSP v. 5.10.00 [94], using the same outgroup species as mentioned above.
For the non-conding nDNA dataset, the approach developed by Wong and Nielsen [111] was used. It is a maximum likelihood method for inferring positive selection in non-coding regions, by comparing non-coding divergence vs. synonymous divergence. This approach assumes that synonymous mutations are neutral and occur at a constant rate within a coding fragment. The ratio of the estimated nucleotide substitution rate in the non-coding region to the estimated synonymous substitution rate from the coding region (ξ), provides a numerical approach to assess whether a non-coding region has evolved by positive selection. The unit of evolution is one nucleotide, and ξ is the nucleotide substitution rate in the non-coding region normalized by the synonymous nucleotide substitution rate in the coding region. Therefore, when a site is subject to neutral selection, ξ = 1. Similarly, ξ > 1 indicates positive selection, while ξ < 1 suggests the occurrence of negative selection.
Three models are implemented in the EvoNC software [111]. The neutral, the two-category, and the three-category models were used in two different tests to determine whether a non-coding region is under positive selection. The Test 1 compares the neutral vs. the three category model, and the Test 2, less conservative, compares the neutral vs. the two-category model [111]. The two mentioned tests were applied to identify non-coding sites under positive selection, with the coding sites of the mtDNA being considered as linked to the ITS1 sites. Therefore, mitochondrial sites were used to estimate the synonymous evolution rate because nuclear and mitochondrial genomes were assumed to be linked to each other and also to Wolbachia genome.
For obtaining ξ, the nucleotide positions must be optimized on a phylogenetic tree which includes the sequences under analysis (i.e. the mitochondrial and nuclear haplotypes concatenated). A matrix of concatenated sequences was built whenever the COI and ITS1 fragments came from the same individual. This matrix had 21 terminals (including N. dissimulator as outgroup) of 1,521 bp in length (683 bp COI + 838 bp ITS1).
The phylogenetic tree was estimated through Bayesian inference. MrModeltest software v. 2.2 [112] was used to infer the most appropriate model of molecular evolution for each dataset based on the Akaike Information Criterion (AIC) [113,114], as suggested by Posada and Buckley [115].
Bayesian phylogenetic analysis was performed using the "Metropolis-coupled Markov chain Monte Carlo" (MCMCMC) algorithm implemented in MrBayes ver. 3.1.2 [116,117]. A partitioned algorithm was used to account for heterogeneity between the two datasets. Program defaults were used for estimation of priors. Two independent analyses were run using a random starting tree with three heated chains and one cold chain over 400,000 generations, with sampling every 100 generations.
The tree space was explored using four chains: one cold chain and three incrementally heated ones, heat being set as 1/(1 + (i -1) T), where i is the chain number (i.e. 1-4) and T is used as default.
Stationarity of the Bayesian analysis was evaluated with the methodologies and statistics implemented in Tracer [118] and AWTY [119,120] and with the standard deviation of the split frequencies. All posterior samples of a run prior to this point were discarded as burn-in. Remaining trees were used to construct a 50% majority-rule consensus tree with mean branch length estimates. The frequency of all observed bipartitions was used to assess the level of support for each node [116,117].
Analyzing demographic structure
Tajima's test DT [103] results can reveal either selective or demographic events. To disentangle the demographic factors for the interpretation of this test, it was necessary to demonstrate that a population bottleneck had occurred. For this purpose we use the program LAMARC [121] to estimate the relative effective population size parameter (θ) and the exponential growth rate (g) by maximum likelihood, as well as the calculation of 95% confidence intervals. Since the maximum likelihood estimation procedure is computationally intensive, 60 individuals were selected randomly from each dataset because results do not change significantly at this sample size [122]. Positive values of g indicate population growth, while negative values indicate shrinkage. The estimation of the parameters was done using the MCMCMC sampling algorithm for ITS1 implemented in LAMARC v. 2.0 [121]. The analysis consisted of two simultaneous searches with heating temperature adjusted automatically with 10 initial chains of 5,000 steps sampled every 20 steps, followed by two final chains of 50,000 steps sampled every 20 steps. Final most likely estimates (MLEs) were calculated using parameter estimates from two replicated analyses.
The evolution model was selected using the program ModelTest [123] as previously described. According to Kuhner et al. [122], the inclusion of unlinked loci reduces the bias in estimating g with a small amount of individuals. As the reproductive mode of N. cervinus makes this recommendation meaningless, the sign of g was analyzed without taking into account its absolute value.
Estimating divergence times
Molecular clock estimates were calculated through the mean divergence time among all nuclear and mitochondrial haplotypes found for N. cervinus. Relative-rate tests were performed to test the equality of evolutionary rates between lineages [124] using MEGA v. 4.0.1 [85]. The divergence times were calculated from the average of the uncorrected pairwise genetic distances among haplotypes with MEGA v. 4.0.1, using the equation T = D/2k (where T is the mean divergence time, D is the mean number of pairwise differences per site, and k is the estimated rate of nucleotide substitution). We decided not to apply substitution models to correct the pairwise distances, because we considered that the levels of divergence among haplotypes within the same species is low enough to deserve a correction. In order to estimate these divergence times, we used a nucleotide substitution rate of 0.85% per site per Myrs for ITS1 [68] and 1.2% per site per Myrs for COI [125].
Additionally, we calculated the expected variation for Wolbachia wsp gene assuming a nucleotide substitution rate of 0.2% [126].
Authors' contributions
MSR carried out the molecular work, analized the data and wrote a first draft of the manuscript. All authors contributed to obtain samples of specimens and to the final analyses and writing of the manuscript.
Supplementary Material
Table S1: Linkage disequilibrium test. Values of the D parameter. The significant disequilibrium linkage after Bonferroni's correction is indicated by the letter B. * p < 0. 005; ** p < 0. 010; *** p < 0. 001
Contributor Information
Marcela S Rodriguero, Email: rodriguero@ege.fcen.uba.ar.
Analía A Lanteri, Email: alanteri@fcnym.unlp.edu.ar.
Viviana A Confalonieri, Email: bibilu@ege.fcen.uba.ar.
Acknowledgements
We thank Pablo Colombo, Guadalupe del Río, Jerson Guedes, Noelia Guzmán, Ariel López, Dardo Martí and Glauber Sturmer for field assistance, and Elin Claridge, María Laura De Wysieki, Mario Elgueta, Ferrán García Marí, Antonio Machado and Adriana Marvaldi for providing some of the samples. We are also indebted to Dr. R. Piccinali and two anonymous reviewers for their critical reading and helpful comments on an earlier version of this manuscript, to C. Tomatis for his valuable suggestions for designing Figure 4 and to L. Paulin for her helpful suggestions concerning the use of Linux plataform. Part of the molecular analyses were carried out by using the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation. This research project was supported by grants from Universidad de Buenos Aires, ANPCyT and CONICET awarded to VAC; MSR was awarded with a doctoral scholarship from CONICET and received a travel grant from University of Buenos Aires. VAC and AAL are members of the scientific research staff of CONICET, Argentina.
References
- Suomalainen E. Significance of parthenogenesis in the evolution of insects. Annu Rev Entomol. 1962;7:349–365. doi: 10.1146/annurev.en.07.010162.002025. [DOI] [Google Scholar]
- Suomalainen E. In: Evolutionary Biology. Dobzhansky T, Hecht M, Steere W, editor. Vol. 3. New York: Plenum Press; 1969. Evolution in Parthenogenetic Curculionidae; pp. 261–296. [Google Scholar]
- Suomalainen E, Saura A, Lokki J. Evolution of parthenogenetic insects. Evol Biol. 1976;9:209–257. [Google Scholar]
- Smith SG, Virkki N. In: Animal Cytogenetics. Volume 3: Insecta 5. John B, editor. Berlin: Gebrüder Borntraeger; 1978. Coleoptera; pp. 236–290. [Google Scholar]
- Lokki J, Saura A. In: Polyploidy biological relevance. Lewis WM, editor. New York: Plenum Press; 1980. Polyploidy in insect evolution; pp. 277–312. [Google Scholar]
- Suomalainen E, Saura A, Lokki J. Cytology and evolution in parthenogenesis. Boca Raton: CRC Press; 1987. [Google Scholar]
- Lanteri AA, Normark BB. Parthenogenesis in the tribe Naupactini (Coleoptera: Curculionidae) Ann Entomol Soc Am. 1995;88:722–731. [Google Scholar]
- Buchanan LL. The species of Pantomorus of America north of Mexico. USDA Misc Publs. 1939;341:1–39. [Google Scholar]
- Marvaldi AE. Eggs and oviposition habits in Entimini (Coleoptera: Curculionidae) Coleopts Bull. 1999;53(2):115–126. [Google Scholar]
- Lachowska D, Rözek M, Holecová M. New data on the cytology of parthenogenetic weevils (Coleoptera, Curculionidae) Genetica. 2008;134(2):235–242. doi: 10.1007/s10709-007-9230-x. [DOI] [PubMed] [Google Scholar]
- Vepsälainen K, Järvinen O. Apomictic parthenogenesis and the pattern of the environment. Amer Zool. 1979;19:739–751. [Google Scholar]
- Gustafsson A. Apomixis in higher plants. I. The causal aspect of apomixis. II. Biotype and species formation. Lund Univ Arssk NF Avd. 1947;43:71–178. [Google Scholar]
- Suomalainen E. Parthenogenesis in animals. Advan Genet. 1950;3:193–253. full_text. [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokki J. Genetic polymorphism and evolution in parthenogenetic animals. VII. The amount of heterozygosity in diploid populations. Hereditas. 1976;83:57–64. doi: 10.1111/j.1601-5223.1976.tb01570.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1987–1990. doi: 10.1126/science.281.5385.1986. [DOI] [PubMed] [Google Scholar]
- Saura A, Lokki J, Suomalainen E. Origin of polyploidy in parthenogenetic weevils. J Theor Biol. 1993;163:449–456. doi: 10.1006/jtbi.1993.1130. [DOI] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia, reproductive parasites of arthropods. Proc R Soc Lond B. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Rodriguero MS, Confalonieri VA, Guedes JCV, Lanteri AA. Wolbachia infection in the tribe Naupactini: association between thelytokous parthenogenesis and infection status. Insect Mol Biol. in press . [DOI] [PubMed]
- Piper RW, Compton SG, Rasplus J-Y, Piry S. The species status of Cathormiocerus britannicus, an endemic, endangered British weevil. Biol Consev. 2001;101:9–13. doi: 10.1016/S0006-3207(01)00048-9. [DOI] [Google Scholar]
- Son Y, Luckhart S, Zhang X, Lieber MJ, Lewis EE. Effects and implications of antibiotic treatment on Wolbachia-infected vine weevil (Coleoptera: Curculionidae) Agr Forest Entomol. 2008;10:147–155. doi: 10.1111/j.1461-9563.2008.00369.x. [DOI] [Google Scholar]
- Charlat S, Hurst GDD, Merçot H. Evolutionary consequences of Wolbachia infections. Trends Genet. 2003;19(4):217–223. doi: 10.1016/S0168-9525(03)00024-6. [DOI] [PubMed] [Google Scholar]
- Engelstädter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst. 2009;40:127–149. doi: 10.1146/annurev.ecolsys.110308.120206. [DOI] [Google Scholar]
- Rokas A, Atkinson RJ, Brown GS, West SA, Stone GN. Understanding patterns of genetic diversity in the oak gallwasp Biorhiza pallida demographic history or a Wolbachia selective sweep? Heredity. 2001;87:294–304. doi: 10.1046/j.1365-2540.2001.00872.x. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Dyer KA, Ahrens M, McAbee K, Jaenike J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics. 2004;168:2049–2058. doi: 10.1534/genetics.104.030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc Lond B. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Syst. 2005;36:621–642. doi: 10.1146/annurev.ecolsys.36.091704.175513. [DOI] [Google Scholar]
- Bull JJ. The evolution of sex determination mechanisms. Menlo Park: Benjamin/Cummings Publ. Co; 1983. [Google Scholar]
- Chadwick CE. A review of Fuller's rose weevil (Pantomorus cervinus (Boh.) (Coleoptera: Curculionidae) J Ent Soc Australia. 1965;2:10–20. [Google Scholar]
- Loiácono MS, Marvaldi AE. In: Bases para el control integrado de los gorgojos de la alfalfa. Lanteri AA, editor. La Plata: De la Campana Ediciones; 1994. Biología y daños ocasionados; pp. 41–48. [Google Scholar]
- Lanteri AA, Guedes JCV, Parra JRP. Weevils injurious for roots of citrus in São Paulo state. Brazil Neotr Entomol. 2002;31(4):561–569. [Google Scholar]
- Lanteri AA. Revisión sistemática del género Asynonychus Crotch (Coleoptera: Curculionidae) Rev Asoc Cienc Nat Lit. 1986;17(2):161–174. [Google Scholar]
- Rodriguero MS. PhD thesis. Universidad de Buenos Aires, Departamento de Ecología, Genética y Evolución; 2009. Origen y consecuencias de la reproducción asexual en una especie de gorgojo de importancia agronómica. [Google Scholar]
- Lunt DH, Zhand DX, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Tavare S. Some probabilistic and statisical problems on the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- Yang Z. Estimating the pattern of nucleotide substitution. J Mol Evol. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Waddell PJ, Steel MA. General time-reversible distances with unequal rates across sites: mixing Γ and inverse Gaussian distributions with invariant sites. Mol Phyl Evol. 1997;8:398–414. doi: 10.1006/mpev.1997.0452. [DOI] [PubMed] [Google Scholar]
- Zhou W, Rousset F, O'Neill SL. Phylogeny and PCR based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Lond B. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccoli M, Pisceda A, Salvato P, Simonato M, Masutti L, Battisti A. Genetic structure and phylogeography of pine shoot beetle populations (Tomicus destruens and T. piniperda, Coleoptera Scolytidae) in Italy. Ann For Sci. 2005;62:361–368. doi: 10.1051/forest:2005031. [DOI] [Google Scholar]
- Mynhardt G, Harris MK, Cognato AI. Population genetics of the pecan weevil (Coleoptera: Curculionidae) inferred from mitochondrial nucleotide data. Ann Entomol Soc Am. 2007;100(4):582–590. doi: 10.1603/0013-8746(2007)100[582:PGOTPW]2.0.CO;2. [DOI] [Google Scholar]
- Anducho-Reyes MA, Cognato AI, Hayes JL, Zúñiga G. Phylogeography of the bark beetle Dendroctonus mexicanus Hopkins (Coleoptera:Curculionidae: Scolytinae) Mol Phyl Evol. 2008;49:930–940. doi: 10.1016/j.ympev.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langor DW, Sperling FAH. Mitochondrial DNA sequence divergence in weevils of the Pissodes strobi species complex (Coleoptera: Curculionidae) Insect Mol Biol. 1997;6:255–265. doi: 10.1046/j.1365-2583.1997.00180.x. [DOI] [PubMed] [Google Scholar]
- Normark BB, Lanteri AA. Incongruence between morphological and mitochondrial-DNA characters suggests hybrid origins of parthenogenetic weevil lineages (genus Aramigus) Syst Biol. 1998;47:459–478. doi: 10.1080/106351598260833. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Lanteri AA, Scataglini MA, Confalonieri VA, Farrell B. Are flightless Galapaganus weevils older than the Galápagos Islands they inhabit? Heredity. 2000;85:20–29. doi: 10.1046/j.1365-2540.2000.00690.x. [DOI] [PubMed] [Google Scholar]
- Scataglini MA, Lanteri AA, Confalonieri VA. Phylogeny of the Pantomorus-Naupactus complex based on morphological and molecular data (Coleoptera: Curculionidae) Cladistics. 2005;21(2):131–142. doi: 10.1111/j.1096-0031.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- Scataglini MA, Lanteri AA, Confalonieri VA. Diversity of boll weevil populations in South America: a phylogeographic approach. Genetica. 2006;126:353–368. doi: 10.1007/s10709-005-1399-2. [DOI] [PubMed] [Google Scholar]
- Brown WM. In: Molecular Evolutionary Genetics. Maclntyre RJ, editor. New York: Plenum Press; 1985. The mitochondrial genome of animals; pp. 95–130. [Google Scholar]
- Mander CV, Phillips CB, Glare TR, Chapman RB. Preliminary assessment of COI and ITS1 sequence variation in Fuller's rose weevil. New Zeal Plant Protection. 2003;56:190–193. [Google Scholar]
- Schön I, Butlin RK, Griffiths HI, Martens K. Slow molecular evolution in an ancient asexual ostracod. Proc R Soc Lond B. 1998;265:235–242. doi: 10.1098/rspb.1998.0287. [DOI] [Google Scholar]
- Simon JC, Delmotte F, Rispe C, Crease T. Phylogenetic relationships between parthenogens and their sexual relatives: the possible routes to parthenogenesis in animals. Biol J Lin Soc. 2003;79:151–163. doi: 10.1046/j.1095-8312.2003.00175.x. [DOI] [Google Scholar]
- Stenberg P, Lundmark M, Knutelski S, Saura A. Evolution of clonality and polyploidy in a weevil system. Mol Biol Evol. 2003;20:1626–1632. doi: 10.1093/molbev/msg180. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Keller G, Ross KG. Effects of Wolbachia on mtDNA variation in two fire ant species. Mol Ecol. 2003;12:1757–1771. doi: 10.1046/j.1365-294X.2003.01864.x. [DOI] [PubMed] [Google Scholar]
- Keller GP, Windsor DM, Saucedo JM, Werren JH. Reproductive effects and geographic distributions of two Wolbachia strains infecting the Neotropical beetle, Chelymorpha alternans Boh (Chrysomelidae, Cassidinae) Mol Ecol. 2004;13:2405–2420. doi: 10.1111/j.1365-294X.2004.02213.x. [DOI] [PubMed] [Google Scholar]
- Marshall JM. The Allonemobius-Wolbachia host-endosymbiont system: evidence for rapid speciation and against reproductive isolation driven by cytoplasmic incompatibility. Evolution. 2004;58:2409–2425. doi: 10.1111/j.0014-3820.2004.tb00871.x. [DOI] [PubMed] [Google Scholar]
- Rand DM. Mitochondrial genomics flies high. Trends Ecol Evol. 2001;16(1):2–4. doi: 10.1016/S0169-5347(00)02036-X. [DOI] [PubMed] [Google Scholar]
- Rand DM. The units of selection on mitochondrial DNA. Annu Rev Ecol Syst. 2001;32:415–448. doi: 10.1146/annurev.ecolsys.32.081501.114109. [DOI] [Google Scholar]
- Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13(4):729–44. doi: 10.1046/j.1365-294X.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23(6):259–63. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Santos C, Montiel R, Arruda A, Alvarez L, Aluja MP, Lima M. Mutation patterns of mtDNA: empirical inferences for the coding region. BMC Evol Biol. 2008;8:167. doi: 10.1186/1471-2148-8-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán N, Lanteri A, Confalonieri V. Variabilidad comparativa de marcadores nucleares y mitocondriales en el gorgojo plaga Naupactus xanthographus (Coleoptera, Curculionidae) [abstract] Libro de Actas del VII Congreso Argentino de Entomología Huerta Grande; 2008. [Google Scholar]
- Parkin EJ, Butlin RK. Within and between individual sequence variation among ITS1 copies in the meadow grasshopper Chorthippus parallelus indicates frequent intrachromosomal gene conversion. Mol Biol Evol. 2004;21:1595–1601. doi: 10.1093/molbev/msh163. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Hauser MT, von Haeseler A, Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- Depaulis F, Mousset S, Veuille M. Power of neutrality tests to detect bottlenecks and hitchhiking. J Mol Evol. 2003;57(1):190–200. doi: 10.1007/s00239-003-0027-y. [DOI] [PubMed] [Google Scholar]
- Hein J, Schierup MH, Wiuf C. Gene genealogies, variation and evolution-a primer in coalescent theory. Oxford: Oxford University Press; 2005. [Google Scholar]
- Caccone A, Amato GD, Powell JR. Rates and patterns of scnDNA and mtDNA divergence within the Drosophila melanogaster subgroup. Genetics. 1988;118:671–683. doi: 10.1093/genetics/118.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargues MD, Klisiowicz DR, Panzera F, Noireau F, Marcilla A, Pérez R, Rojas MG, O'Connor JE, González-Candelas F, Galvão C, Jurberg J, Carcavallo RU, Dujardin JP, Mas-Coma S. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Inf Gen Evol. 2006;6(1):46–62. doi: 10.1016/j.meegid.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Saia SEMG, Pessenda LCR, Gouveia SEM, Aravena R, Bendassolli JA. Last glacial maximum (LGM) vegetation changes in the Atlantic Forest, southeastern Brazil. Quatern Int. 2008;184:195–201. doi: 10.1016/j.quaint.2007.06.029. [DOI] [Google Scholar]
- Ledru M-P, Mourguiart P, Riccomini C. Related changes in biodiversity, insolation and climate in the Atlantic rainforest since the last interglacial. Palaeogeogr Palaeocl Palaeoecol. 2009;271:140–152. doi: 10.1016/j.palaeo.2008.10.008. [DOI] [Google Scholar]
- Parvizi P, Benlarvi M, Ready PD. Mitochondrial and Wolbachia markers for the sandfly Phlebotomus papatasi: little population differentiation between peridomestic sites and gerbil burrows in Isfahan province, Iran. Med Vet Entomol. 2003;17(4):351–362. doi: 10.1111/j.1365-2915.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- Paland S, Lynch M. Transitions to asexuality result in excess amino acid substitutions. Science. pp. 990–992. [DOI] [PubMed]
- Jiggins FM, Bentley JK, Majerus MEN, Hurst GDD. How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc R Soc Lond B. 2001;268:1123–1126. doi: 10.1098/rspb.2001.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclau P. La conservación de los recursos naturales y el hombre en la selva paranaense. Bol Técnico Fund Vida Silv Arg. 1994;20:1–139. [Google Scholar]
- Reiss RA, Schwert DP, Ashworth AC. Field preservation of Coleoptera for molecular genetics analyses. Environ Entomol. 1995;24:716–719. [Google Scholar]
- Normark BB. Phylogeny and Evolution of Parthenogenesis in the Aramigus tessellatus Complex (Coleoptera: Curculionidae) PhD thesis. Cornell University, Department of Ecology and Evolutionary Biology. 1994.
- Vrain TC, Wakarchuk DC, Levesque AC, Hamilton RI. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundam Appl Nematol. 1992;15:563–573. [Google Scholar]
- Cherry T, Szalanski AL, Todd TC, Powers TO. The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae) J Nematol. 1997;29:23–29. [PMC free article] [PubMed] [Google Scholar]
- Braig HR, Zhou W, Dobson SL, O'Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180(9):2373–2378. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16 S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibity in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalanski AL, Owens CB. Genetic variation of the Southern corn rootworm (Coleoptera: Chrysomelidae) Fla Entomol. 2003;86(3):329–333. doi: 10.1653/0015-4040(2003)086[0329:GVOTSC]2.0.CO;2. [DOI] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0.1. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Sorenson MD, Quinn TW. Numts: A challenge for avian systematics and population biology. Auk. 1998;115:214–221. [Google Scholar]
- Zhang DX, Hewitt GM. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol. 2003;12:563–584. doi: 10.1046/j.1365-294X.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang DX, Hewitt GM. Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes. Mol Biol Evol. 2000;17(3):406–15. doi: 10.1093/oxfordjournals.molbev.a026320. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang DX, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: evolution's misplaced witness. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/S0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregation sites. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Goloboff PA. NONA v 2.0. Published by the Author. 1999. http://www.cladistics.com/aboutNona.htm Accesed on July 15, 2009.
- Nixon KC. Winclada ver 10.008. Published by the author. 2002. http://www.Cladistics.com/about_winc.htm Accesed on July 15, 2009.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage I General considerations: heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE. Il calcolo delle assicurazioni su gruppi di teste. Studi in Onore del Professore Salvatore Ortu Carboni. Rome, Italy. 1935. pp. 13–60.
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubbl R Inst Sup Scienze Econom Comm di Firenze. 1936;8:3–62. [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. In: Oxford surveys in evolutionary biology. Futuyma D, Antonovics J, editor. Vol. 7. New York: Oxford University Press; 1990. Gene genealogies and then coalescent process; pp. 1–44. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Gen. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Wong WS, Nielsen R. Detecting selection in non-coding regions of nucleotide sequences. Genetics. 2004;167:949–958. doi: 10.1534/genetics.102.010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v.2. Program distributed by the author, 2004. Evolutionary Biology Center, Uppsala University. 2009. Accesed on July 16.
- Akaike H. In: Second International Symposium on Information Theory. Petrov BN, Csaki F, editor. Budapest: Akademia Kiado; 1973. Information theory and an extension of the maximum likelihood principle; pp. 267–281. [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in Phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES v.3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer Release 1.4. 2007. http://beast.bio.ed.ac.uk/Tracer Accesed on July 16, 2009.
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004. http://ceb.csit.fsu.edu/awty Accesed on July 16, 2009. [DOI] [PubMed]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (Are We There Yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24(4):581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22(6):768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimates of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Tajima F. Simple methods for testing molecular clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccone A, Sbordoni V. Molecular biogeography of cave life: A study using mitochondrial DNA from Bathysciine beetles. Evolution. 2001;55:122–130. doi: 10.1111/j.0014-3820.2001.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Wenseleers T. Conflict from cell to colony. Ph.D. thesis. University of Leuven, Department of Biology. 2001.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Linkage disequilibrium test. Values of the D parameter. The significant disequilibrium linkage after Bonferroni's correction is indicated by the letter B. * p < 0. 005; ** p < 0. 010; *** p < 0. 001