Abstract
Research on animal personality can be approached from both a phenotypic and a genetic perspective. While using a phenotypic approach one can measure present selection on personality traits and their combinations. However, this approach cannot reconstruct the historical trajectory that was taken by evolution. Therefore, it is essential for our understanding of the causes and consequences of personality diversity to link phenotypic variation in personality traits with polymorphisms in genomic regions that code for this trait variation. Identifying genes or genome regions that underlie personality traits will open exciting possibilities to study natural selection at the molecular level, gene–gene and gene–environment interactions, pleiotropic effects and how gene expression shapes personality phenotypes. In this paper, we will discuss how genome information revealed by already established approaches and some more recent techniques such as high-throughput sequencing of genomic regions in a large number of individuals can be used to infer micro-evolutionary processes, historical selection and finally the maintenance of personality trait variation. We will do this by reviewing recent advances in molecular genetics of animal personality, but will also use advanced human personality studies as case studies of how molecular information may be used in animal personality research in the near future.
Keywords: personality, ecological genetics, genomics, micro-evolution, balancing selection, maintenance of variation
1. Introduction
Individuals in many animal species differ consistently in suites of behavioural traits (Réale et al. 2007; Gosling 2008), comparable to how humans differ in personality (John et al. 2008). Personality can be seen as an emergent phenomenon, where consistent variation in behavioural expression becomes apparent through the reaction to mild challenges. More broadly, animal personality is defined as a suite of individual differences in behaviour that are consistent over time or contexts (Sih & Bell 2008). Quantitative genetic studies have found that between 20 and 50 per cent of the phenotypic variation in animal personality traits has a genetic basis (Van Oers et al. 2005; Van Oers & Sinn in press), whereas the residual irreversible and reversible phenotypic variance are assigned to development and learning (e.g. Quinn & Cresswell 2005; Arnold et al. 2007; Brydges et al. 2008). Personality is found to be an important factor influencing fitness (Smith & Blumstein 2008) and personality traits are known to be under natural (Réale et al. 2007) and sexual selection (Van Oers et al. 2008). One of the main questions that still remains unresolved is why variation in personality exists and how this is maintained. A process known to actively maintain genetic polymorphism is balancing selection (Turelli & Barton 2004). Different forms of balancing selection are considered to maintain variation in personality traits (Penke et al. 2007). For example, overdominance and antagonistic pleiotropy are examples of genetic mechanisms that cause selection to be balanced (Roff 1997). Antagonistic pleiotropy signifies that genetic variants have a positive effect on one trait, but a negative effect on another trait (Roff & Fairbairn 2007), often resulting in trade-offs. However, molecular processes alone are not enough to explain maintenance in trait variation. Therefore alternatives such as spatial and/or temporal environmental heterogeneity and frequency-dependent selection may also contribute to balancing selection (Dall 2004).
Knowledge of the molecular mechanisms underlying personality traits will help us in answering questions regarding the origin of, correlations between, micro-evolutionary processes behind and historical selection on personality traits, and finally the maintenance of variation in personality (see also Bell & Aubin-Horth 2010). Although molecular genetic research on animal personality is still in its infancy, the development of new methodology might bring us a big step further in pinpointing the actual genes that are responsible for consistent variation in and correlations between behavioural traits. In this paper, we will highlight how recent advances in molecular genetic techniques can help us in studying molecular genetic variation in personality traits from an ecological and evolutionary viewpoint. Our aim is to show that investigating genetic variation, the basis for evolutionary change, not only helps to unravel the constraints and physiological mechanisms underlying personality variation, but can also help us to study the evolution of behavioural syndromes and personality traits.
2. Identifying genomic variation underlying personality
There are two general strategies to pinpoint the regions of the genome that are of interest for complex traits such as personality traits. First of all there are hypothesis-free scanning approaches, like genome-wide quantitative trait locus (QTL) or association-mapping, where linkage or association is tested between variation in genomic polymorphisms and variation in the trait of interest (Slate 2005). Depending on marker density and recombination history of the sample, regions can be identified that cover from a single gene to several hundreds of genes. A hypothesis-driven and non-mutually exclusive approach is the candidate gene approach. Here, information from other species or behaviours is used to specifically test for associations between polymorphisms in candidate genes and personality traits (Comings et al. 2000; Savitz & Ramesar 2004).
(a). Genome-wide approach
Genome-wide approaches provide us with unbiased methods to identify genes related to personality traits. Until recently, obtaining large-scale genomic data has been a limiting step for the progress of these approaches on non-model species. However, new high-throughput genomics technologies such as next-generation sequencing techniques have now hugely decreased the unit time and cost of obtaining sequence data in ecologically important species (Ellegren 2008a,b; Mardis 2008).
(i). QTL mapping
QTL mapping is based on the presence of individual phenotypic data in a pedigreed population and individual genotypic data of genetic markers that are distributed over the whole organism's genome (Slate 2005). The results from these analyses are then used to infer the genetic architecture underlying the trait of interest (Erickson et al. 2004). QTL analysis of genetic markers usually results in a candidate chromosomal region linked to the phenotype covering several dozens to hundreds of genes. Subsequently, this region is narrowed based on haplotype sharing or the identification of more polymorphic markers at these specific QTL sites. Here positional or functional candidate genes are identified for further genetic studies with the aim of identifying those loci that are of major importance for phenotypic trait variation. In other words, QTL analysis can be seen as a linkage (co-segregation) analysis that is based on pedigree information (one or many pedigrees from natural populations or crossing experiments). Mapping populations consist either of inbred line crosses, crosses between outbred populations or natural populations with known pedigree structure (Lynch & Walsh 1998; Slate 2005).
To give an example from human genetics, Gillespie et al. (2008) measured the three Eysenckian personality domains in a population of 1280 adolescent Australians. These individuals consisted of 82 monozygotic and 421 dizygotic twin families with their offspring genotyped at 757 microsatellite markers. In a multivariate variance components analysis, they found links between the three Eysenck personality traits (extraversion, neuroticism and psychoticism) and genomic regions that included the serotonin receptor (HTR2a) and the ADHD4 genes.
QTL studies on animal personality traits have mainly been limited to studies on rodents using controlled crosses between lines or strains (e.g. Gershenfeld et al. 1997; Hovatta & Barlow 2008). More recently, personality has also been found to be an important factor influencing animal wellbeing in farm animals (Koolhaas et al. 2001; Christiansen & Forkman 2007; Rodenburg et al. 2008), leading to behaviour genetic studies identifying QTLs for behavioural traits in livestock. For example, Gutierrez-Gil et al. (2008) identified 29 QTL regions in a cross between two cattle populations measured for flight distance and social separation. In total, these regions explained only a small fraction of the phenotypic variation, ranging from 4 to 8 per cent. The most notable candidate gene found in one QTL region, located at the distal end of chromosome 29, was the dopamine receptor D4 (DRD4) gene (see below), showing that DRD4 is probably one of the most important genes involved in variation of personality traits, in this case flight distance to a human approacher.
Slate (2005) published a timely review of the prospects for QTL studies in natural populations. This paper provides a good guideline of what techniques can be used to perform QTL studies with non-model organisms, in the context of ecological and evolutionary issues. For most species for which personality data have been collected in natural populations, advances have been hampered by the lack of pedigree information as well as the lack of sufficient numbers of markers to be able to construct genetic maps. Animal models that use complex pedigrees to estimate genetic parameters (Kruuk 2004) are now also used in personality studies on natural populations (Quinn et al. 2009). Moreover, genomic polymorphism data for high-quality genetic maps are now becoming available for many non-model species. A whole-genome linkage map of the zebra finch Taeniopygia guttata based on about 2000 single-nucleotide polymorphism (SNP) markers has been constructed (Backström et al. 2010), and tens of thousands of SNPs have been identified for the great tit Parus major (Van Bers et al. 2010), to name just two dominant bird species used in personality research over the last two decades. QTL and association studies in natural populations therefore come within reach.
(ii). Association studies
Genome-wide association (GWA) studies use high-throughput genotyping technologies to assay the variation in several hundreds of thousands of SNPs and relate them to the trait of interest (Risch 2000; de Bakker et al. 2005). An important difference with QTL studies is that association studies use population-based data with abundant recombination history and thereby have the potential to fine-map the functional genomic region or to localize the functional polymorphism itself. This allows the direct identification of ‘genes’ with potentially known function in model species that are causing variation in the trait of interest. Up to now, GWA studies related to personality are rare and have only been conducted on humans (De Moor et al. 2009). The first GWA study on all five human personality factors (i.e. neuroticism, extraversion, openness to experience, conscientiousness and agreeableness) used a sample of 3972 individuals from an isolated population on Sardinia, Italy that was genotyped on 362 129 SNPs (Terracciano et al. 2008). A few of the most promising SNPs that were identified per factor successfully replicated in two other independent samples. The authors highlight two important conclusions that can be drawn from this pioneer study. First, personality traits are influenced by many genes that each explain only small amounts of variation (1–2%) and these polymorphisms are only identified when sample sizes are large enough. Second, genetic effects are most probably found when specific phenotypes are measured, rather than when pooled together into broader factors or principal component analysis (Terracciano et al. 2008). Behavioural patterns, like boldness or risk-taking behaviours, should therefore be broken down into smaller individual behaviours. This issue points back to one of the challenges in behaviour genetics—the definition and quantification of behaviour (Sokolowski 2001). This second point might especially be of great interest to animal personality researchers since it could indicate that specific genes play a role in determining variation in single personality traits, but additional genes modify the correlations among traits.
The fact that GWA studies are mostly limited to human personality studies is caused by the high number of individuals and markers needed for these kinds of analyses. With decreasing sequencing and genotyping costs, these methods will become available for the non-model species with which ecologists and evolutionary biologists work in the near future. Especially when groups combine their efforts in consortia, developing arrays and chips of SNPs and other polymorphisms, GWA studies on personality traits in natural populations become feasible. Whether GWA studies will make other approaches, such as candidate gene approaches, superfluous in the future, remains uncertain. For example, GWA studies often make use of polymorphisms with high minor allele frequencies (MAF), and they will therefore not pick up possibly important genes with lower MAF (Wilkening et al. 2009).
(b). Candidate gene approach
A major advantage of studies on the genetic basis of personality differences in natural populations is that candidates for trait loci can be nominated on the basis of knowledge of similar phenotypes in model species such as humans, domestic fowl or mice, thus circumventing the tedious process of unprejudiced genome-wide approaches. Candidate gene studies require little prior sequence information and are therefore well suited for evolutionary analyses in natural populations of non-model species (Fitzpatrick et al. 2005).
In humans, among the most probable genetic candidates for personality variation are various polymorphisms within the DRD4 gene and the serotonin transporter gene (SERT; Savitz & Ramesar 2004). Polymorphisms within the DRD4 gene have been found to account for about 3 per cent of the variation in novelty seeking in humans (Munafo et al. 2008b). Studies looking at SERT have found a relationship between a polymorphism in a regulatory sequence for this gene and anxiety-related traits (Eley & Plomin 1997; Gordon & Hen 2004). However, the evidence for an association is inconsistent when slightly different measures of the trait (harm avoidance, neuroticism, etc.) are used (Munafo et al. 2009). The SERT gene might nevertheless play a role in anxiety but its effects might be rather subtle, for example, on amygdala reactivity, which mediates anxiety only under certain circumstances (Munafo et al. 2008a). Other, not so well-studied genes with possible effects on variation in personality include the monoamine oxidase A gene, the dopamine receptor D2 gene, the serotonin receptor genes 5-HT2c and HTR2a, and the tyrosine hydroxylase gene (reviewed in Savitz & Ramesar 2004).
Several studies on non-human animals have looked at the relation between personality traits and the candidate genes identified in human studies. An association between exploratory behaviour and the DRD4 homologue has been detected in species ranging from apes (Shimada et al. 2004) to dogs (Ito et al. 2004), fish (Boehmler et al. 2007) and birds (Fidler et al. 2007). In the latter, Fidler et al. (2007) found that great tits artificially selected for divergent levels of exploratory behaviour differed in the allele frequency of an exonic SNP in the DRD4 gene. This association was confirmed in a natural population, where the levels of exploratory behaviour differed for birds with different genotypes (Fidler et al. 2007). This suggests that the association between DRD4 and this facet of personality is very general across vertebrates. However, when tested across samples of four different great tit populations, the association was significant in only one sample with an estimated effect size of around 5 per cent (Korsten et al. 2010). This could indicate that the DRD4 polymorphism is only linked to the functional variant in some but not all populations, or that the association is dependent on the environment or other population-specific characteristics (Korsten et al. 2010).
Using the candidate gene approach is accompanied by some challenges. First, complex traits like personality traits are expected to be influenced by numerous genes, most of them having only small effects. The approach is therefore always biased towards genes with higher effect sizes. Secondly, most genes are expected to be involved in epistatic interactions. The genetic background in natural populations is, however, variable and this might make the detection of single-locus associations complicated. Genomic regions that are more important in interaction with other loci will therefore only be detected by advanced analysis methods in random scans of the whole genome (Reif & Lesch 2003; Cordell 2009). Thirdly, there is a general difficulty of replication of results and there are often population differences in the associations. These difficulties may partly be due to the generally small effect sizes of genetic polymorphisms influencing complex traits. These challenges are, however, no reason to neglect candidate gene approaches as a starting point, but highlight that only efforts combining these with genome-wide approaches are effective in revealing the genetic architecture of personality.
3. Use of genome information for evolutionary studies on personality traits
In this section, we will describe how genome information can be used to highlight the evolutionary forces that currently act or historically have acted on personality variation, and how we can use genome information to study between-population and between-species differences in personality.
(a). Evolutionary history of genetic personality correlates
Identifying genes or genomic regions influencing personality trait variation represents a first step in describing the genetic architecture of personality. Investigating the historical and potential future evolutionary dynamics of the identified genomic region is an important second step towards understanding the evolution and maintenance of personality variation. With evolutionary dynamics or evolutionary history, we indicate the time path since the establishment of the functional polymorphism. This encompasses the selective forces that have acted on the functional polymorphism and its linked markers.
There are basically two different ways to analyse the evolutionary dynamics of personality traits: (i) investigating correlations between fitness measures and personality variants or genotypes underlying these variants; (ii) analysing the genomic region that was found to be associated with personality variation in terms of genetic structure, genetic diversity patterns and footprints of selection. The first approach will give an idea about the current fitness consequences and potential future evolutionary trajectories of different personality variants, and is ideally performed across major environmental clines for the species under study, because of an expected interactive effect between genotype and environment upon fitness (Van Oers et al. 2005; Ellegren & Sheldon 2008; for a review see Dingemanse et al. 2010a,b). The second approach allows estimation of the selection history and the age of the underlying genetic variants within the associated genomic region and provides an idea of the origin and selective forces that have shaped personality trait variation in the past. In the following, we will describe the second approach in the context of personality research.
After genetically mapping a behavioural trait, one is often confronted with the fact that not only a single genetic polymorphism (marker locus) is found to be associated with the trait of interest, but rather a couple of adjacent polymorphisms show similar strengths of association (Ioannidis et al. 2009). This is mostly owing to linkage disequilibrium among the loci in the genomic region under study. In order to delineate the associated genomic region and to perform tests for selection (see below), it is important to analyse the genetic variation of all loci surrounding the associated markers up to a distance where linkage disequilibrium decreases to a negligible level. The best way to capture all genetic variation in a genomic region without missing rare variants is sequencing all alleles in the whole region with novel techniques enabling high-throughput sequencing in large population samples. Methods such as parallel tagged sequencing (Meyer et al. 2008) or array capture approaches (Hodges et al. 2009) are suitable to produce a high number of target sequences in population samples. Moreover, sequencing has the advantage over genotyping each marker separately of directly resulting in haplotypes, where allelic phase of a large number of neighbouring loci is known. This supersedes error-prone phase estimation by population-based methods and leads to precise linkage disequilibrium estimates and more reliable haplotype gene tree reconstructions.
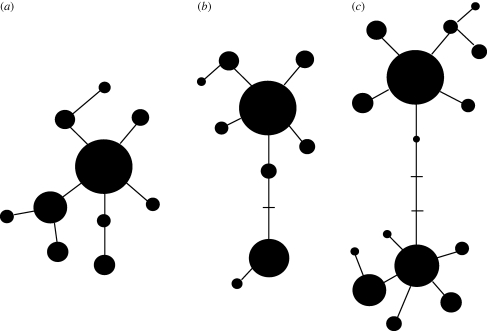
Knowing the linkage disequilibrium structure can help to assess the recombination history of a genomic region (Mueller 2004). Along with modelling approaches to estimate recombination rates (McVean et al. 2002; Li & Stephens 2004), genomic fragments can be identified, which are largely free of historical recombination. Such fragments can be seen as evolutionary units where all loci on them share the same phylogenetic history. Reconstructing the phylogenetic relationship of all haplotypes within recombination-free fragments, and the comparison between such reconstructed trees, has the potential to infer the selection history (Bamshad & Wooding 2003). For instance, more than one distinct and diverse haplotype group with common haplotypes in each would indicate long-lasting balancing selection, whereas a single (or less diverse) common haplotype distantly related to the remaining diverse haplotype group would indicate recent directional selection on this outlier haplotype (figure 1).
Figure 1.
Sketches of expected haplotype trees under (a) neutral evolution (b) recent positive selection (partial selective sweep) and (c) long-lasting balancing selection (on two distinct haplotypes). The haplotype trees show the relatedness between distinct haplotypes (black circles). The size of each circle represents the population frequency of the haplotype. The lines link the haplotypes assuming the lowest number of mutational steps. Missing intermediate haplotypes are represented by bars. The age of haplotypes can be inferred from the surrounding haplotype diversity (number of satellite haplotypes).
Other tests for selection are based on allele frequency or haplotype frequency spectra. There is a tendency for an excess of intermediate-frequency polymorphisms and, hence, positive Tajima's D (Tajima 1989) after long-lasting balancing selection. However, a signal of a recent strong increase in allele frequency (positive directional selection) may still be visible if at least one allele in a balanced polymorphism is young (shorter than expected neutral coalescent time). All scenarios of positive selection are likely to lead to the well-explored footprints of selective sweeps (for reviews see Anisimova & Liberles 2007; Jensen et al. 2007; Thornton et al. 2007). Since the locus that is associated with behavioural trait variation is still polymorphic, and its alleles have not yet reached fixation, we expect to observe the pattern of a partial sweep. Using variants of the long-range haplotype test (Sabeti et al. 2002; Mueller & Andreoli 2004; Voight et al. 2006) or tests based on the haplotype frequency spectrum (Watterson 1978; Depaulis & Veuille 1998; Zeng et al. 2007), the footprint of a partial sweep can be detected with high power. Methods that model the genetic differentiation between subpopulations can also be used to detect loci under directional selection and, to a lesser extent, loci under balancing selection (Beaumont & Balding 2004).
The DRD4 gene is certainly the best explored personality gene in terms of evolutionary dynamics. In humans, a 48 bp tandem repeat polymorphism in exon 3 of the DRD4 gene is reported to be associated with both novelty seeking behaviour and attention deficit/hyperactivity disorder in several sampled populations (Ebstein et al. 1996; Savitz & Ramesar 2004). A meta-analysis did not find a significant overall effect of the tandem repeat polymorphism on approach-related traits, but rather between-study heterogeneity (Munafo et al. 2008b). It has often been argued that such heterogeneity is based on gene × environment interactions or population-specific epistatic effects (Munafo & Flint 2009). In significant studies, often, the second most frequent allele of the DRD4 tandem repeat locus, the 7R allele, is associated with higher levels of novelty seeking (Savitz & Ramesar 2004). There is, however, strong geographical variation in the frequency of the 7R allele, indicating an adaptive value of this allele. East Asians generally possess low proportions of 7R alleles (1% or less), most African, native North American and European populations have intermediate frequencies (around 20%), whereas South American Indians can have high proportions (up to 78%; Chen et al. 1999). This pattern has been associated with population migration distances in prehistoric times. Across six different migration routes, the populations that remained near their putative origins showed a lower proportion of 7R alleles than those that migrated farther away (Chen et al. 1999). It was argued that increased exploratory behaviour is adaptive in migratory societies because it allowed for more successful exploitation of resources in the novel environments (Chen et al. 1999).
The adaptive value of the 7R allele in prehistoric migration is in line with the interpretation that the 7R allele originated as a rare mutational event, and increased to high frequency by positive selection in some populations (but see Wang et al. 2004; Hattori et al. 2009). It is hypothesized that the 7R allele arose from the most common 4R allele by multiple low probability steps of mutation and recombination (Ding et al. 2002). Strong linkage disequilibrium between the 7R allele and surrounding DRD4 polymorphisms indicate the relative young age of the 7R allele in comparison to the other major alleles (4R and 2R). A comparison of the intra-allelic variability estimates the age (of the most recent common ancestor) of the 7R alleles at about 45 000 years (Wang et al. 2004). Moreover, the site frequency spectrum of extended 7R haplotypes indicates historical positive selection (Wang et al. 2004). By contrast, the ancestral 4R haplotypes have the fewest amino acid changing variants, implying purifying selection (Ding et al. 2002). Interestingly, the overall site frequency spectrum across all haplotypes and the fact of geographically dependent high frequencies of 2R, 4R and 7R alleles are consistent with a balanced selection system (Wang et al. 2004). Such multi-allelic adaptive genetic variation is probably common. Its detection would only require allele-specific analyses of selection (see also Pitcher & Neff 2006).
(b). Genetic covariation between personality traits
Estimates of genetic correlations are fundamental to understand the evolution of behavioural constructs like personality or behavioural syndromes. As known from quantitative genetic studies, heritabilities of and therefore genetic correlations between personality traits may vary across different environments (Dingemanse et al. 2010a,b; Van Oers & Sinn 2010). The underlying molecular mechanisms behind these genetic correlations are mostly unknown. A QTL mapping study on chickens found that variation in feather pecking (FP) behaviour of juveniles was not explained by the same QTLs for the same behaviour as adults (Buitenhuis et al. 2005). A QTL for open-field behaviour in juvenile chickens did match the one for adult FP behaviour, indicating the presence of closely linked genes for open-field behaviour in young laying hens and FP in adult laying hens (Buitenhuis et al. 2005).
Although direct evidence is scarce, there are, however, some ideas about the genetic architecture of complex traits. Recent GWA studies revealed many associated loci at potentially regulatory sites with small effects and only a few loci, often at coding regions, with moderate effects (Flint & Mackay 2009). As the causal link between a genetic variant and behavioural variation is mediated by a network of coregulated genes and physiological processes, pleiotropic effects of single genes are likely to be the rule rather than the exception (Weiss 2008; Flint & Mackay 2009). Similar pleiotropic effects at many loci in a genetic network could produce the genetic covariation as seen in behavioural syndromes. However, more plausible seems to be the existence of a few ‘master genes’ orchestrating the covariation of behavioural traits (see also Aubin-Horth et al. 2009). Such strong effects postulated for master genes have been observed as epistatic interactions and suggest pervasive non-additive effects among loci (Shao et al. 2008). There are concepts that such hidden genetic variation is released in response to selection owing to environmental dynamics (Le Rouzic & Carlborg 2007). One might hypothesize that these sometimes hidden epistatic effects underlie the covariation between personality traits and its variation across different environments.
(c). Comparative genomics
Studies have started to compare phenotypic (Bell 2005; Dingemanse et al. 2007) and molecular genetic (Korsten et al. 2010) variation in personality between natural populations. Differences in trait values between individuals within a population are supposed to have the same value as differences in trait value between populations, permitting a direct comparison among the relative trait values of individuals to one another. A very interesting but challenging next step is to perform comparisons across species with different phylogenies, ecologies and social systems (Uher 2008). The diversity across species would on the one hand be the motivation to undertake multiple species comparisons, since this might inform us about the evolutionary trace of personality variation. On the other hand, this also entails particular challenges in methodology and some caution is required when comparing the same behavioural test conducted on different species or populations. Species may exhibit different or even unique personality traits and the behavioural response towards a standardized challenge may be species-specific. Hence, it is hard to distinguish whether trait variation between two species is caused by variance within a trait, differences in trait expression, or because one is simply measuring two different behavioural traits.
Several behavioural approaches have been suggested to compare personality traits of different species, mostly based on factor analyses within species (Capitanio 2004). A novel solution was presented by Uher (2008), where she introduced a bottom-up approach to characterize and compare personalities of different species. All observable behaviours naturally occurring in a species are thereby measured and grouped into units of behaviours that belong to a certain situation. These units are then reduced by merging all those that represent similar behavioural traits measured in different situations. Repeatable units or domains are then reserved and form the species-specific personality structure (Uher 2008). However, a great drawback for studies interested in the evolutionary background of personality are the practical limitations, since for most species it is often impossible to measure all behaviours in detail on a large enough sample set (Van Oers 2008).
An alternative way around the difficulty of measuring species differences in personality traits is the use of comparative genomics (see also Bell & Aubin-Horth 2010). It is increasingly recognized that comparative genomics, where sequences from two or more species are aligned and compared, is a powerful tool for detecting regions that evolve under negative, positive or balancing selection, indicative of functionality. By examining genome sequences from multiple species, comparative genomics offers new insight into genome evolution and the way natural selection moulds DNA sequence evolution (Ellegren 2008b). Adaptive evolution can be inferred from, for example, protein-coding sequences showing an increased rate of non-synonymous substitutions in divergence compared with presumed neutral sequence data, or a high frequency of derived alleles (Mitchell-Olds et al. 2007).
Only genetic sites that show signs of adaptive evolution in genomic comparisons between species and are still polymorphic within at least one of the species are interesting for studies on personality variation. Genetic regions that show adaptive signatures in comparative studies can still be polymorphic when the selective sweep is very recent or when a balancing selection process maintains the genetic variance. This specific point has been discussed in more detail by Penke et al. (2010) for SNPs within the ADRB2 gene, which is associated with cognitive ability. A derived allele that was under positive selection in a human–chimpanzee comparison had gone to fixation in some human populations, though the ancestral allele had been shown to be of positive effect at later age. This was not the case in a Scottish population, where the protective effects on old-age cognitive ability were high enough to maintain the genetic variability (Penke et al. 2010). In the long run, antagonistic pleiotropic trade-offs that involve such polymorphisms under positive selection are likely to be evolutionary unstable (Roff & Fairbairn 2007). The general appearance of many personality domains in various species therefore indicates that personality polymorphisms are under some form of balancing selection.
Apart from comparing polymorphisms within candidate genes between species (see above), a next step could be to use cross-species QTL concordance as a tool for QTL dissection. This technique is, for example, used with mouse emotionality and human neuroticism (Willis-Owen & Flint 2007; Fullerton et al. 2008), but could also be used for comparing QTL results from model species with non-model species. A possible limitation of this method is that it seems likely that the genetic determinants of traits may not have been flawlessly preserved throughout evolution (Willis-Owen & Flint 2007), and it is therefore to be expected that the number of loci to be found will be highly dependent on the genetic distance of the two compared species. Comparative genomics might thereby be of help to identify general rules and patterns across species without possessing the difficulties of comparative behavioural measurements. This is true for the genetic basis of variation in the personality traits themselves, but also for the genetic correlation among these traits.
4. Outlook
Studies on animal personality traits within the field of molecular genetics and genomics now need to make the step to natural populations. While this was already feasible for studies using candidate genes, this now also comes within reach for genome-wide mapping studies. Compilations of polymorphisms are currently built up for natural populations of several non-model species and will serve as a basis for linkage and association studies. With these tools more and more functional polymorphisms will be detected. The detection and evolutionary analysis of genomic loci associated with personality traits will certainly lead to an understanding of why personality variation in natural populations is maintained and why genetic correlations are present in some cases and not in others. The subsequent comparison of evolutionary trajectories across different populations and species will be informative for the analysis of personality evolution under different ecological settings.
Acknowledgements
We thank Lars Penke and one anonymous reviewer for their valuable comments on an earlier version of the manuscript. K.v.O. was supported by a NGI-HORIZON grant.
Footnotes
One contribution of 16 to a Theme Issue ‘Evolutionary and ecological approaches to the study of personality’.
References
- Anisimova M., Liberles D. A.2007The quest for natural selection in the age of comparative genomics. Heredity 99, 567–579 10.1038/sj.hdy.6801052 (doi:10.1038/sj.hdy.6801052) [DOI] [PubMed] [Google Scholar]
- Arnold K. E., Ramsay S. L., Donaldson C., Adam A.2007Parental prey selection affects risk-taking behaviour and spatial learning in avian offspring. Proc. R. Soc. B 274, 2563–2569 10.1098/rspb.2007.0687 (doi:10.1098/rspb.2007.0687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N., Letcher B. H., Hofmann H. A.2009Gene-expression signatures of Atlantic salmon's plastic life cycle. Gen. Comp. Endocrinol. 163, 278–284 10.1016/j.ygcen.2009.04.021 (doi:10.1016/j.ygcen.2009.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N., et al. 2010The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20, 485–495 10.1101/gr.101410.109 (doi:10.1101/gr.101410.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M., Wooding S. P.2003Signatures of natural selection in the human genome. Nat. Rev. Genet. 4, 99–111 10.1038/nrg999 (doi:10.1038/nrg999) [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Balding D. J.2004Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13, 969–980 10.1111/j.1365-294X.2004.02125.x (doi:10.1111/j.1365-294X.2004.02125.x) [DOI] [PubMed] [Google Scholar]
- Bell A. M.2005Behavioural differences between individuals and two populations of stickleback (Gasterosteus aculeatus). J. Evol. Biol. 18, 464–473 10.1111/j.1420-9101.2004.00817.x (doi:10.1111/j.1420-9101.2004.00817.x) [DOI] [PubMed] [Google Scholar]
- Bell A. M., Aubin-Horth N.2010What can whole genome expression data tell us about the ecology and evolution of personality? Phil. Trans. R. Soc. B 365, 4001–4012 10.1098/rstb.2010.0185 (doi:10.1098/rstb.2010.0185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmler W., Carr T., Thisse C., Thisse B., Canfield V. A., Levenson R.2007D4 Dopamine receptor genes of zebrafish and effects of the antipsychotic clozapine on larval swimming behaviour. Genes Brain Behav. 6, 155–166 10.1111/j.1601-183X.2006.00243.x (doi:10.1111/j.1601-183X.2006.00243.x) [DOI] [PubMed] [Google Scholar]
- Brydges N. M., Colegrave N., Heathcote R. J. P., Braithwaite V. A.2008Habitat stability and predation pressure affect temperament behaviours in populations of three-spined sticklebacks. J. Anim. Ecol. 77, 229–235 10.1111/j.1365-2656.2007.01343.x (doi:10.1111/j.1365-2656.2007.01343.x) [DOI] [PubMed] [Google Scholar]
- Buitenhuis A. J., et al. 2005Quantitative trait loci for behavioural traits in chickens. Livestock Prod. Sci. 93, 95–103 10.1016/j.livprodsci.2004.11.010 (doi:10.1016/j.livprodsci.2004.11.010) [DOI] [Google Scholar]
- Capitanio J. P.2004Personality factors between and within species. In Macaque societies (eds Tieryy B., Singh M., Kaufmanns W.), pp. 13–33 Cambridge, UK: Cambridge University Press [Google Scholar]
- Chen C. S., Burton M., Greenberger E., Dmitrieva J.1999Population migration and the variation of Dopamine D4 Receptor (DRD4) allele frequencies around the globe. Evol. Hum. Behav. 20, 309–324 10.1016/S1090-5138(99)00015-X (doi:10.1016/S1090-5138(99)00015-X) [DOI] [Google Scholar]
- Christiansen S. B., Forkman B.2007Assessment of animal welfare in a veterinary context—a call for ethologists. Appl. Anim. Behav. Sci. 106, 203–220 10.1016/j.applanim.2007.01.004 (doi:10.1016/j.applanim.2007.01.004) [DOI] [Google Scholar]
- Comings D. E., et al. 2000A multivariate analysis of 59 candidate genes in personality traits: the temperament and character inventory. Clin. Genet. 58, 375–385 10.1034/j.1399-0004.2000.580508.x (doi:10.1034/j.1399-0004.2000.580508.x) [DOI] [PubMed] [Google Scholar]
- Cordell H. J.2009Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 10.1038/nrg2579 (doi:10.1038/nrg2579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall S. R. X.2004Behavioural biology: fortune favours bold and shy personalities. Curr. Biol. 14, R470–R472 10.1016/j.cub.2004.06.011 (doi:10.1016/j.cub.2004.06.011) [DOI] [PubMed] [Google Scholar]
- de Bakker P. I. W., Yelensky R., Pe'er I., Gabriel S. B., Daly M. J., Altshuler D.2005Efficiency and power in genetic association studies. Nat. Genet. 37, 1217–1223 10.1038/ng1669 (doi:10.1038/ng1669) [DOI] [PubMed] [Google Scholar]
- de Moor M. H. M., et al. 2009Meta-analysis of genome-wide association results in >10 000 individuals for the big five personality traits. Behav. Genet. 39, 643–643 [Google Scholar]
- Depaulis F., Veuille M.1998Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15, 1788–1790 [DOI] [PubMed] [Google Scholar]
- Ding Y. C., et al. 2002Evidence of positive selection acting at the human dopamine receptor D4 gene locus. Proc. Natl Acad. Sci. USA 99, 309–314 10.1073/pnas.012464099 (doi:10.1073/pnas.012464099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemanse N. J., Wright J., Kazem A. J. N., Thomas D. K., Hickling R., Dawnay N.2007Behavioural syndromes differ predictably between 12 populations of three-spined stickleback. J. Anim. Ecol. 76, 1128–1138 10.1111/j.1365-2656.2007.01284.x (doi:10.1111/j.1365-2656.2007.01284.x) [DOI] [PubMed] [Google Scholar]
- Dingemanse N. J., Dochterman N., Wright J.2010aA method for exploring the structure of behavioural syndromes to allow formal comparison within and between data sets. Anim. Behav. 79, 439–450 10.1016/j.anbehav.2009.11.024 (doi:10.1016/j.anbehav.2009.11.024) [DOI] [Google Scholar]
- Dingemanse N. J., Kazem A. J. N., Réale D., Wright J.2010bBehavioural reaction norms: animal personality meets individual plasticity. Trends Ecol. Evol. 25, 81–89 10.1016/j.tree.2009.07.013 (doi:10.1016/j.tree.2009.07.013) [DOI] [PubMed] [Google Scholar]
- Ebstein R. P., et al. 1996Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of novelty seeking. Nat. Genet. 12, 78–80 10.1038/ng0196-78 (doi:10.1038/ng0196-78) [DOI] [PubMed] [Google Scholar]
- Eley T. C., Plomin R.1997Genetic analyses of emotionality. Curr. Opin. Neurobiol. 7, 279–284 10.1016/S0959-4388(97)80017-7 (doi:10.1016/S0959-4388(97)80017-7) [DOI] [PubMed] [Google Scholar]
- Ellegren H.2008aComparative genomics and the study of evolution by natural selection. Mol. Ecol. 17, 4586–4596 10.1111/j.1365-294X.2008.03954.x (doi:10.1111/j.1365-294X.2008.03954.x) [DOI] [PubMed] [Google Scholar]
- Ellegren H.2008bSequencing goes 454 and takes large-scale genomics into the wild. Mol. Ecol. 17, 1629–1631 10.1111/j.1365-294X.2008.03699.x (doi:10.1111/j.1365-294X.2008.03699.x) [DOI] [PubMed] [Google Scholar]
- Ellegren H., Sheldon B. C.2008Genetic basis of fitness differences in natural populations. Nature 452, 169–175 10.1038/nature06737 (doi:10.1038/nature06737) [DOI] [PubMed] [Google Scholar]
- Erickson D. L., Fenster C. B., Stenoien H. K., Price D.2004Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13, 2505–2522 10.1111/j.1365-294X.2004.02254.x (doi:10.1111/j.1365-294X.2004.02254.x) [DOI] [PubMed] [Google Scholar]
- Fidler A. E., Van Oers K., Drent P. J., Kuhn S., Mueller J. C., Kempenaers B.2007Drd4 gene polymorphisms are associated with personality variation in a passerine bird. Proc. R. Soc. B 274, 1685–1691 10.1098/rspb.2007.0337 (doi:10.1098/rspb.2007.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick M. J., Ben-Shahar Y., Smid H. M., Vet L. E. M., Robinson G. E., Sokolowski M. B.2005Candidate genes for behavioural ecology. Trends Ecol. Evol. 20, 96–104 10.1016/j.tree.2004.11.017 (doi:10.1016/j.tree.2004.11.017) [DOI] [PubMed] [Google Scholar]
- Flint J., Mackay T. F. C.2009Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19, 723–733 10.1101/gr.086660.108 (doi:10.1101/gr.086660.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton J. M., et al. 2008Human-mouse quantitative trait locus concordance and the dissection of a human neuroticism locus. Biol. Psychiatry 63, 874–883 10.1016/j.biopsych.2007.10.019 (doi:10.1016/j.biopsych.2007.10.019) [DOI] [PubMed] [Google Scholar]
- Gershenfeld H. K., Neumann P. E., Mathis C., Crawley J. N., Li X. H., Paul S. M.1997Mapping quantitative trait loci for open-field behavior in mice. Behav. Genet. 27, 201–210 10.1023/A:1025653812535 (doi:10.1023/A:1025653812535) [DOI] [PubMed] [Google Scholar]
- Gillespie N. A., Zhu G., Evans D. M., Medland S. E., Wright M. J., Martin N. G.2008A genome-wide scan for eysenckian personality dimensions in adolescent twin sibships: psychoticism, extraversion, neuroticism, and lie. J. Pers. 76, 1415–1445 10.1111/j.1467-6494.2008.00527.x (doi:10.1111/j.1467-6494.2008.00527.x) [DOI] [PubMed] [Google Scholar]
- Gordon J. A., Hen R.2004Genetic approaches to the study of anxiety. Annu. Rev. Neurosci. 27, 193–222 10.1146/annurev.neuro.27.070203.144212 (doi:10.1146/annurev.neuro.27.070203.144212) [DOI] [PubMed] [Google Scholar]
- Gosling S. D.2008Personality in non-human animals. Soc. Pers. Psychol. Compass 2, 985–1001 10.1111/j.1751-9004.2008.00087.x (doi:10.1111/j.1751-9004.2008.00087.x) [DOI] [Google Scholar]
- Gutierrez-Gil B., Ball N., Burton D., Haskell M., Williams J. L., Wiener P.2008Identification of quantitative trait loci affecting cattle temperament. J. Hered. 99, 629–638 10.1093/jhered/esn060 (doi:10.1093/jhered/esn060) [DOI] [PubMed] [Google Scholar]
- Hattori E., Nakajima M., Yamada K., Iwayama Y., Toyota T., Saitou N., Yoshikawa T.2009Variable number of tandem repeat polymorphisms of DRD4: re-evaluation of selection hypothesis and analysis of association with schizophrenia. Eur. J. Hum. Genet. 17, 793–801 10.1038/ejhg.2008.247 (doi:10.1038/ejhg.2008.247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E., Rooks M., Xuan Z. Y., Bhattacharjee A., Gordon D. B., Brizuela L., McCombie W. R., Hannon G. J.2009Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 4, 960–974 10.1038/nprot.2009.68 (doi:10.1038/nprot.2009.68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta I., Barlow C.2008Molecular genetics of anxiety in mice and men. Ann. Med. 40, 92–109 10.1080/07853890701747096 (doi:10.1080/07853890701747096) [DOI] [PubMed] [Google Scholar]
- Ioannidis J. P. A., Thomas G., Daly M. J.2009Genome-wide association studies validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10, 318–329 10.1038/nrg2544 (doi:10.1038/nrg2544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., et al. 2004Allele frequency distribution of the canine dopamine receptor D4 gene exon III and I in 23 breeds. J. Vet. Med. Sci. 66, 815–820 10.1292/jvms.66.815 (doi:10.1292/jvms.66.815) [DOI] [PubMed] [Google Scholar]
- Jensen J. D., Wong A., Aquadro C. F.2007Approaches for identifying targets of positive selection. Trends Genet. 23, 568–577 10.1016/j.tig.2007.08.009 (doi:10.1016/j.tig.2007.08.009) [DOI] [PubMed] [Google Scholar]
- John O. P., Naumann L. P., Soto C. J.2008Paradigm shift to the integrative big-five trait taxonomy: history, measurement, and conceptual issues. In Handbook of personality: theory and research (eds John O. P., Robins R. W., Pervin L. A.), pp. 114–158 New York, NY: Guilford Press [Google Scholar]
- Koolhaas J. M., De Boer S. F., Buwalda B., Van der Vegt B. J., Carere C., Groothuis A. G. G.2001How and why coping systems vary among individuals. In Coping with challenge: welfare in animals including humans (ed. Broom D. M.), pp. 197–209 Dahlem, Berlin: Dahlem University Press [Google Scholar]
- Korsten P., et al. 2010Association between DRD4 gene polymorphism and personality variation in great tits: a test across four wild populations. Mol. Ecol. 19, 832–843 10.1111/j.1365-294X.2009.04518.x (doi:10.1111/j.1365-294X.2009.04518.x) [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B.2004Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. Lond. B 359, 873–890 10.1098/rstb.2003.1437 (doi:10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A., Carlborg O.2007Evolutionary potential of hidden genetic variation. Trends Ecol. Evol. 23, 33–37 10.1016/j.tree.2007.09.014 (doi:10.1016/j.tree.2007.09.014) [DOI] [PubMed] [Google Scholar]
- Li N., Stephens M.2004Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B.1998Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates, Inc [Google Scholar]
- Mardis E. R.2008Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402 10.1146/annurev.genom.9.081307.164359 (doi:10.1146/annurev.genom.9.081307.164359) [DOI] [PubMed] [Google Scholar]
- McVean G., Awadalla P., Fearnhead P.2002A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Stenzel U., Hofreiter M.2008Parallel tagged sequencing on the 454 platform. Nat. Protoc. 3, 267–278 10.1038/nprot.2007.520 (doi:10.1038/nprot.2007.520) [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T., Willis J. H., Goldstein D. B.2007Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 8, 845–856 10.1038/nrg2207 (doi:10.1038/nrg2207) [DOI] [PubMed] [Google Scholar]
- Mueller J. C.2004Linkage disequilibrium for different scales and applications. Brief. Bioinform. 5, 355–364 10.1093/bib/5.4.355 (doi:10.1093/bib/5.4.355) [DOI] [PubMed] [Google Scholar]
- Mueller J. C., Andreoli C.2004Plotting haplotype-specific linkage disequilibrium patterns by extended haplotype homozygosity. Bioinformatics 20, 786–787 10.1093/bioinformatics/btg481 (doi:10.1093/bioinformatics/btg481) [DOI] [PubMed] [Google Scholar]
- Munafo M. R., Flint J.2009Replication and heterogeneity in gene × environment interaction studies. Int. J. Neuropsychopharmacol. 12, 727–729 10.1017/S1461145709000479 (doi:10.1017/S1461145709000479) [DOI] [PubMed] [Google Scholar]
- Munafo M. R., Brown S. M., Harkless K. C.2008aSerotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol. Psychiatry 63, 852–857 10.1016/j.biopsych.2007.08.016 (doi:10.1016/j.biopsych.2007.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo M. R., Yalcin B., Willis-Owen S. A., Flint J.2008bAssociation of the dopamine D4 receptor (DRD4) gene and approach-related personality traits: meta-analysis and new data. Biol. Psychiatry 63, 197–206 10.1016/j.biopsych.2007.04.006 (doi:10.1016/j.biopsych.2007.04.006) [DOI] [PubMed] [Google Scholar]
- Munafo M. R., Freimer N. B., Ng W., Ophoff R., Veijola J., Miettunen J., Jrvelin M. R., Taanila A., Flint J.20095-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. J. Med. Chem. 150B, 271–281 10.1002/ajmg.b.30808 (doi:10.1002/ajmg.b.30808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke L., Denissen J. J. A., Miller G. F.2007The evolutionary genetics of personality. Eur. J. Pers. 21, 549–587 10.1002/per.629 (doi:10.1002/per.629) [DOI] [Google Scholar]
- Penke L., Muñoz Maniega S., Houlihan L. M., Murray C., Gow A. J., Clayden J. D., Bastin M. E., Wardlaw J. M., Deary I. J.2010White matter integrity in the splenium of the corpus callosum is related to successful cognitive aging and partly mediates the protective effect of an ancestral polymorphism in ADRB2. Behav. Genet. 40, 146–156 10.1007/s10519-009-9318-4 (doi:10.1007/s10519-009-9318-4) [DOI] [PubMed] [Google Scholar]
- Pitcher T. E., Neff B. D.2006MHC class IIB alleles contribute to both additive and nonadditive genetic effects on survival in Chinook salmon. Mol. Ecol. 15, 2357–2365 10.1111/j.1365-294X.2006.02942.x (doi:10.1111/j.1365-294X.2006.02942.x) [DOI] [PubMed] [Google Scholar]
- Quinn J. L., Cresswell W.2005Personality, anti-predation behaviour and behavioural plasticity in the chaffinch Fringilla coelebs. Behaviour 142, 1377–1402 10.1163/156853905774539391 (doi:10.1163/156853905774539391) [DOI] [Google Scholar]
- Quinn J. L., Patrick S. C., Bouwhuis S., Wilkin T. A., Sheldon B. C.2009Heterogeneous selection on a heritable temperament trait in a variable environment. J. Anim. Ecol. 78, 1203–1215 10.1111/j.1365-2656.2009.01585.x (doi:10.1111/j.1365-2656.2009.01585.x) [DOI] [PubMed] [Google Scholar]
- Réale D., Reader S. M., Sol D., McDougall P. T., Dingemanse N. J.2007Integrating temperament in ecology and evolutionary biology. Biol. Rev. 82, 291–318 10.1111/j.1469-185X.2007.00010.x (doi:10.1111/j.1469-185X.2007.00010.x) [DOI] [PubMed] [Google Scholar]
- Reif A., Lesch K. P.2003Toward a molecular architecture of personality. Behav. Brain Res. 139, 1–20 10.1016/S0166-4328(02)00267-X (doi:10.1016/S0166-4328(02)00267-X) [DOI] [PubMed] [Google Scholar]
- Risch N. J.2000Searching for genetic determinants in the new millennium. Nature 405, 847–856 10.1038/35015718 (doi:10.1038/35015718) [DOI] [PubMed] [Google Scholar]
- Rodenburg T. B., Komen H., Ellen E. D., Uitdehaag K. A., van Arendonk J. A. M.2008Selection method and early-life history affect behavioural development, feather pecking and cannibalism in laying hens: a review. Appl. Anim. Behav. Sci. 110, 217–228 10.1016/j.applanim.2007.09.009 (doi:10.1016/j.applanim.2007.09.009) [DOI] [Google Scholar]
- Roff D. A.1997Evolutionary quantitative genetics. New York, NY: Chapmann & Hall [Google Scholar]
- Roff D. A., Fairbairn D. J.2007The evolution of trade-offs: where are we? J. Evol. Biol. 20, 433–447 10.1111/j.1420-9101.2006.01255.x (doi:10.1111/j.1420-9101.2006.01255.x) [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., et al. 2002Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 10.1038/nature01140 (doi:10.1038/nature01140) [DOI] [PubMed] [Google Scholar]
- Savitz J. B., Ramesar R. S.2004Genetic variants implicated in personality: a review of the more promising candidates. Am. J. Med. Genet. Part B Neuropsychiatric Genet. 131B, 20–32 10.1002/ajmg.b.20155 (doi:10.1002/ajmg.b.20155) [DOI] [PubMed] [Google Scholar]
- Shao H., et al. 2008Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl Acad. Sci. USA 105, 19 910–19 914 10.1073/pnas.0810388105 (doi:10.1073/pnas.0810388105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M. K., Inoue-Murayama M., Ueda Y., Maejima M., Murayama Y., Takenaka O., Hayasaka I., Ito S.2004Polymorphism in the second intron of dopamine receptor D4 gene in humans and apes. Biochem. Biophys. Res. Commun. 316, 1186–1190 10.1016/j.bbrc.2004.03.006 (doi:10.1016/j.bbrc.2004.03.006) [DOI] [PubMed] [Google Scholar]
- Sih A., Bell A. M.2008Insights for behavioral ecology from behavioral syndromes. Adv. Study Behav. 38, 227–281 10.1016/S0065-3454(08)00005-3 (doi:10.1016/S0065-3454(08)00005-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J.2005Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14, 363–379 10.1111/j.1365-294X.2004.02378.x (doi:10.1111/j.1365-294X.2004.02378.x) [DOI] [PubMed] [Google Scholar]
- Smith B. R., Blumstein D. T.2008Fitness consequences of personality: a meta-analysis. Behav. Ecol. 19, 448–455 10.1093/beheco/arm144 (doi:10.1093/beheco/arm144) [DOI] [Google Scholar]
- Sokolowski M. B.2001Drosophila: genetics meets behaviour. Nat. Rev. Genet. 2, 879–890 10.1038/35098592 (doi:10.1038/35098592) [DOI] [PubMed] [Google Scholar]
- Tajima F.1989Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A., et al. 2008Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry, 15, 647–656 10.1038/mp.2008.113 (doi:10.1038/mp.2008.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. R., Jensen J. D., Becquet C., Andolfatto P.2007Progress and prospects in mapping recent selection in the genome. Heredity 98, 340–348 10.1038/sj.hdy.6800967 (doi:10.1038/sj.hdy.6800967) [DOI] [PubMed] [Google Scholar]
- Turelli M., Barton N. H.2004Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and GxE interactions. Genetics 166, 1053–1079 10.1534/genetics.166.2.1053 (doi:10.1534/genetics.166.2.1053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher J.2008Comparative personality research: methodological approaches. Eur. J. Pers. 22, 427–455 10.1002/per.680 (doi:10.1002/per.680) [DOI] [Google Scholar]
- Van Bers N. E. M., Van Oers K., Kerstens H. H. D., Dibbits B. W., Crooijmans R. P. M. A., Visser M. E., Groenen M. A. M.2010Genome-wide SNP detection in the great tit Parus major using high throughput sequencing. Mol. Ecol. 19(Suppl. 1), 89–99 10.1111/j.1365-294X.2009.04486.x (doi:10.1111/j.1365-294X.2009.04486.x) [DOI] [PubMed] [Google Scholar]
- Van Oers K.2008Animal personality, behaviours or traits: what are we measuring? Eur. J. Pers. 22, 457–474 [Google Scholar]
- Van Oers K., Sinn D. L.In press The quantitative and molecular genetics of animal personality. In Animal personalities: behavior, physiology, and evolution (eds Carere C., Maestripieri D.), Chicago, IL: University of Chicago Press [Google Scholar]
- Van Oers K., Sinn D. L.2010Towards a basis for the phenotypic gambit: advances in the evolutionary genetics of animal personality. In From genes to behavior (eds Inoue-Murayama M., Kawamura S., Inoue E., Weiss A.), pp. 165–183 Tokyo, Japan: Springer [Google Scholar]
- Van Oers K., De Jong G., Van Noordwijk A. J., Kempenaers B., Drent P. J.2005Contribution of genetics to the study of animal personalities: a review of case studies. Behaviour 142, 1185–1206 10.1163/156853905774539364 (doi:10.1163/156853905774539364) [DOI] [Google Scholar]
- Van Oers K., Drent P. J., Dingemanse N. J., Kempenaers B.2008Personality is associated with extra-pair paternity in great tits (Parus major). Anim. Behav. 76, 555–563 10.1016/j.anbehav.2008.03.011 (doi:10.1016/j.anbehav.2008.03.011) [DOI] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X. Q., Pritchard J. K.2006A map of recent positive selection in the human genome. PLoS Biol. 4, e72. 10.1371/journal.pbio.0040072 (doi:10.1371/journal.pbio.0040072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., et al. 2004The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am. J. Hum. Genet. 74, 931–944 10.1086/420854 (doi:10.1086/420854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A.1978Homozygosity test of neutrality. Genetics 88, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K. M.2008Tilting at quixotic trait loci (QTL): an evolutionary perspective on genetic causation. Genetics 179, 1741–1756 10.1534/genetics.108.094128 (doi:10.1534/genetics.108.094128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S., Chen B. W., Bermejo J. L., Canzian F.2009Is there still a need for candidate gene approaches in the era of genome-wide association studies? Genomics 93, 415–419 10.1016/j.ygeno.2008.12.011 (doi:10.1016/j.ygeno.2008.12.011) [DOI] [PubMed] [Google Scholar]
- Willis-Owen S. A. G., Flint J.2007Identifying the genetic determinants of emotionality in humans; insights from rodents. Neurosci. Biobehav. Rev. 31, 115–124 10.1016/j.neubiorev.2006.07.006 (doi:10.1016/j.neubiorev.2006.07.006) [DOI] [PubMed] [Google Scholar]
- Zeng K., Shi S., Wut C. I.2007Compound tests for the detection of hitchhiking under positive selection. Mol. Biol. Evol. 24, 1898–1908 10.1093/molbev/msm119 (doi:10.1093/molbev/msm119) [DOI] [PubMed] [Google Scholar]