Abstract
Cationic antimicrobial host defense peptides (HDPs) combat infection by directly killing a wide variety of microbes, and/or modulating host immunity. HDPs have great therapeutic potential against antibiotic-resistant bacteria, viruses and even parasites, but there are substantial roadblocks to their therapeutic application. High manufacturing costs associated with amino acid precursors have limited the delivery of inexpensive therapeutics through industrial-scale chemical synthesis. Conversely, the production of peptides in bacteria by recombinant DNA technology has been impeded by the antimicrobial activity of these peptides and their susceptibility to proteolytic degradation, while subsequent purification of recombinant peptides often requires multiple steps and has not been cost-effective. Here we have developed methodologies appropriate for large-scale industrial production of HDPs; in particular, we describe (i) a method, using fusions to SUMO, for producing high yields of intact recombinant HDPs in bacteria without significant toxicity; and (ii) a simplified 2-step purification method appropriate for industrial use. We have used this method to produce seven HDPs to date (IDR1, MX226, LL37, CRAMP, HHC-10, E5 and E6). Using this technology, pilot-scale fermentation (10 L) was performed to produce large quantities of biologically active cationic peptides. Together, these data indicate that this new method represents a cost-effective means to enable commercial enterprises to produce HDPs in large-scale under Good Laboratory Manufacturing Practice (GMP) conditions for therapeutic application in humans.
Keywords: Antimicrobial peptide, Host-defense peptide, bacterial expression system
Introduction
The increase in resistance of bacterial strains to existing antibiotics is a major public health concern (WHO (2008) World Health Statistics 2008. World Health Organization Press), and has spurred intensive efforts to develop new classes of antibiotics that are effective against antibiotic-resistant strains, with limited success [37]. The anti-infective properties of HDPs, evolutionary conserved biodefence molecules found in all species of life [43], are mediated by direct antimicrobial activities, modulation of host immune responses, or both. Although initially termed antimicrobial peptides (AMPs) due to their ability to kill microbes directly, the immunomodulatory properties associated with many of these peptides have resulted in their designation as host defense peptides (HDPs) [13]. These are characterized by being short (12–50 amino acids), having an overall positive charge of +2 to +9, and sufficient hydrophobicity to allow for interaction with or traversing of membranes of target cells [21] [18].
HDPs are elements of the host innate immune defense system. The upregulation and/or release of HDPs is induced upon infection with bacteria or viruses. The more than 1000 naturally occurring HDPs are divided into 4 broad structural classes, which represent amphipathic α-helixes (e.g. cathelicidins), β-sheets with 2–4 disulfide bridges (α and β defensins and protegrins), extended structures (indolicidin), and beta-loop peptides (brevinin) [19]. Intensive research has led to the development of synthetic peptides with enhanced antimicrobial activities based on their natural counterparts as well as IDRs (innate defense regulators), which do not need to kill microbes, but rather stimulate the host immune defenses to facilitate clearance of an infection in vivo [12, 13, 20, 36].
HDPs have received increased interest as antimicrobial therapeutics, because of their antimicrobial activity against a variety of pathogenic bacteria, including those that are resistant to conventional antibiotics [2]. HDPs display minimal inhibitory concentrations (MIC) as low as 0.25 µg/mL in vitro [28], and their capacity to engender resistance is lower than that of conventional antibiotics. This likely occurs either because HDPs nonspecifically interact with bacterial membranes or because they act on multiple targets [13, 19, 41]. Nevertheless, constitutive or induced resistance to HDPs has been reported in several pathogenic bacteria, and can be recapitulated in vitro (for an overview see [22, 32, 41]).
Several groups have taken advantage of advances in modeling and QSAR (Quantitative Structure Activity Relationships) methods [9, 33], as well as an understanding of how endogenous peptides affect innate immune defenses [6, 11, 42], to develop synthetic HDPs and IDRs not found in Nature. QSAR is a mathematical relation between the biological activity of a molecular system and its geometric and chemical characteristics. Most structure-function studies provide a working conceptual model of bioactive models. In summary, QSAR studies attempt to find a consistent relationship between biological activity and molecular properties, so that these “rules” can be used to evaluate the activity of new compounds.
Synthetic peptides with antimicrobial activity and/or immunomodulatory capabilities have proven remarkably effective when used in animal models of infection against diverse pathogens including MRSA and malaria among others [1, 5, 30].
Despite their promise as therapeutics, clinical trials with endogenous or synthetic HDPs have been limited, with none yet approved for use in humans [2], although other peptide drugs have been approved such as the HIV fusion inhibitor Fuzeon. One important contributing factor is that the costs of manufacturing are high, making the price per dose quite expensive. Current production methodologies center on solid phase peptide synthesis, but require expensive precursor components. Several reports have introduced methods for producing HDPs in bacteria or yeast [8, 17, 26, 31, 44], but to date, scale-up has not proven cost-effective. Even when production is possible, purification requires multiple biochemical steps that are expensive and significantly reduce yield.
Here, we describe a procedure to produce HDPs on a large scale in bacteria. We have chosen seven cationic peptides for expression:
LL-37 (# 1) is a 37-amino acid human cathelicidin peptide, which has strong immunomodulatory activity and weaker direct antimicrobial activity due to inhibition by physiological salt concentrations. By contrast, CRAMP (# 2) is the mouse homolog of human LL-37 with 60% identity and somewhat more potent direct antimicrobial activity. IDR-1 (# 3) is the prototype of the IDR peptide class since it has no direct antimicrobial activity but protects in animal infections by modulating the innate immune response and is now in phase I clinical trials (seq: KSRIVPAIPVSLL) [36].
MX-226 (# 4) is the most clinically advanced antimicrobial peptide that has shown statistically significant efficacy in Phase III clinical trials as an antimicrobial and in Phase II trials as an anti-inflammatory.
E5 (also called sub2, # 5) and E6 (also called sub3, # 6) are 12-amino acid peptides optimized through substitutions into the bovine bactenecin peptide [15] while HHC-10 (# 7) is a promising broad spectrum 9-amino acid candidate that emerged from a QSAR modeling approach [9].
The technique for HDP production in bacteria relies on generating a fusion protein between the peptide and SUMO that both protect the bacteria from the toxicity of the peptides, and the peptides from host proteolytic enzymes. The sumoase protease Ulp1, which specifically cleaves after the C-terminal Gly-Gly residues in SUMO is then used to release HDP sequences at the C-terminus of the fusion protein [4, 27, 38]. Finally, we have devised a simple two-step purification protocol. The procedure is readily amenable to cost-effective industrial-scale GMP production of HDPs and IDRs.
Material and Methods
Bacterial strains and plasmids
E.coli strain BL21 (DEplys) was used as a host for cloning and gene expression of various smtp3-HDP fusion constructs. E.coli was grown at 37°C in LB media for cloning and at 30°C in LB media for expression; kanamycin (25 µg/mL) was added during growth of plasmid containing strains. The high-density fermentation media was prepared as described [24].
The vector pET28b was used for cloning and expression of the target genes. The vector allows for expression of exogenous proteins and peptides under the control of the T7 promoter (Novagen, Madison, WI). Sumo-pET28a and sumoase-pET28b were kindly provided by Dr. X. Cheng (Emory University). All recombinant DNA manipulation was performed using standard protocols.
Construction of expression vectors for expression of cationic peptide fusions
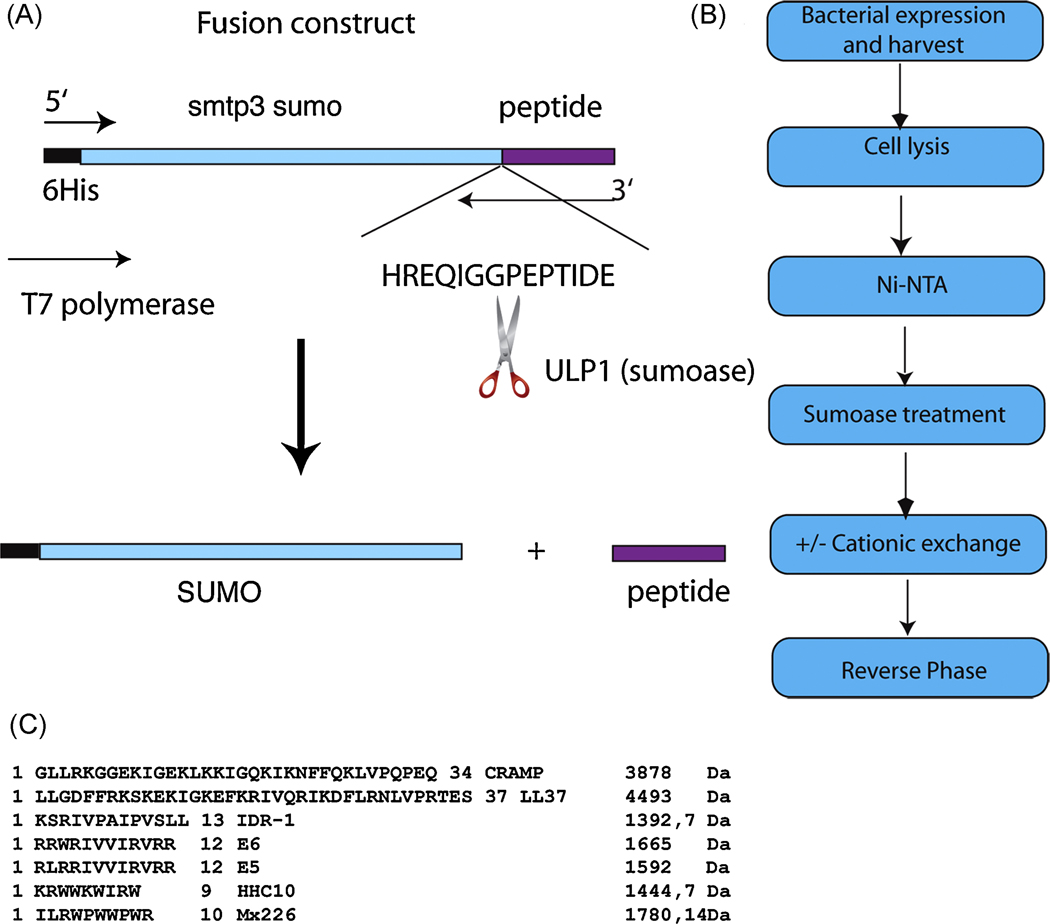
The overall scheme for construction of SUMO-HDP fusion proteins is illustrated in Figure 1A. Briefly, the yeast smtp3 gene, encoding sumo, including a 5’ 6xHis-linker was cloned into pET28a using the Nco1 and Nde1 sites. Peptide sequences are presented in Fig. 1C. The smtp3 gene was modified and shortened at the 3’ end to allow for a seamless fusion product after the Gly-Gly target sequence (permitting release of the peptide by sumoase Ulp1) using primers listed in Table 1.
Figure 1.
A. Schematic outline of the cloning strategy for in frame translation of the fusion product SUMO-peptide. Enhanced is the amino acid sequence at the cleavage site, showing the GG recognition sequence for sumoase followed by a generic peptide sequence.
B. Schematic outline of the purification protocol after expression of the fusion protein. Purification is achieved with 2 chromatographic steps; the cationic exchange chromatography is optional if a large RPC column size is less desired and cost-restrictive.
C. Amino acid sequence and the corresponding mass (Da) for all the peptides tested with this methodology.
Table 1.
Primer sequences for all the HDP genes cloned and fused to smtp3 (sumo). The restriction sites are shown in bold, the HDP gene sequence is shown in lower case and the smtp3 (sumo) fusion part is underlined.
| Primer name | Primer sequence |
|---|---|
| 5’ cgcgCCATGGGGcatcatcatcatcatcatTCGGACTCAGAAGTC 3’ | 5’His-sumo-Nco1s |
| 5’ GCGCGCCCATATGctacagcagggacaccgggatcgccggcacgatgcgggatttCACCAATCTGTTC 3’ | 3’ Idr1-Ndeas |
| 5’ GCGCGCCCATATGctaccagcggatccatttccaccagcgtttACCACCAATCTGTTC 3’ | 3’ HHC10-Ndeas |
| 5’ CGCGCCCATATGctactagcggcggacgcggatgacgacgatgcgccagcggcgACCACCAATCTGTTC 3’ | 3’ sub3Ndeas |
| 5’ CGCGCCCATATGctactagcggcggacgcggatgacgacgatgcggcggaggcgACCACCAATCTGTTC 3’ | 3’ sub2Ndeas |
| 5’ GCGCGCCCATATGctatttgcggcgccacggccaccacggccagcgaaggatACCACCAATCTGTTC 3’ | 3’ Mx226Ndeas |
| 5’ GAACAGATTGGTGGTctgctgggtgatttcttccgg 3’ | 5’ sumoLL37linker |
| 5’ ccggaagaaatcacccagcagACCACCAATCTGTTC3’ | 3’ sumoLL37linker |
| 5’ GCGCGCCCATATGctaggactctgtcctgggtac 3’ | 3’ LL37Ndeas |
All chosen HDP genes encode the active form of the corresponding peptides. The resulting PCR product fuses the cationic peptide gene 3’ in frame with the GG cleavage site of smtp3 (sumo), leaving no residual amino acids after cleavage (Figure 1A). The PCR product was restricted and ligated into the restricted vector, generating his-sumo-hdp-pET28b. For LL-37 and CRAMP, the corresponding genes were cloned from a cDNA isolated either from human bone marrow cDNA library (LL-37) or from primary mast cells harvested from C57Bl/6 mice (data not shown). Starting from the cDNA, the active part of the gene for LL-37 was isolated via 2 step PCR amplification protocol with primers 1, 7, 8 and 9. The resulting PCR construct was cloned into pET28b using the generated Nco1-Nde1 fragments, and E. coli BL21 (DElys, RIL) were transformed with the ligated construct. Positive clones were selected after colony PCR analysis, and tested for expression.
Expression and high-density fermentation of positive strains
For small-scale expression, 5 mL overnight cultures of E. coli cells (BL21 DE(plys)) harboring the vector sumo-AMP/HDP-pET28a were incubated for 12h at 37°C. Subsequently 2 mL were used to inoculate 500 mL LB supplemented with kanamycin (25 µg/mL). The cultures were incubated at 30°C in shaking flasks, and expression was induced at an OD600 of 0.5 with IPTG at a final concentration of 0.4 mM. Cells were harvested 4h after induction and lysed in PBS + 300 mM NaCl + 10 mM imidazole using sonication. Lysed extract was cleared by centrifugation at 13,000 × g for 20 min. Large-scale production of fusion protein was achieved using high-density fermentation in a 10 L fermentor with high-glucose feed as described previously [24, 39].
Purification of expressed fusion proteins
The catalytic domain of sumoase was purified as described previously [27] and frozen as concentrated 50 % glycerol stocks in the same concentration as described. The enzyme is very specific for SUMO, and no non-specific activity was observed; depending on the volume of the sample, 5–10 µL of sumoase, corresponding to 400 U, were added for complete cleavage of the fusion protein.
After cell lysis by sonication, the cleared supernatant was applied to a Ni-NTA gravity column and allowed to pass through. The column was washed with 10 × column volumes of PBS + 300 mM NaCl + 20 mM Imidazole and subsequently with 10 × column volumes of PBS + 300 mM NaCl + 40 mM Imidazole. Bound proteins were eluted with 2–3 column volumes of PBS + 300 mM NaCl + 250 mM Imidazole.
The eluted fusion protein was cleaved either overnight at 4°C or for 1h at RT using 400 U of sumoase and the cleaved peptide further purified and separated from its fusion partner using a C2/C18 or Resource reversed phase chromatography column and a 0.1 %TFA/acetonitrile gradient; peptides eluted between 25 and 35 % acetonitrile (Figure 1B). Purification was monitored using Tricine-SDS-PAGE [35].
Characterization of the purified recombinant peptides
Purified peptides were lyophilized and compared to the synthetic peptides using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) to assess the correct size after cleavage and Edman degradation to verify the correct amino acid content. All MALDI-TOF spectra were collected in positive ion mode and the collected data was within the acceptable accuracy of the instrument’s capabilities. For the antimicrobial peptides, MICs were determined in vitro using the modified microtiter broth dilution method [40] and compared to established MICs for the corresponding chemically synthesized peptides. For immunomodulatory peptides, induction of the cytokines TNFα and IL6 as well as the chemokine MCP-1 were assessed by enzyme linked immunoabsorbant assay (ELISA) following stimulation of human peripheral blood mononuclear cells (PBMC) with peptides for 16h. PBMCs from different human donors were isolated as previously described [29] in accordance with University of British Columbia ethical approval and guidelines, and the cytokine induction was compared to the results with the synthetic peptide, as described previously [36]. Peptide concentration at 300 µg/mL was used in all of the ELISA assays. Bacterial contaminants were removed using the ProteoSpin kit (Norgen) according to the manufacturer’s guidelines.
Results
Expression of cationic peptides in bacteria
We sought to develop a cost-effective means to express cationic peptides in E. coli with high yield and then purify them. To express peptides in E. coli, the cDNAs encoding them were cloned in frame and 3’ to a gene encoding His-tagged sumo. Whereas expression of the genes alone under the same promoter control resulted in few if any colonies (data not shown), adding the His-sumo to the 5’ end of the peptide gene resulted in normal bacterial growth with concomitant fusion protein expression in the soluble fraction. Therefore, we concluded that the His-SUMO served to block the antimicrobial activity of the fused peptides.
Several cationic peptides of different peptide lengths were chosen for expression using the SUMO fusion system (Figure 1C). These included LL-37, CRAMP, MX226, IDR-1, E5 and E6 and HHC-10.
All constructs tested expressed high levels of the fusion peptide both in small scale (50 mL) as well as 1L expression levels (data not shown). The identification of the intact fusion peptides provided evidence that the SUMO-peptide fusion was protected against cleavage by endogenous proteases. On small scale, increase in peptide length did not influence the overall expression yield of the fusion protein, but increasing peptide length will enhance the yield of the pure peptide as a result of a more favorable ratio of peptide to SUMO within the fusion construct.
High-density fermentation
Two of the peptides, one immunomodulatory peptide (IDR-1) and one antibacterial (E6) were chosen for high-density fermentation with glucose feed in a 10 L pilot scale. We decided on IDR-1 because it represents the prototype of the immunomodulatory peptides [36] and has entered phase 1 clinical trial with Inimexpharma; and on E6 as one of the first published short peptides resulting from a bioinformatics approach [15]. Both fermentations yielded around 1.5 kg wet biomass, of which 6% encompassed SUMO-IDR-1 and 4.3 % SUMO-E6 (Figures 3B and 4D). Under the conditions used, expression of IDR-1 appeared at near optimal levels based on the fusion protein amounts present in the soluble fraction, whereas E6 did not. Although no bacterial lysis occurred during the fermentation of either peptide, we anticipate that each construct would likely require optimization to achieve maximum yield.
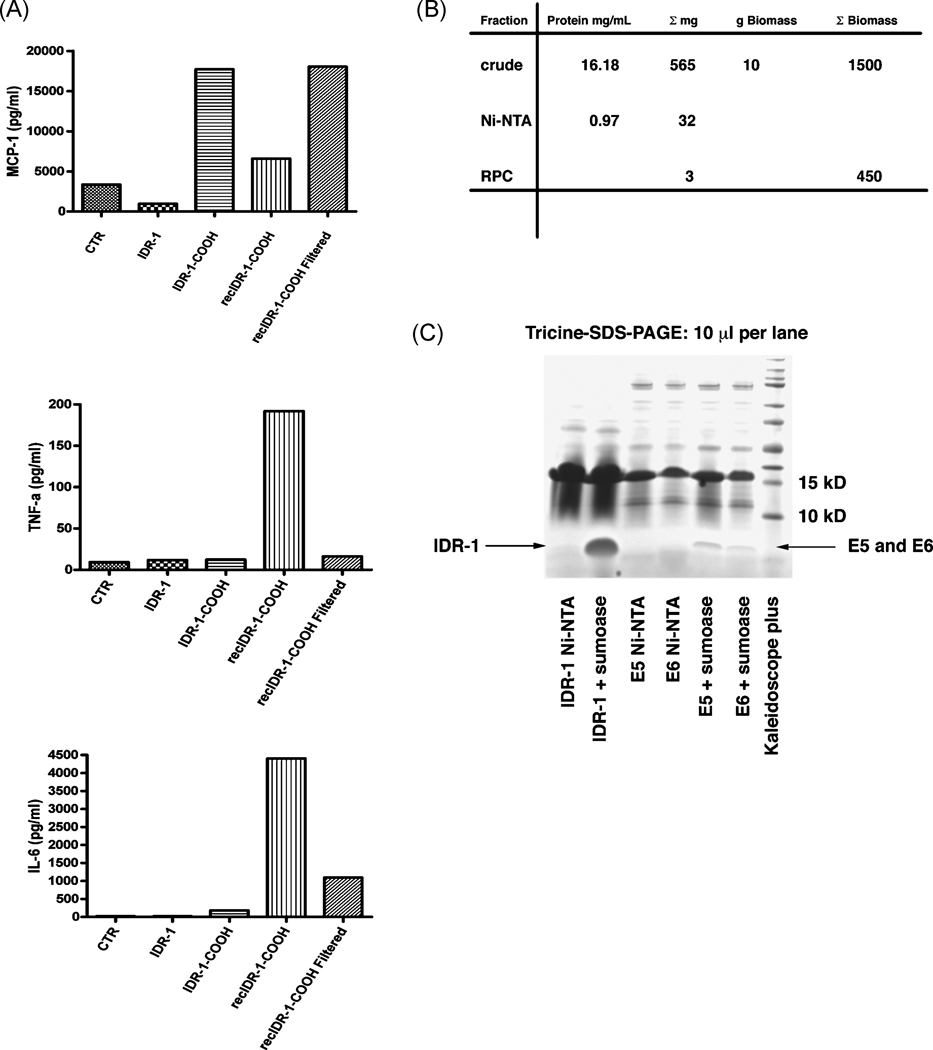
Figure 3.
A. Cytokine release of PBMCs from different human donors after treatment with either recombinant or synthetic IDR-1. Cytokines tested are IL-6, TNFα as well as the chemokine MCP-1.
B. Purification table for IDR-1 showing the yield of pure IDR-1 after an initial pilot purification of 10 g of biomass from the fermentation as well as a projection of possible yields from processing the total biomass of this fermentation.
C. Tricine-SDS-PAGE comparing cleaved peptide yields of a high-density fermentation against a 1L shaking flask expression. The amounts shown for E5 and E6 are representative for all the peptides tested in shaking flasks expression.
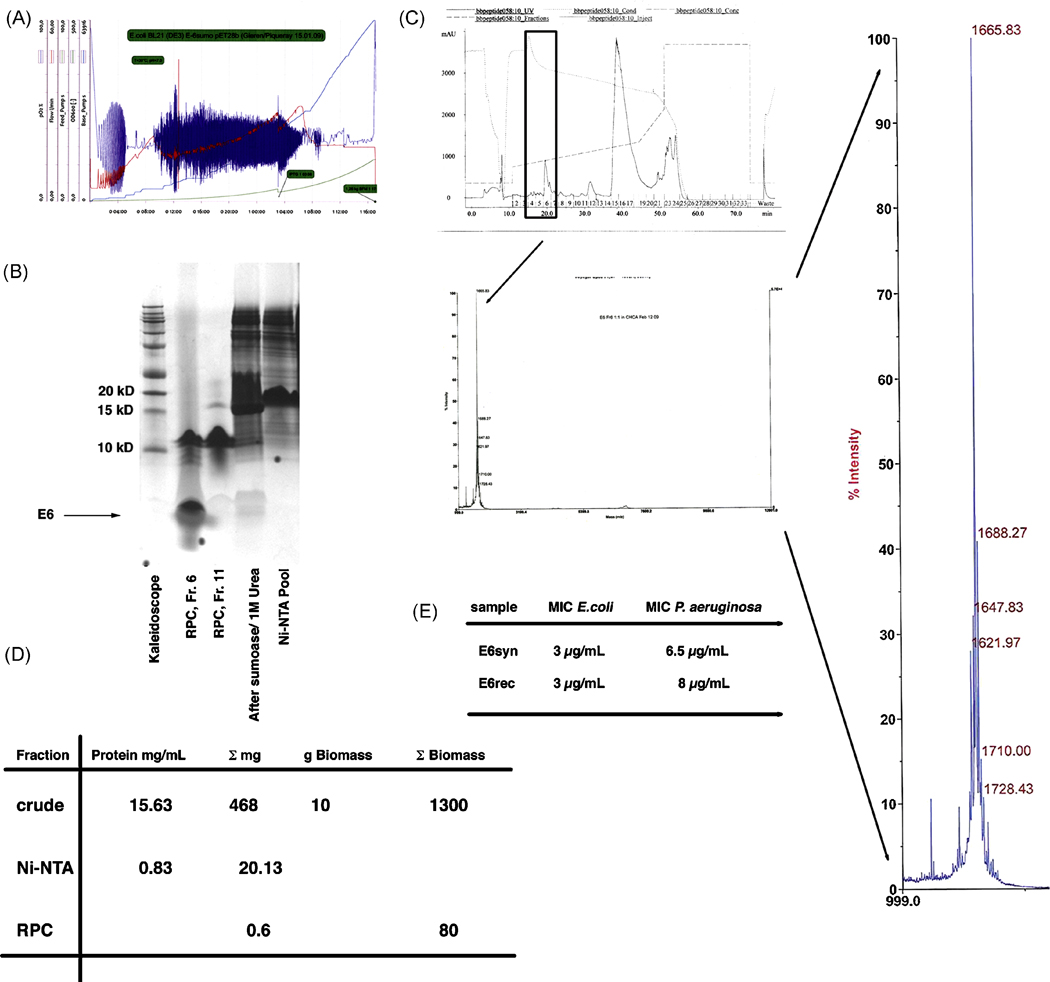
Figure 4.
A. Fermentation profile of a 10 L high-density fermentation of SUMO-E6 using glucose-feed and IPTG induction for 14h. The fermentation yielded 1.3 kg biomass. The total biomass is comparable to the one achieved using an immunomodulatory peptide, indicating that expression of an antimicrobial peptide is not deleterious to the bacterial host.
B. Tricine-SDS-PAGE of the purification steps used to achieve pure E6. Lane 2 showing RPC fraction # 6 corresponds to the fraction sent off for MALDI (see figure 4C).
C. RP-chromatography profile of the purification of E6 and the subsequent determination of its mass. MALDI analysis determined the correct mass of 1665 Da for this peptide with no further contamination present up to 30 kDa. The MS profile of E6 was magnified for better visualization.
D. Purification table showing the overall yield for E6 purification.
E. Direct comparison of MIC for both synthetic and recombinant E6 using E. coli K12 and P. aeruginosa PA014. The recombinant E6 showed identical MIC for E. coli and a slightly higher MIC for P. aeruginosa, again indicating that no contaminant protein is present, since then the amount of E6 would be lower than the one for the synthetic peptide.
Purification of Peptides
In the first step, the His-SUMO fusion peptides were purified using a Ni-NTA sepharose column. The peptides were then released from the fusion by cleavage with the SUMO-specific protease sumoase, which recognizes the 3-dimensional fold around a GG sequence at the SUMO-peptide boundary (Figure 1A). The released peptide was further purified using RPC-C2/C18-FPLC or Resource RPC-FPLC. A C8 linker on the RPC column was shown not to be effective in separating sumo from the peptide.
Purification and cleavage of the peptide was successful for all peptides tested on a small scale (see below for selected examples). As shown in Fig. 2 and 4, we achieved excellent purification from the scale-up investigation of IDR-1 and E6. To do this, 10 g of the harvested biomass from the fermenter was lysed, cleared and applied to an affinity chromatography column in purification step 1 using Ni-NTA (Figure 1B). The fusion protein was eluted from the column with 250 mM imidazole and then cleaved with ~400 U of sumoase for 1 h at RT. SUMO-IDR-1 accounted for 6 % of the total protein produced, which gave an estimated yield of 0.48 g/L fermentation (Figure 2D, Figure 3D). For peptides with high tryptophan content, it was necessary to add 1 M urea to the cleavage reaction to ensure precise cleavage of the peptide from the fusion partner SUMO (Figure 4B and 5A).
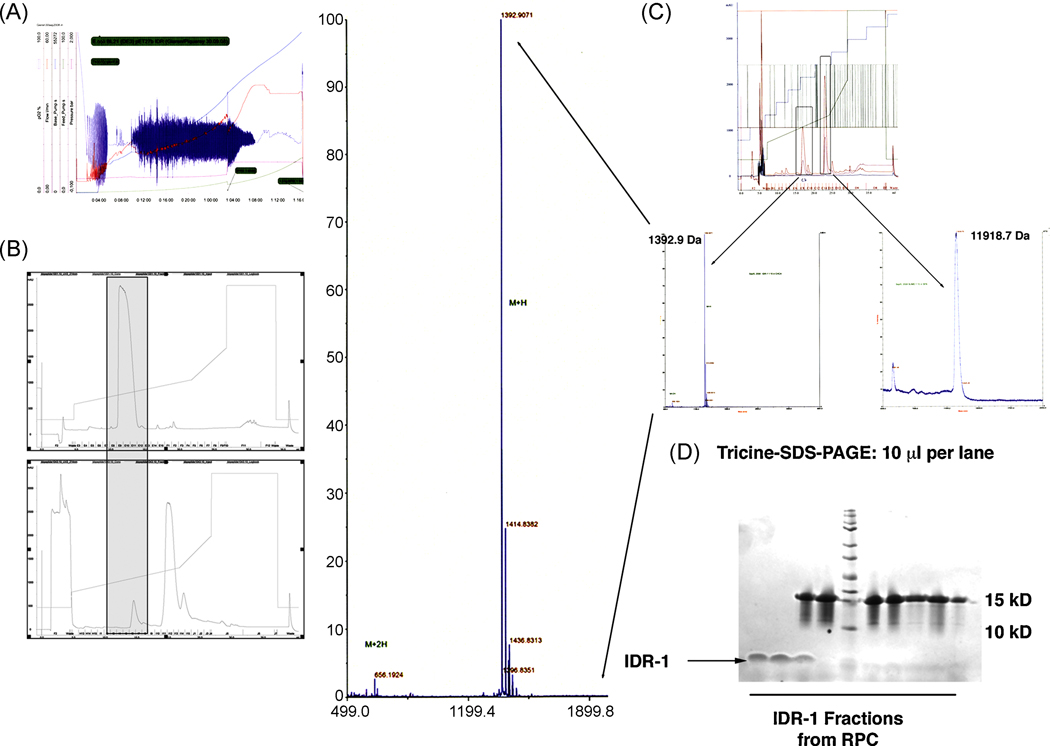
Figure 2.
A. Fermentation profile of a 10 L high-density fermentation of SUMO-IDR-1 using glucose-feed and IPTG induction for 14h. The fermentation yielded 1.5 kg biomass.
B. Comparison of the RP-chromatography profile for both the synthetic (upper panel) and recombinant IDR-1 peptide (lower panel). Both peptides eluted at the same percent of acetonitrile.
C. MALDI analysis of the purified IDR-1 peptide as well as the remaining his-SUMO protein after cleavage. The determined mass for IDR- 1 was identical to the synthetic peptide and the correct mass for his-sumo revealed 100 % cleavage using its protease sumoase. The MS profile for IDR-1 was magnified for better visualization.
D. Tricine-SDS-PAGE separation of the proteins from the recovered fractions from RP-chromatography shown in figure 2B. As can be seen in lanes 1 and 2, pure IDR-1 was recovered from fractions 11 and 12 followed by elution of SUMO in lanes 3–10). Lane 5 shows the Kaleidoscope Marker Plus (BioRAD).
Figure 5.
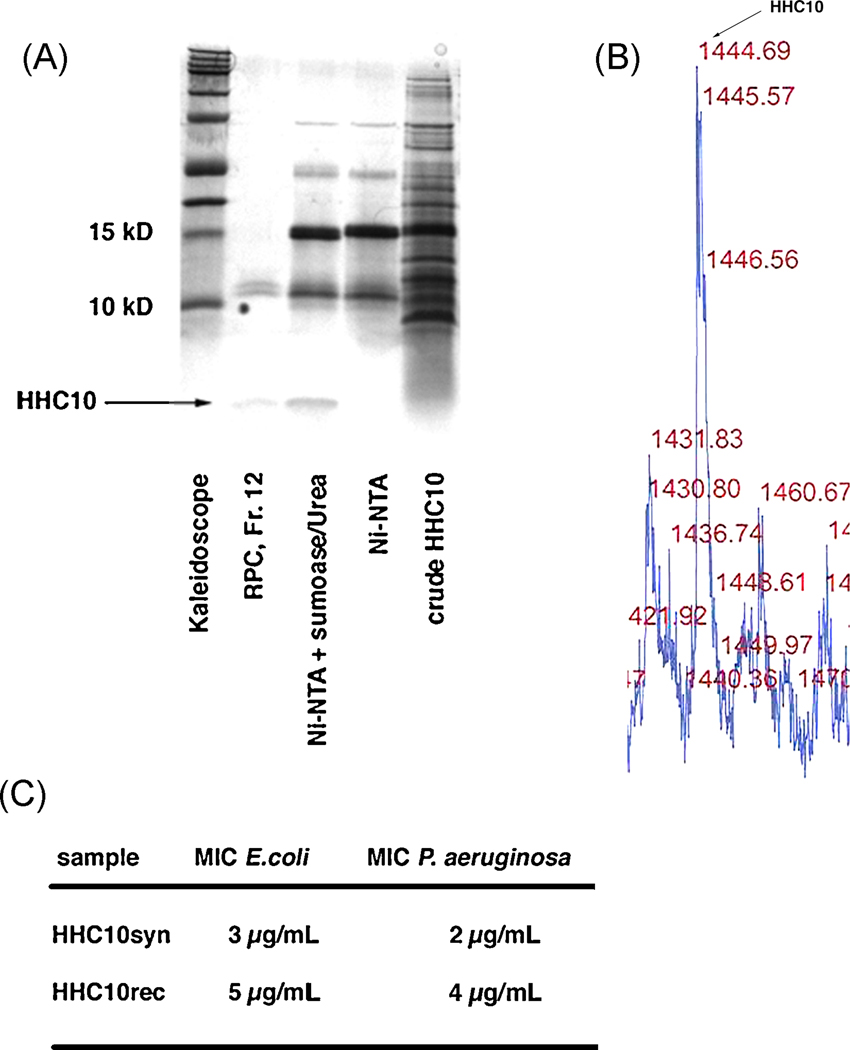
A. Tricine-SDS-PAGE of the purification steps used to achieve pure HHC-10 from a 1 L shaking flask expression, fraction 12 in lane 2 corresponds to the sample used for mass determination. Again, no larger protein contaminants could be found, indicating that the higher bands visible in the gel are possibly aggregates of HHC-10, which could be avoided in a different solvent.
B. MALDI profile for HHC-10 showing the correct mass of 1444 Da.
C. Direct comparison of MIC for both synthetic and recombinant HHC-10 using E. coli K12 and P. aeruginosa PA014. The recombinant HHC10 showed a slightly higher MIC for E. coli and P. aeruginosa.
Following cleavage, proteins were successfully separated using reversed phase chromatography to produce homogenously pure peptide (Figure 2D and Figure 4C). RP-chromatography was needed to separate SUMO and sumoase from the peptide, because repeated application onto an affinity Ni-NTA did not result in successful separation of the two (data not shown). Our data suggest that the overall positive charge of the peptide (between +4 and +6) may induce electrostatic interactions with negatively charged residues within SUMO (overall charge -5), perhaps annealing the two proteins during a non-hydrophobic separation procedure. Indeed it is likely that these interactions play an important role in both neutralization of biological activity and protection from endogenous proteases.
Expression and purification of other peptides
Two other cationic peptides, HHC-10 (Figure 5A) and MX-226 (data not shown), were successfully expressed in shaking flask cultures as a fusion and successful cleavage with sumoase and purification achieved. 1 M urea was required to ensure correct cleavage at the desired position on SUMO (Figure 5A), a procedure found necessary for Trp-containing peptides. We also successfully expressed and purified larger HDPs including CRAMP and LL-37 (36 and 37 aa long) with this methodology (data not shown).
Identification of purified peptides
The identity of the isolated peptides was confirmed by mass spectrometry (Figures 2C and 4C) using MALDI-TOF to determine the exact mass of purified peptides. IDR-1 showed the exact molecular weight (MW) of 1391.9 Da, which corresponded to its theoretical mass (cf. Figures 1C and 2C). N-terminal sequencing of the IDR-1 in the gel confirmed the correct first 5 amino acids of IDR-1 (KSRIV, data not shown), indicating that cleavage occurred at the right position after the Gly-Gly sequence in SUMO with no additional amino acids introduced to the peptide. The other band observed (Figure 2C) at a molecular mass of 11918.7 Da, was identical to the mass of the separated SUMO recovered from reverse phase chromatography (see Figure 2C, second peak) suggesting that our sumoase cleavage protocol achieved 100% cleavage [34]. Likewise, the measured mass of 1664.83 Da for E6 corresponded to its theoretical peptide mass (Figure 4C, see Figure 1C for comparison). Mass determination for the short, Trp-rich peptide HHC-10 after purification showed the predicted mass of 1443.69 Da (Figure 1C and 5B). In this case, as mentioned above, correct mass of the purified, cleaved peptide could only be achieved through addition of 1 M urea to help unfold the short hydrophobic stretch of this Trp-rich peptide adjacent to the cleavage site for sumoase. Although sumoase is thought to be highly specific [27, 34], we have observed that introducing a stretch of several tryptophan residues close to the cleavage site most likely prevents access to the site through steric hindrance and causes random cleavage within the fusion protein. Addition of 1 M urea prevented such unspecific cleavage and led to the release of the full-length peptide.
Peptide yields
The amount of IDR-1 shown in Figure 2D corresponds to 1% of the total IDR-1 recovered from a single chromatography run. With available equipment, a total of 5 runs were needed to process the 10 g biomass obtained from a single purification using the C2/C18 RPC column. However this was reduced to 2 runs using the Resource RPC column and therefore can readily be adapted to larger scale. To determine yield, the purified peptide was weighed on a fine scale after lyophilization. However, these measurements can only be considered a rough estimate, since residual salt concentrations left attached to the peptide could affect the weight. IDR-1 concentration was subsequently determined using amino acid analysis of the pure peptide and showed about 50% residual salt within the peptide, which is similar to synthetically produced peptides. Overall, 3 mg of pure IDR-1 was obtained from 10 g of biomass (Figure 2D and 3B). In an optimized process, we expect to achieve 0.08 g/L fermentation of pure IDR-1. For E6 the purification of the fermented biomass yielded 0.6 mg from 10 g wet cells (Figure 4D). Despite similar fermentations the heterologous expression of sumo-E6 only accounted for 4.3 % of the total protein in E.coli. Tricine-SDS-PAGE analysis of E6 indicated several bands around 10 kDa, but thorough mass analysis up to 30 kDa did not reveal any peak other than the one for E6. These data suggest that significant aggregation might be occurring. Alternatively, LPS may have become attached to the E6 peptide, since the MW for LPS in E. coli is around 5–10 kDa.
Characterization of the IDR-1, E6 and HHC-10
For the HDPs E6 and HHC-10 we determined the minimal inhibitory concentration (MIC) for growth of E. coli and Pseudomonas aeruginosa PA014, and compared these values to those obtained with the corresponding synthetic peptide (Fig. 4E and 5C). Using both bacteria, MICs for the recombinant, homogenous peptide E6 were identical to the corresponding synthetic peptides, given that MICs are operatively considered to be accurate to within two-fold.
For the immunomodulatory peptide IDR-1 the efficacy of the recombinant peptide was compared to the synthetic peptides using cytokine/chemokine ELISAs to determine the stimulation of a pro-inflammatory response on PBMC from a variety of human donors. TNFα was used as an indicator of inflammatory cytokine stimulation and possible contamination with bacterial components, because synthetic IDR-1 does not trigger a TNFα response. Initially, the recombinant peptide yielded higher values for IL-6 and TNFα release than two different lots of synthetic peptide. This difference could not be attributed to differences in apparent concentration between the purified and synthetic peptides. We concluded that a bacterial contaminant was still present in the IDR-1 preparation, although the amount varied strongly with the donor. We therefore further purified the recombinant peptide using a LPS removal kit (Norgen), successfully removing LPS as the contaminant and achieving the same TNFα response for the recombinant peptide compared to the synthetic peptide (Figure 3A). On the other hand, the IL-6 response and production of MCP-1 as an indicator for chemokine induction was still higher between the recombinant and synthetic IDR-1 (Figure 3A), so a synthetic control IDR-1 peptide lacking amidation (IDR-1-COOH) was included and indicated that the increased MCP-1 and IL6 responses were due to the lack of amidation on the recombinant peptide. Most synthetic peptides include a C-terminal amidation as part of the synthesis process, but bacteria cannot produce peptides with a C-terminal amidation, thus post-translational modifications of bacterially produced peptides via a chemical route might be necessary for therapeutic development.
Discussion
To explore the pharmaceutical and therapeutic potential of antimicrobial peptides, a cost-effective and scalable method for production of active and effective HDPs is required. Procedures to express HDPs as recombinant peptides have encountered difficulties associated with cytotoxicity to the bacterial host, and, as a consequence, difficulty in scale up and low yields following purification [8, 31, 44], especially when large fusion proteins are chosen [16]. Even when successful, the requirement for processing of fusion peptides (often requiring chemicals or costly enzymes) and multistep purification of peptides has shown reduced yields, has not proven cost effective, and has met with intermittent success depending on the peptide.
In this study we have described a procedure for high yield production of several antimicrobial or immunomodulatory peptides using a fusion protein partner that has been well established to ensure high-level soluble expression of fusion proteins, even with proteins that are otherwise difficult to express (e.g. MMP13, [27]. Using this system, we have found that the fusion protein accounts for 10 to 25 % of total protein, and yielded 3 mg of pure peptide from 10 g of biomass.
Unlike previous methods, expression of the SUMO-peptide fusion does not have to be forced into the insoluble fractions, which, on an industrial scale, can be costly due to the requirement for urea and guanidium chloride to solubilize the inclusion bodies [14, 23, 46]. Moreover, the fusion protein remains in the soluble fraction without lethal effects to the host bacterial cells. Thus, proteolytic degradation of the fusion protein and release of the peptide does not readily occur. Generally, there are several options available to cleave a peptide from its fusion partner, utilizing both chemical and enzymatic routes of cleavage. For chemical cleavage, two different methods have been explored. Cyanogen bromide, which cleaves C-terminally after methionine, has been used extensively, but is inefficient in its cleavage [14]. Second, cleavage of the acid-labile peptide bond between asparagines and proline using hydrochloric acid [46], leaves a proline overhang at the N-terminus of a peptide and requires neutralization of the acid, properties that are neither generally nor easily applicable on an industrial scale.
Enzymatically, several proteases, such as thrombin, Factor Xa or enterokinase, will cleave at their recognition sites, once these are introduced within the linker between the fusion protein and the peptide during cloning [45]. However, these enzymes are expensive and not feasible for industrial scale purification. Sumoase is unique in that it recognizes only residues within SUMO as its cleavage substrate and cleavage occurs precisely after the Gly-Gly in the SUMO sequence [27], leaving no unwanted amino acids at the N-terminus of the peptide. Another important advantage is that this protease is produced cheaply using the T7 driven pET system and can be easily purified with Ni-NTA affinity chromatography in a single step [27]. Also, 400 U of enzyme were used for complete cleavage [27], which seems substantial, but a 0.5 L culture yields 10,000 × that amount, suggesting that the enzyme is actually quite cost-effective.
A few labs have recently used the SUMO fusion system for cost-effective antimicrobial peptide expression and demonstrated its efficacy [7, 25], however, we have shown that successful endotoxin removal is critical when considering their use for therapeutic purposes. Removal of endotoxins has proven to be a challenge and several routes had to be taken until the unfavorable TNFα response was prevented. Our data indicates that the bacterially produced peptides have to be an exact match to the established synthetic peptides, if they are to be successful therapeutics. As we have demonstrated, this is especially true for the peptides’ immunomodulatory potential, a peptide property that is gaining more and more importance in therapeutic applications [2]. We were able to produce peptides of varying lengths ranging from 37 amino acids for LL-37 down to 9 amino acids for HHC-10 with excellent MIC values. To the best of our knowledge, nobody has achieved bulk production of such small peptides in bacteria, especially not peptides that are already tested in clinical trials (IDR-1 (www.inimexpharma.com) and MX-226 (www.migenix.com)), whereas other recent publications have achieved expression and purification of peptides in the range of 30 amino acids and more [7, 25].
Our data also indicate that posttranslational modifications of peptides can alter their cytokine profile, as demonstrated in Figure 3A. Amidation of the C-terminus of IDR-1, as it is common practice for synthetic peptides, resulted in less cytokine response than the free C-terminus IDR-1 samples, both synthetic and recombinant. Peptide amidation can occur in mammals, since most hormonal peptides and neuropeptides are amidated through peptidylglycine α-amidating monooxygenase PAM, which is essential for their activity [3, 10]. Amidation also protects from C-terminal peptide degradation, which prolongs the half-life of peptides in serum. So far, amidation has not been reported for endogenous human antimicrobial peptides.
Correct determination of peptide concentration following chromatography can only be achieved through amino acid determination, because salts used in the purification tend to attach to the peptides. This is especially crucial when using cationic exchange chromatography, where high concentrations of sodium chloride are used to elute the peptide. We have found salt to be present in the lyophilized peptide even after RP-chromatography as the last step, which does not involve salt.
Taken together, this expression system allows for large-scale production of HDPs, which retain activity similar to peptides synthesized by chemical means. The system can produce both immunomodulatory and antimicrobial peptides, apparently independent of amino acid sequence, length, or charge in industrial scale quantities. Even small peptides of just 9 amino acid in length can be produced with acceptable yields. Purification is achieved in two steps, which are easily scalable for industrial application. In short, we provide evidence that this system will enable cost effective production of HDPs under GMP conditions in support of therapeutic applications for these molecules against infectious diseases.
Research Highlights
Novel bacterial expression system for host defense peptides
Scalable, simplified 2 step purification
Large-scale expression of peptides as small as 9 amino acids in length
Successful application of this method to peptides already in clinical trials
Suitable for large-scale production of antimicrobial peptides and host-defense peptides under GMP for therapeutic use
ACKNOWLEDGEMENTS
We appreciate the financial support from the Foundation for the National Institutes of Health and Canadian Institutes for Health Research through the Grand Challenges in Global Health Initiative to REWH. REWH is a recipient of a Canada Research Chair while JK hold a fellowship from the Canadian Cystic Fibrosis Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andes D, Craig W, Nielsen A, Kristensen HH. In vivo Pharmacodynamic Characterization of a Novel Plectasin Antibiotic, NZ2114, in a Murine Infection Model. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.01584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bommarius B, Kalman D. Antimicrobial and host defense peptides for therapeutic use against multidrug-resistant pathogens: new hope on the horizon. IDrugs. 2009;12:376–380. [PubMed] [Google Scholar]
- 3.Bradbury AF, Smyth DG. Peptide amidation. Trends Biochem Sci. 1991;16:112–115. doi: 10.1016/0968-0004(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 4.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carballar-Lejarazu R, Rodriguez MH, de la Cruz Hernandez-Hernandez F, Ramos-Castaneda J, Possani LD, Zurita-Ortega M, et al. Recombinant scorpine: a multifunctional antimicrobial peptide with activity against different pathogens. Cell Mol Life Sci. 2008;65:3081–3092. doi: 10.1007/s00018-008-8250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carretero M, Escamez MJ, Garcia M, Duarte B, Holguin A, Retamosa L, et al. In vitro and in vivo wound healing-promoting activities of human cathelicidin LL-37. J Invest Dermatol. 2008;128:223–236. doi: 10.1038/sj.jid.5701043. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Zhu F, Cao Y, Qiao S. Novel expression vector for secretion of cecropin AD in Bacillus subtilis with enhanced antimicrobial activity. Antimicrob Agents Chemother. 2009;53:3683–3689. doi: 10.1128/AAC.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YQ, Zhang SQ, Li BC, Qiu W, Jiao B, Zhang J, et al. Expression of a cytotoxic cationic antibacterial peptide in Escherichia coli using two fusion partners. Protein Expr Purif. 2008;57:303–311. doi: 10.1016/j.pep.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Cherkasov A, Hilpert K, Jenssen H, Fjell CD, Waldbrook M, Mullaly SC, et al. Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem Biol. 2009;4:65–74. doi: 10.1021/cb800240j. [DOI] [PubMed] [Google Scholar]
- 10.Czyzyk TA, Ning Y, Hsu MS, Peng B, Mains RE, Eipper BA, et al. Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev Biol. 2005;287:301–313. doi: 10.1016/j.ydbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Gilliet M, Lande R. Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr Opin Immunol. 2008;20:401–407. doi: 10.1016/j.coi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Hamill P, Brown K, Jenssen H, Hancock RE. Novel anti-infectives: is host defence the answer? Curr Opin Biotechnol. 2008;19:628–636. doi: 10.1016/j.copbio.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 14.Haught C, Davis GD, Subramanian R, Jackson KW, Harrison RG. Recombinant production and purification of novel antisense antimicrobial peptide in Escherichia coli. Biotechnol Bioeng. 1998;57:55–61. doi: 10.1002/(sici)1097-0290(19980105)57:1<55::aid-bit7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 15.Hilpert K, Volkmer-Engert R, Walter T, Hancock RE. High-throughput generation of small antibacterial peptides with improved activity. Nat Biotechnol. 2005;23:1008–1012. doi: 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- 16.Hong I, Kim YS, Choi SG. Simple purification of human antimicrobial peptide dermcidin (MDCD-1L) by intein-mediated expression in E.coli. J Microbiol Biotechnol. 2010;20:350–355. [PubMed] [Google Scholar]
- 17.Hsu KH, Pei C, Yeh JY, Shih CH, Chung YC, Hung LT, et al. Production of bioactive human alpha-defensin 5 in Pichia pastoris. J Gen Appl Microbiol. 2009;55:395–401. doi: 10.2323/jgam.55.395. [DOI] [PubMed] [Google Scholar]
- 18.Jenssen H, Fjell CD, Cherkasov A, Hancock RE. QSAR modeling and computer-aided design of antimicrobial peptides. J Pept Sci. 2008;14:110–114. doi: 10.1002/psc.908. [DOI] [PubMed] [Google Scholar]
- 19.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenssen H, Hancock RE. Therapeutic potential of HDPs as immunomodulatory agents. Methods Mol Biol. 2010;618:329–347. doi: 10.1007/978-1-60761-594-1_20. [DOI] [PubMed] [Google Scholar]
- 21.Jenssen H, Lejon T, Hilpert K, Fjell CD, Cherkasov A, Hancock RE. Evaluating different descriptors for model design of antimicrobial peptides with enhanced activity toward P. aeruginosa. Chem Biol Drug Des. 2007;70:134–142. doi: 10.1111/j.1747-0285.2007.00543.x. [DOI] [PubMed] [Google Scholar]
- 22.Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 23.Kim JM, Jang SA, Yu BJ, Sung BH, Cho JH, Kim SC. High-level expression of an antimicrobial peptide histonin as a natural form by multimerization and furin-mediated cleavage. Appl Microbiol Biotechnol. 2008;78:123–130. doi: 10.1007/s00253-007-1273-5. [DOI] [PubMed] [Google Scholar]
- 24.Korz DJ, Rinas U, Hellmuth K, Sanders EA, Deckwer WD. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J Biotechnol. 1995;39:59–65. doi: 10.1016/0168-1656(94)00143-z. [DOI] [PubMed] [Google Scholar]
- 25.Li JF, Zhang J, Song R, Zhang JX, Shen Y, Zhang SQ. Production of a cytotoxic cationic antibacterial peptide in Escherichia coli using SUMO fusion partner. Appl Microbiol Biotechnol. 2009;84:383–388. doi: 10.1007/s00253-009-2109-2. [DOI] [PubMed] [Google Scholar]
- 26.Li Y. Carrier proteins for fusion expression of antimicrobial peptides in Escherichia coli. Biotechnol Appl Biochem. 2009;54:1–9. doi: 10.1042/BA20090087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 28.Mookherjee N, Hancock RE. Cationic host defence peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell Mol Life Sci. 2007;64:922–933. doi: 10.1007/s00018-007-6475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mookherjee N, Wilson HL, Doria S, Popowych Y, Falsafi R, Yu JJ, et al. Bovine and human cathelicidin cationic host defense peptides similarly suppress transcriptional responses to bacterial lipopolysaccharide. J Leukoc Biol. 2006;80:1563–1574. doi: 10.1189/jlb.0106048. [DOI] [PubMed] [Google Scholar]
- 30.Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 31.Piers KL, Brown MH, Hancock RE. Recombinant DNA procedures for producing small antimicrobial cationic peptides in bacteria. Gene. 1993;134:7–13. doi: 10.1016/0378-1119(93)90168-3. [DOI] [PubMed] [Google Scholar]
- 32.Pranting M, Negrea A, Rhen M, Andersson DI. Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob Agents Chemother. 2008;52:2734–2741. doi: 10.1128/AAC.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pripp A, Isaksson T, Stepaniak L, Sorhaug T, Ardo Y. Quantitative structure activity relationship modelling of peptides and proteins as a tool in food science. Trends in Food Science and Technology. 2005;16:484–494. [Google Scholar]
- 34.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, et al. An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol. 2007;25:465–472. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- 37.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 38.Tadas P, Sanders C, Butt TR. SUMO Protocols. New York: Humana Press; 2009. SUMO Fusion Technology for Enhanced Protein Production in Prokaryotic and Eukaryotic Expression Systems; pp. 303–317. [DOI] [PubMed] [Google Scholar]
- 39.Tishkov VI, Galkin AG, Fedorchuk VV, Savitsky PA, Rojkova AM, Gieren H, et al. Pilot scale production and isolation of recombinant NAD+- and NADP+-specific formate dehydrogenases. Biotechnol Bioeng. 1999;64:187–193. [PubMed] [Google Scholar]
- 40.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 41.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 42.Yu J, Mookherjee N, Wee K, Bowdish DM, Pistolic J, Li Y, et al. Host defense peptide LL-37, in synergy with inflammatory mediator IL-1beta, augments immune responses by multiple pathways. J Immunol. 2007;179:7684–7691. doi: 10.4049/jimmunol.179.11.7684. [DOI] [PubMed] [Google Scholar]
- 43.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Falla T, Wu M, Fidai S, Burian J, Kay W, et al. Determinants of recombinant production of antimicrobial cationic peptides and creation of peptide variants in bacteria. Biochem Biophys Res Commun. 1998;247:674–680. doi: 10.1006/bbrc.1998.8848. [DOI] [PubMed] [Google Scholar]
- 45.Zhou QF, Luo XG, Ye L, Xi T. High-level production of a novel antimicrobial peptide perinerin in Escherichia coli by fusion expression. Curr Microbiol. 2007;54:366–370. doi: 10.1007/s00284-006-0466-y. [DOI] [PubMed] [Google Scholar]
- 46.Zorko M, Japelj B, Hafner-Bratkovic I, Jerala R. Expression, purification and structural studies of a short antimicrobial peptide. Biochim Biophys Acta. 2009;1788:314–323. doi: 10.1016/j.bbamem.2008.10.015. [DOI] [PubMed] [Google Scholar]