Summary
Activation induced cytidine deaminase (AID) initiates antibody gene diversification by creating U:G mismatches. However, AID is not specific for antibody genes. Off-target lesions can activate oncogenes or cause chromosome translocations. Despite its importance in these transactions little is known about how AID finds its targets. To address this, we performed an shRNA screen. We found that Spt5, a factor associated with stalled RNA polymerase II (Pol II) and single stranded DNA (ssDNA), is required for class switch recombination (CSR). Spt5 interacts with AID, it is required for the association of AID and Pol II, and for AID recruitment to Ig switch regions and non-Ig targets. ChIP-seq experiments reveal that Spt5 co-localizes with AID and stalled Pol II. Further, Spt5 accumulation at sites of Pol II stalling is predictive of AID-induced mutation. We propose that AID is targeted to sites of Pol II stalling in part via its association with Spt5.
Introduction
AID is a cytidine deaminase that initiates immunoglobulin somatic hypermutation (SHM) and class switch recombination (CSR) (Muramatsu et al., 2000; Muramatsu et al., 1999; Revy et al., 2000). It does so by deaminating cytidine residues in ssDNA (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003; Sohail et al., 2003). The resulting U:G mismatches can be processed by several different DNA repair pathways to produce mutations or DNA double-strand breaks (Di Noia and Neuberger, 2007; Peled et al., 2008).
In addition to diversifying the antibody repertoire by SHM and CSR, AID also contributes to malignant transformation by initiating chromosome translocations (Ramiro et al., 2006; Ramiro et al., 2004; Robbiani et al., 2008, Nussenzweig and Nussenzweig, 2010) and by producing mutations in non-Ig genes such as Bcl-6 (Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998). Although the comparative frequency of mutation at non-Ig genes is low, AID mutates 25% of the genes transcribed in germinal center B cells, where it is normally expressed (Liu et al., 2008). Furthermore, even low levels of mutation are sufficient to produce substrates for translocation (Robbiani et al., 2008; Robbiani et al., 2009). Consistent with the breadth of genes found mutated by AID in germinal center B cells, AID over-expression in transgenic mice leads to extensive translocation of non-Ig genes and cancer (Robbiani et al., 2009). In addition, AID deregulation has been associated with H. pylori infection and gastric cancer (Matsumoto et al., 2007), and with translocation in prostate malignancy (Lin et al., 2009). Finally, AID is also of interest because it has been implicated as a cytosine demethylase involved in reprogramming pluripotent cells (Bhutani et al., 2010; Morgan et al., 2004; Popp et al., 2010; Rai et al., 2008).
Although the precise mechanism which targets AID to Ig genes is unknown, AID induced mutations are associated with transcription and are most prevalent in a 2 kb region beginning downstream of the promoter (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008; Storb et al., 2007). Transcription is also required for CSR, suggesting that RNA polymerase II (Pol II) might facilitate AID access to target DNA (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer-Nordgren and Sirlin, 1986; Stavnezer et al., 2008; Storb et al., 2007; Yancopoulos et al., 1986). This idea was confirmed by the observation that transcriptional regulatory elements are essential to both hypermutation and CSR (reviewed in (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008; Storb et al., 2007)). Consistent with these findings, AID is associated with Pol II (Nambu et al., 2003). In E. coli and in in vitro assays, transcription liberates ssDNA, the substrate for AID (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003; Sohail et al., 2003). In more complex systems, transcription is also required for AID to access chromatinized substrates (Shen et al., 2009); however, the role of transcription in SHM and CSR is not completely understood.
AID is a relatively small enzyme composed of 198 amino acids (Muramatsu et al., 1999). It preferentially deaminates cytosine residues embedded in WRCY consensus sequences (where W=adenosine/thymine, R=purine, and Y=pyrimidine) (Rogozin and Kolchanov, 1992). This preference is dictated in part by the composition of the active site (Wang et al., 2010). However, WRCY motifs are present throughout the genome and cannot fully account for AID target choice. While several AID co-factors have been reported, including replication protein A (RPA), protein kinase-Ar1α, and CTNNBL1, none of these are known to impart specificity to AID (Basu et al., 2005; Chaudhuri et al., 2004; Conticello et al., 2008; McBride et al., 2006; Pasqualucci et al., 2006).
Here we report that Spt5, a factor normally associated with stalled or paused Pol II, is required for CSR. Spt5 is required for AID recruitment to switch regions, for switch region mutation, and for AID association with Pol II. Furthermore, genes that accumulate Spt5 also accumulate AID and suffer AID-dependent mutations.
Results
shRNA Screen for CSR in CH12 Cells
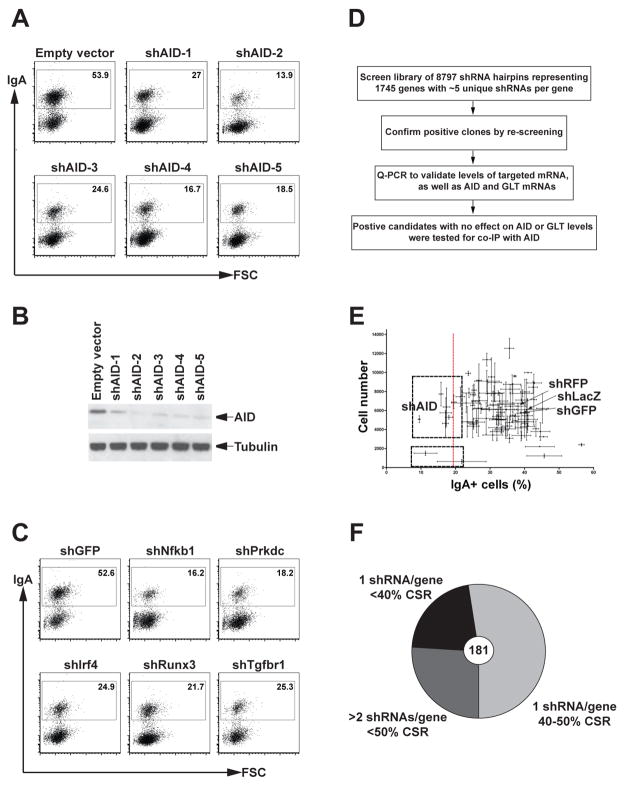
To identify factors required for CSR, we developed a lentiviral-based shRNA screening strategy using the murine B cell line, CH12. This cell line expresses AID and undergoes CSR to IgA in response to stimulation with interleukin 4 (IL-4), CD40 ligation and transforming growth factor β (TGFβ) (Nakamura et al., 1996). AID is limiting for CSR in these cells because its knockdown by specific shRNA results in reduction of CSR in a manner consistent with the decrease in AID protein levels (Figure 1A and B). In addition, shRNA-induced knockdown of other known regulators of the reaction result in the expected decrease in CSR (Figure 1C). Therefore, the level of CSR in CH12 cells is limited by the amount of AID and its co-factors suggesting that CSR can be used as an assay for additional factors that might be required for AID function in these cells. To screen for such factors, we developed an shRNA screen for CSR in CH12 cells.
Figure 1. Lentiviral-based shRNA screen in CH12 cells.
CSR is sensitive to AID depletion. Flow cytometry plots of CH12 cells infected with five unique shRNAs to AID (shAID1-5) and empty vector control. Numbers indicate the percentage of IgA positive cells.
(B) AID protein levels in whole cells extracts from the same cells shown in (A). Western blots were probed with an anti-AID antibody and anti-tubulin as a loading control.
(C) Representative flow cytometry plots of CH12 cells infected with shRNAs against genes involved in CSR: Nfkb1 (NFκB p50 subunit), Prkdc (DNAPKcs catalytic subunit), Irf4, Runx3 and Tgfbr1 (TFGβ receptor 1) (Table S3).
(D) Schematic of the experimental approach used for the screen.
(E) Representative data from a single plate of shRNAs analyzed in triplicate. Error bars show the standard deviation obtained from the three replicate plates for %IgA+ cells (X axis) and cell numbers (Y axis). Negative (LacZ, GFP and RFP) and positive (AID) controls shRNAs are indicated. The dotted red line shows the position corresponding to 50% of the averaged negative control CSR value. Two sets of clones with <50% CSR are boxed. The upper box consists of viable clones that are considered as positive hits. The lower box contains clones that were discarded due to poor viability.
(F) Pie chart showing the distribution of 181 selected hits as a function of the number of shRNAs per gene and their effect on CSR as calculated based on the percentage reduction of CSR compared to the averaged negative control values as shown in (E). See also Tables S2 and S3.
We assembled an shRNA lentiviral library containing 8,797 hairpins representing 1,745 genes selected primarily on the basis of their expression in CH12 cells (Table S1) and germinal center B cells (Klein et al., 2003; Moffat et al., 2006; Root et al., 2006) (Table S2). Factors directly involved in transcription, or in co-transcriptional and post-transcriptional events, such as mRNA processing, turnover and export, and DNA repair factors, kinases and phosphatases were preferentially retained (reviewed in (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008; Storb et al., 2007)). Finally, we added several DNA repair factors and transcription-associated factors that were not selected based on their expression, but that might be required based on the literature (Table S2).
The recombinant lentiviruses were prepared and screened in a 96-well format in triplicate (Moffat et al., 2006; Root et al., 2006), and CSR and viability for each sample was evaluated by flow cytometry (Figure 1D). Each plate contained three negative control shRNAs (shLacZ, shGFP and shRFP) and a positive control AID shRNA (Figure 1E, shAID). Positive hits were defined as viable shRNA-expressing clones that exhibited at least 50% reduction in CSR compared to the controls (Figures 1E). Positive hits were re-arrayed and re-screened in triplicate. The screen uncovered 181 hits of which 28 were previously shown to be involved directly or indirectly in CSR (Figure 1F and Table S3).
We tested the candidate hairpins for knockdown of the target mRNA and their effects on AID mRNA, and μ-and α–germline transcripts (GLTs). We focused on those genes that did not alter AID mRNA or μ-and α–GLTs and assayed for association of the corresponding protein with AID by co-immunoprecipitation.
Spt5 is required for CSR in CH12 and primary B cells
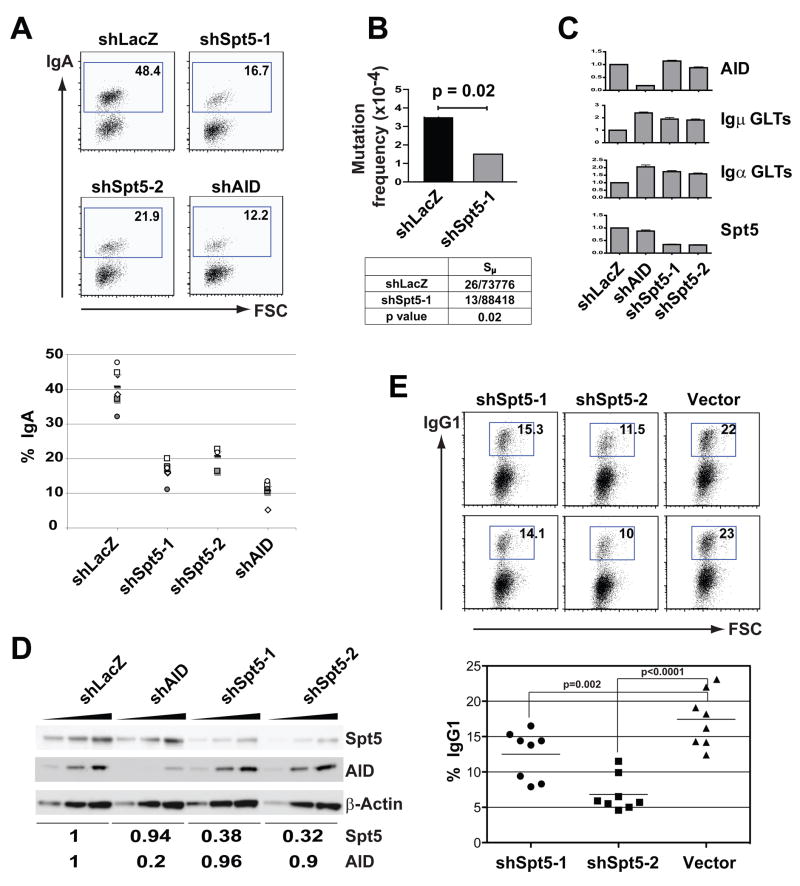
Suppressor of Ty 5 homolog (Spt5), a transcription elongation factor associated with paused Pol II, was selected for further analysis (reviewed in (Gilmour, 2009; Lis, 2007; Peterlin and Price, 2006)). Two unique shRNAs targeting Spt5 decreased CSR (Figure 2A), and the decrease was specific as determined by complementation with an Spt5 cDNA lacking the sequence targeted by shSpt5-1 (Spt5Δ), but not by a cDNA with intact target sites (Figure S1). Spt5 knockdown also decreased switch region hypermutation (Figure 2B), but did not alter the steady state levels of AID, or μ- or α–germline mRNA (Figure 2C), or cell division as measured by CFSE dye dilution (Figure S2). Finally, CH12 cells expressing these shRNAs showed decreased Spt5 protein, whereas AID protein levels were unaltered (Figure 2D).
Figure 2. Spt5 is required for CSR and switch region mutation in CH12 cells.
(A) Upper panel shows representative flow cytometry plots of CH12 infected with two unique shRNAs to Spt5 (shSpt5-1 and shSpt5-2) and controls (shLacZ and shAID) and stimulated to undergo CSR. Numbers indicate percentage of IgA positive cells. The graph in the lower panel summarizes the data from 4–6 independent experiments.
(B) Decreased switch region mutation after Spt5 knockdown in CH12 cells. Upper panel represents the mutation frequency and corresponding p value from control (shLacZ) and shSpt5-1 infected cells stimulated to undergo CSR for 48 hrs. The table in the lower panel summarizes the mutation analysis (represented as unique mutations/nucleotides sequenced).
(C) Graphs show Q-PCR analysis for Spt5, AID, Igα and Igμ germline (GLT) mRNA levels in activated CH12 cells infected as in (A) with the indicated shRNAs. The data summarizes three independent experiments with standard deviation indicated as error bars. In all cases, shLacZ was assigned an arbitrary value of 1.0.
(D) Western blot analysis of Spt5 and AID protein levels in WCEs from activated CH12 cells infected with the indicated shRNAs. Threefold serial dilutions of WCEs were loaded. β-Actin was used as a loading control. Numbers below the blots represent normalized band intensities for Spt5 and AID with the shLacZ lanes assigned an arbitrary value of 1.
(E) Spt5 is required for CSR in primary B cells. Representative flow cytometry plots of B cells stimulated with LPS + IL-4 and infected with retroviruses expressing shSpt5-1, shSpt5-2 or LMP vector alone. Efficiency of switching was determined by gating on GFP-positive cells. Numbers indicate percentage of IgG1 positive cells. The graph in the lower panel summarizes the data from three independent experiments.
See also Figures S2 and S3.
Similarly, primary B cells treated with LPS and IL-4 and infected with retroviruses directing the synthesis of shRNAs specific for Spt5 showed decreased Spt5 protein (Figure S3) and a concomitant decrease in CSR to IgG1 (Figure 2E). We conclude that Spt5 is required for CSR in primary B cells.
Spt5 associates with AID in fibroblasts and primary B cells
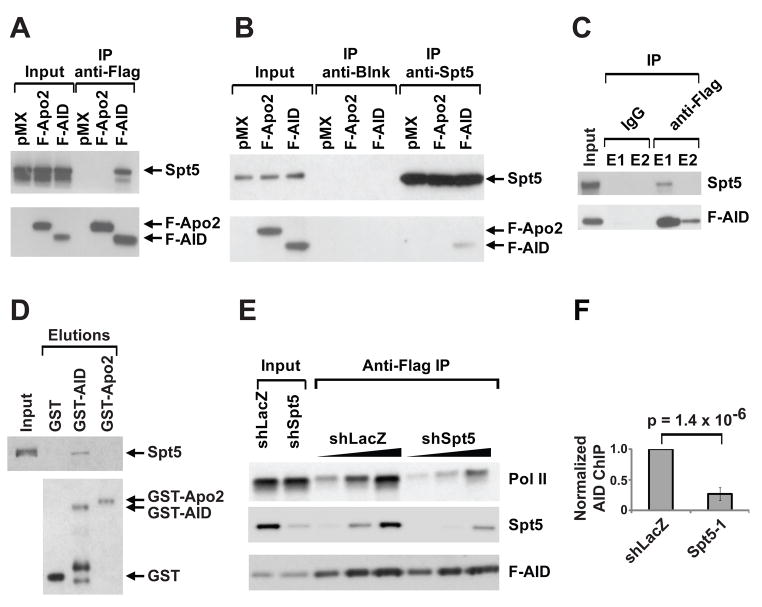
Since both Spt5 (Wada et. al., 1998) and AID (Nambu et al., 2003) associate with Pol II, we asked if Spt5 is also associated with AID. Endogenous Spt5 was co-precipitated from 293T cells transfected with Flag-tagged AID (F-AID) using anti-Flag antibodies (Figure 3A). Conversely, F-AID was co-precipitated by anti-Spt5 antibodies from the same cells under identical conditions (Figure 3B). In contrast, APOBEC-2, a closely related deaminase, did not co-precipitate with Spt5 in either direction (Figure 3A and 3B). Finally, endogenous Spt5 was also co-immunoprecipitated with F-AID from activated B cells isolated from F-AID “knock in” mice (AIDF/F mice) that express physiological levels of AID, and undergo near-normal levels of CSR (Figure 3C and Figure S4). DNA or RNA was not required for the Spt5-AID interaction since the extracts were treated with Benzonase, a nuclease that digests all nucleic acids. We conclude that Spt5 is associated with AID in transfected fibroblasts and activated B cells.
Figure 3. Spt5 interacts with AID in fibroblasts and primary B cells.
(A) Anti-Flag immunoprecipitates from whole cell extracts (WCEs) from 293T cells transfected with Flag-tagged AID (F-AID), or Flag-tagged Apobec2 (F-Apo2) or pMX vector probed with anti-Flag or anti-Spt5 antibodies as indicated.
(B) Anti-Spt5 immunoprecipitates from WCEs from 293T cells transfected as in (A). Blots were probed as in (A). Anti-Blnk was used as an isotype control.
(C) Anti-Flag immunoprecipitates from WCEs from cultured splenic AIDF/F B cells. Blots were probed as in (A). E1 and E2 represent first and second elutions with Flag peptide respectively.
(D) Bacterially expressed GST-AID, GST-APOBEC2 (GST-Apo2) or GST alone were bound to glutathione sepharose beads and incubated with purified recombinant Spt5-Spt4 heterodimer (DSIF). Bound material was eluted with glutathione and analyzed by SDS-PAGE and blotted using antibodies against Spt5 and GST. The input lane for DSIF represents 1% of the amount used in the reaction.
(E) Anti-Flag immunoprecipitates from WCEs of CH12 cells transfected with F-AID and depleted of Spt5 by shSpt5-1. shLacZ is used as a control. Blots were probed as in (A) and with anti-Pol II.
(F) ChIP analysis for AID occupancy in Sμ regions of CH12 cells infected with shSpt5-1 or shLacZ control. Data represents a total of 7 experiments using two different anti-AID antibodies (Chaudhuri et al., 2004; McBride et al., 2006). For each experiment, shLacZ was assigned an arbitrary value of 1. The p value is indicated.
See also Figures S4 and S5.
AID and Spt5 can associate in vitro
Since Spt5 can directly associate with Pol II in vitro (Yamaguchi et al., 1999a), we asked if this was the case for the interaction between Spt5 and AID. To test this idea, bacterially-expressed GST-AID was captured on glutathione sepharose beads and incubated with purified recombinant Spt5. Only a fraction of the Spt5 was specifically bound to GST-AID, but there was no binding to GST-APOBEC2 or GST (Figure 3D). Thus, AID and Spt5 can interact in vitro, but the association is weak and other factors or post-translational modifications likely facilitate this association in vivo. Consistent with this idea, extracts prepared in the presence of phosphatase inhibitors showed increased AID-Spt5 association (Figure S5A). Although AID activity in vivo is enhanced by phosphorylation of serine 38 (S38) or threonine 140 (T140) (Chaudhuri et al., 2004; McBride et al., 2006; McBride et al., 2008), neither S38A nor T140A mutations alter the interaction of AID with Spt5 (Fig S5B).
Pol II association with AID is dependent on Spt5
Spt5 binds to Pol II and induces stalling in vitro (Yamaguchi et al., 1999a) and in vivo (Lis, 2007; Rahl et. al., 2010). In addition, Spt5 also functions as an adaptor that links several co-transcriptional activities to the Pol II machinery (see Discussion). To determine whether Spt5 is required for AID association with Pol II, we depleted Spt5 from CH12 cells expressing F-AID and examined the effects on the association between AID and Pol II. Whereas both Pol II and Spt5 are normally co-precipitated with F-AID, the association between AID and Pol II was decreased in Spt5-depleted cells when compared to the shLacZ control, suggesting that the AID-Pol II interaction (Nambu et al., 2003) is dependent on Spt5 (Figure 3E). In contrast, Pol II depletion did not alter the AID-Spt5 interaction suggesting that Pol II is not essential for this association (Figure S5C). We conclude that Spt5 serves as an adaptor that recruits AID to Pol II.
AID recruitment to Ig switch regions is dependent on Spt5
To determine whether AID recruitment to the Ig switch region is dependent on Spt5, we performed quantitative PCR-based ChIP analysis with two different anti-AID antibodies (Chaudhuri et al., 2004; McBride et al., 2006). Spt5 depletion resulted in significant reduction of AID occupancy in the switch region (Figure 3F, p = 1.4 × 10−6). We conclude that Spt5 is required for AID recruitment to the Ig switch region in B cells undergoing CSR.
Spt5 is associated with stalled Pol II in B cells
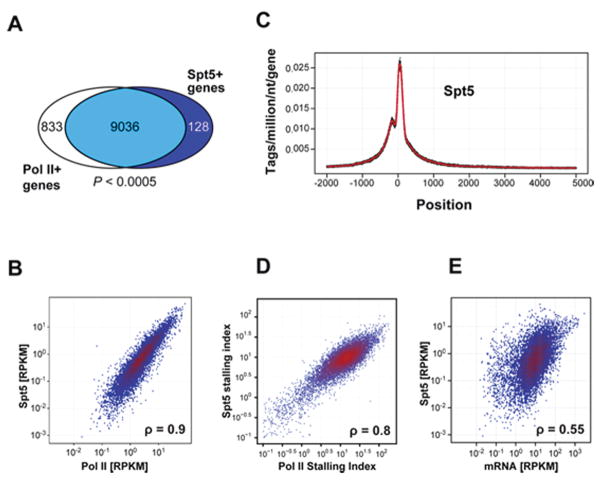
AID mutates Ig genes and up to 25% of the expressed genes in germinal center B cells (Liu et al., 2008; Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998). To determine whether Spt5 localization in the genome of activated B cells coincides with AID-dependent mutation, we performed genome-wide chromatin immunoprecipitation and sequencing (ChIP-seq) with antibodies against Spt5 and Pol II. Spt5 was found throughout the genome of activated B cells undergoing CSR (Table S4). As in other cell types that have been assayed for Spt5 localization, this protein was also concentrated at promoter regions co-incident with Pol II peaks in activated B cells (Gilmour, 2009; Lis, 2007; Peterlin and Price, 2006; Rahl et al., 2010) (Figure 4A–C).
Figure 4. ChIP-Seq analysis of Spt5 genomic occupancy.
(A) Venn diagram showing overlap between genes recruiting Spt5 and Pol II using ChIP-Seq data from LPS+IL4 activated B cells (Table S4). There was a significant association between the presence of Spt5 and Pol II at genes (Pearson’s Chi-square test; P < 0.0005).
(B) Correlation between Spt5 and Pol II density per gene. For each gene that recruited above-background amounts of Pol II and Spt5, the number of sequence tags aligning between -1 Kb upstream of the transcriptional start site to its transcriptional termination site were normalized per gene length (in Kb), per million aligned reads (reads per Kb per million, RPKM) and shown as a hexagonal binning plot. Spearman’s correlation coefficient (ρ) is indicated.
(C) Spt5 profile at all Spt5+ genes from −2 Kb to +5 Kb of the TSS. Data was normalized as reads per million per nucleotide. Dots represent densities at individual nucleotides and the line a 10 nucleotide moving average.
(C) Correlation between the stalling index calculated based on Pol II or Spt5 occupancy (see Material and Methods). Spearman’s correlation coefficient (ρ) is indicated.
(E) Comparative analysis of transcript levels (determined by mRNA-Seq, (Kuchen et al., 2010)) and Spt5 recruitment at all Spt5+ genes. Spearman’s correlation coefficient (ρ) is indicated. See also Tables S4 and S5
Spt5 is a stalling factor in vitro (Wada et al., 1998; Yamaguchi et al., 1999b) and associated with stalled Pol II in various cell types in vivo (Rahl et. al., 2010; Zeitlinger et. al. 2007). The amount of PolII stalling can be quantitated by calculating a stalling or traveling index (Is), which is a ratio of the Pol II density at promoter regions compared to the gene body (Zeitlinger et. al. 2007; see Materials and Methods). Genes with Is > 3 are considered stalled whereas those with Is < 1 are considered elongating genes (Zeitlinger et. al. 2007). Stalling is widespread in the B cell genome (5,594 genes, 61%, Table S4), and in addition, the Pol II and Spt5 stalling indices were significantly correlated (Spearman’s correlation coefficient, ρ =0.8), consistent with previous observations in other cell types (Nachaev et. al., 2010; Rahl et. al., 2010) (Figure 4D, and Yamane et. al., submitted). Most strikingly, AID occupancy in activated B cells is also tightly correlated with Spt5 (see below, and Yamane et. al., unpublished data and data deposition information to be added in proof).
To determine how Spt5 accumulation relates to mRNA levels, we compared the density of Spt5 sequence reads to B cell mRNA-seq levels (both measured as reads per kbp per million sequences (RPKM) (Figure 4D, and (Kuchen et al., 2010)). Although there was some correlation between Spt5 and mRNA levels (Figure 4E, ρ = 0.55), there was a 1- to 2-log variation in mRNA levels for genes accumulating similar levels of Spt5. Thus, in B cells, as in other cells (Nechaev et. al., 2010; Rahl et. al., 2010), Spt5 (or Pol II) accumulation is not necessarily equivalent to cellular mRNA levels.
Spt5 genomic occupancy is predictive of AID-dependent mutation
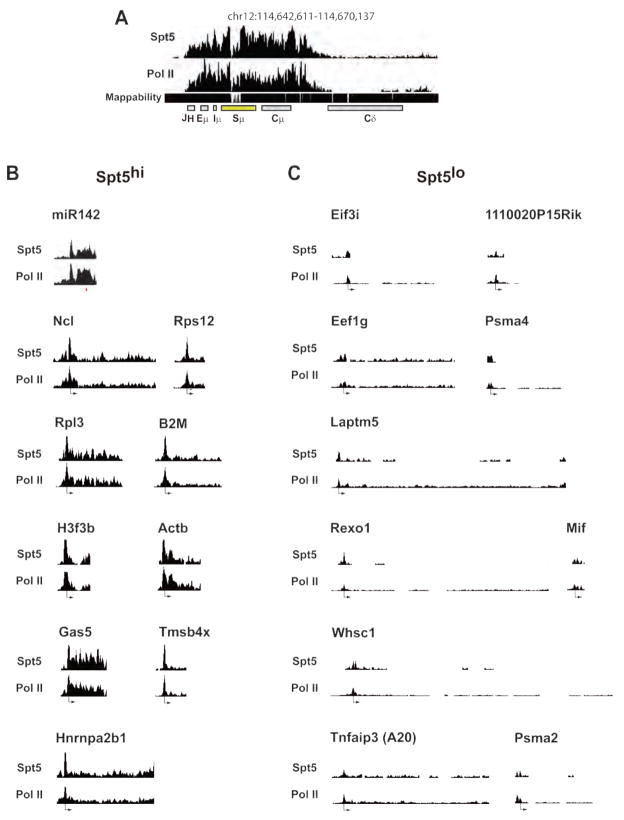
Upon genome-wide analysis of Spt5 occupancy in the promoter proximal region (−1 to 2 kb relative to the transcriptional start site (TSS)), we found that Iμ bore the greatest tag count (Fig 5A and Table S5). The IgVH region could not be mapped because each B cell has a unique rearrangement; however, a strong Spt5 signal was found from the IgH enhancer region through the switch region (Figure 5A). Mir142, a robust AID target (Robbiani et al., 2009), is also embedded in a region of high Spt5 accumulation (Figure 5B and Table S6). In contrast, Taci, Whsc1, H2Eα, A20, Anxa4 and Wdfy3, all of which are expressed in activated B cells (Kuchen et al., 2010), but are not mutated (Liu et al., 2008; Robbiani et al., 2009), do not accumulate Spt5 (Figure 5C and Tables S4 and S5).
Figure 5. ChIP-seq profiles of Spt5 on selected genes.
(A, B and C) Pol II and Spt5 reads per million plotted in 100 bp windows across (A) the Igμ locus, (B) Spt5hi and (C) Spt5lo genes. The axes scales are identical for all histograms. Tag mappability (shown below) was calculated based on the percentage of 36 nt sequences that uniquely aligned to the genomic site with a 10 bp window resolution. Only windows with a significant enrichment compared to a random background model are shown. The location of the TSS for each gene is indicated. The histograms cover the length of the gene. Whsc1 and Tnfaip3 were previously sequenced (Robbiani et. al., 2009). All profiles were generated using the UCSC genome browser.
See also Tables S4 and S5.
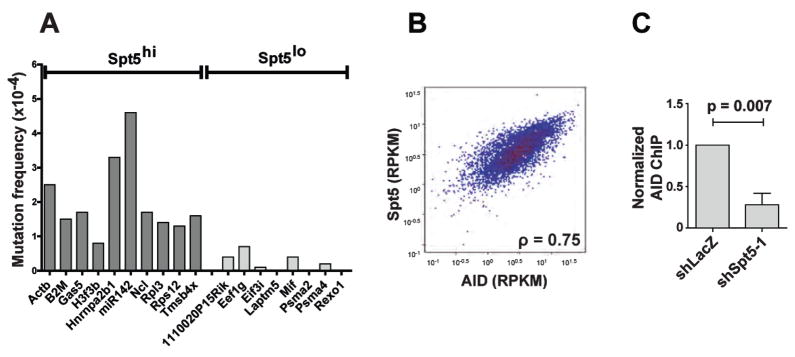
To determine whether Spt5 accumulation is predictive of mutations, we sequenced 10 genes that ranked within the top 5% of genes analyzed for Spt5 tag density (Spt5hi), measured as the density of sequence tags or reads per million base-pairs (TPM), in the promoter-proximal region (Table S5, Figures 5B and 6, and (Kuchen et al., 2010)). As controls, we sequenced 8 highly expressed genes (Kuchen et al., 2010) that had a ~4–6 fold lower Spt5 tag density (Spt5lo) in the same region (Figures 5C and 6, and Table S5). For each selected gene, a region starting around the TSS, corresponding to the peak of Spt5, and extending ~500–600bp downstream was sequenced (Figures 5, 6 and S6). Because the rate of mutation at non-Ig genes is normally very low unless repair is impaired (Liu et al., 2008; Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998), we used B cells derived from transgenic mice over-expressing AID from the Igκ promoter (IgκAID) (Robbiani et al., 2009). These mice display elevated levels of AID protein with concomitant increases in CSR and somatic mutation; nevertheless, they retain AID targeting specificity (Robbiani et al., 2009). All 10 Spt5hi genes (Table S5) were mutated with frequencies from 4.6 × 10−4 for miR142 to 0.8 × 10−4 for H3f3b (Figure 6A and S6). In contrast, none of the 8 Spt5lo genes (Table S5) were mutated above background levels (Figure 6A and S6).
Figure 6. Spt5 occupancy is predictive of AID-dependent somatic mutations.
(A) Graphical representation of somatic mutation analysis for Spt5hi and Spt5lo genes from IgκAID and AID−/− splenic B cells (see Figure 5B and 5C). Mutations in the AID−/− control is subtracted in each case (see Figure S6) and mutation frequencies indicated.
(B) Correlation between Spt5 and AID read density per gene. For each gene that recruited above-background amounts of AID and Spt5, the number of sequence tags aligning between -1 Kb upstream of the transcriptional start site to its transcriptional termination site were normalized per gene length (in Kb), per million aligned reads (reads per Kb per million, RPKM) and shown as a hexagonal binning plot. The Spearman’s correlation coefficient (ρ) is indicated.
(C) ChIP analysis for AID occupancy at the Gas5 gene in CH12 cells infected with shSpt5-1 or shLacZ control. Data represents a total of 4 experiments using two different anti-AID antibodies. For each experiment, shLacZ was assigned an arbitrary value of 1. The p value is indicated. See also Figure S6.
To determine whether genes occupied by Spt5 correspond to sites of AID recruitment, we compared Spt5 and AID ChIP-seq data (Figure 6B and Yamane et. al., unpublished data and data deposition information to be added in proofs). Strikingly, the tag density for AID per gene (measured as reads per kilobase per million (RPKM)) was uniformly and directly proportional to the tag density of Spt5 (ρ = 0.75, Figure 6B and Yamane et. al., unpublished data and deposition information to be added in proof s). To determine if AID recruitment to non-Ig genes was dependent on Spt5, we performed ChIP for AID localization at the Gas5 gene, a stalled gene (Table S4) which accumulates AID-mediated mutation (Figure 6A). As shown in Figure 6C, AID recruitment to Gas5 is impaired upon Spt5 depletion. We conclude that Spt5 and AID accumulation coincide genome-wide and that high density Spt5 occupancy is predictive of AID-mediated mutation.
Discussion
Genetic and biochemical evidence indicate that AID initiates SHM, CSR and chromosome translocation by deaminating cytidine residues in ssDNA that are exposed during transcription (Chaudhuri and Alt, 2004; Di Noia and Neuberger, 2007; Nussenzweig and Nussenzweig, 2010; Peled et. al., 2008; Stavnezer et. al., 2008). AID initiated processes are therefore limited by regulators of transcription initiation such as PTIP, which facilitates Pol II access to specific switch regions by regulating their H3K4 methylation (Daniel et. al., 2010). However, active transcription is not sufficient to allow AID access to DNA, and cannot explain why AID-mediated lesions are found primarily in the promoter proximal region of only some transcribed genes. Since Pol II stalling is a feature of promoter-proximal regions, the observation that Spt5, a stalling factor, associates with AID and is required for AID localization to target genes, provides a molecular explanation for the pattern of mutation.
Inducible transcription of genes carrying paused Pol II is an important mechanism for regulating gene expression (Gilmour, 2009; Lis, 2007; Peterlin and Price, 2006; Bai et. al., 2010; Core et al., 2008; Guenther et al., 2007; Lefebvre et. al., 2002; Muse et al., 2007; Zeitlinger et al., 2007; Bentley and Groudine, 1986; Krumm et al., 1992;Raschke et al., 1999;Kao et al., 1987). Pausing is typically found downstream of promoters and is associated with permanganate sensitivity, which is indicative of the presence of ssDNA (Giardina et al., 1992).
Spt5 is required for Pol II stalling in vitro and in vivo
Spt5 was originally identified as an elongation factor in a yeast suppressor screen (Swanson et. al., 1991). It was subsequently purified biochemically as a heterodimeric complex with Spt4 called 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity inducing factor (DSIF) (Wada et al., 1998; Yamaguchi et al., 1999b). DSIF, in association with negative elongation factor (NELF), binds to Pol II and induces pausing in vitro (Wada et al., 1998; Yamaguchi et al., 1999a). Genome-wide ChIP studies have established a strong correlation between Spt5 and Pol II stalling in vivo (Rahl et al., 2010). These and related studies showed that the presence of Pol II in promoter regions does not necessarily correlate with transcription (Bai et. al., 2010; Gilmour, 2009; Lefebvre et. al., 2002; Lis, 2007; Nachaev et. al. 2010; Peterlin and Price, 2006; Rahl et al., 2010). Consistent with these studies, we find only a partial correlation between Spt5 or Pol II occupancy and mRNA levels in activated B cells (Figure 4E), and importantly, that shRNA knockdown of Spt5 did not decrease AID mRNA, or Igμ or Igα sterile transcripts (Figures 2C and 2D).
Current models suggest that the stalled Pol II complex is re-activated by inductive signals that recruit the P-TEFβ kinase, which phosphorylates Pol II and Spt5, thereby releasing NELF from the complex and activating transcription (Kim and Sharp, 2001; Marshall et al., 1996; Marshall and Price, 1995; Wada et al., 1998; Yamada et al., 2006). Phosphorylated Spt5 remains associated with Pol II throughout the elongation phase. Spt5 also engages in interactions with various co-transcriptional factors thereby serving as an adaptor linking these factors to the transcriptional machinery. Spt5 links Pol II to splicing factors (Pei and Shuman, 2002), capping enzyme (Wen and Shatkin, 1999), the exosome complex (Andrulis et al., 2002), transcription coupled repair factors (Ding et al., 2010), NFkB, and E-box proteins (Amir-Zilberstein and Dikstein, 2008). Our data suggest that Spt5 also facilitates the interaction of AID with Pol II (Figure 3E) and thereby targets this enzyme to genomic loci accumulating paused Pol II (Figures 3F, 4D, 6B and Yamane et. al., unpublished data and data deposition information to be added in proofs).
Stalled Pol II in the Ig locus
In activated B cells, the Ig locus is unique in having a large domain of densely packed Spt5 and Pol II molecules extending several kilobases (Figure 5A and Tables S4 and S5). The idea that Pol II pausing might be linked to mutation (Peters and Storb, 1996) was proposed based on the characteristics of Ig hypermutation, and the position of hypermutation relative to transcriptional start sites (reviewed in (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008; Storb et al., 2007)). A mutator factor, MuF, was hypothesized to associate with Pol II and generate mutations when Pol II is paused during elongation (Peters and Storb, 1996). More recently, detailed analyses of transcription and Pol II occupancy in the switch regions have confirmed that transcription is indeed impeded throughout the switch regions, most likely due to the presence of G-rich repetitive sequence elements that facilitate DNA distortion and formation of R loops (Daniels and Lieber, 1995; Rajagopal et al., 2009; Ronai et al., 2007; Tian and Alt, 2000; Wang et al., 2009; Yu et al., 2003). Altogether, this makes the Ig locus an ideal substrate for targeted mutation by AID because: (1) Spt5 facilitates association between AID and Pol II, (2) the stalled Pol II molecules provide an abundance of ssDNA for AID, and (3) the reduced rate of elongation provides AID with increased time of residence at the target.
Finally, in addition to the switch region, several genes mutated by AID were already known to have paused Pol II at sites corresponding to regions that are somatically mutated including c-myc (Bentley and Groudine, 1986; Krumm et al., 1992), Pim1 (Rohwer et al., 1996), and Igκ (Raschke et al., 1999). Our experiments provide a mechanistic explanation for the association between Pol II stalling and AID-mediated somatic mutation. In addition, they reveal the full spectrum of AID targets, including genes such as Gas5, which also undergoes reciprocal translocation in B cell lymphomas (Nakamura et al., 2008).
Concluding remarks
Although our findings demonstrate a mechanism by which AID gains access to the promoter proximal region of genes, several questions remain about how antibody diversification is mediated. In particular, AID recruitment is only the first of several steps required to bring about CSR and SHM. Following its recruitment to DNA, AID must gain access to target DNA. Although Spt5 acts as an adaptor for AID, localizing it to paused Pol II and associated ssDNA, this may not be sufficient. AID mutates both DNA strands, and paused Pol II exposes only the non-transcribed strand (Giardina et al., 1992; Gilmour, 2009; Lis, 2007; Peterlin and Price, 2006). In addition, the association between AID and paused Pol II does not explain why repair differs between Ig and non-Ig genes, and between different non-Ig AID targets (Liu et al., 2008). Hence, the mechanisms governing post-AID recruitment events required for CSR and SHM remain to be elucidated. AID and Spt5 can interact directly in vitro but the interaction is weak suggesting that additional factors or posttranslational modifications may be required. Nevertheless, our data suggests that Spt5 links Pol II and AID, thereby providing a mechanistic explanation for the well-established correlation between AID and transcription. The association between Spt5 and AID also explains intrinsic features of hypermutation and CSR, including the enrichment of mutation in the promoter-proximal regions, which correspond to sites of Pol II stalling (Nechaev et. al., 2010; Rahl et. al. 2010; Zeitlinger et. al. 2007).
In conclusion, we propose that AID utilizes the phenomenon of Pol II stalling, which is widespread in the B cell genome, and is particularly prominent on Ig loci, to gain access to its target genes across the genome.
Experimental Procedures
Library preparation
The lentiviral shRNA library (Table S2) was prepared, titered, arrayed and validated as described (Moffat et al., 2006; Root et al., 2006; http://www.broadinstitute.org/rnai/trc/lib).
Library screening
The lentiviral library was screened in a 96-well plate format in triplicate, starting from the infection stage through to flow cytometry analysis (schematically represented in Figure 1C). Each plate contained negative control viruses targeting LacZ, GFP and RFP and a positive control shRNA targeting AID. Cells were infected, selected, and stimulated to undergo CSR followed by FACS analysis (details in Supplemental Information).
shRNA knockdown in primary B cells
The hairpin sequences for shSpt5-1 and shSpt5-2 (Table S2) were cloned into the LMP retroviral vector (Open Biosystems) and transfected into BOSC23 cells to produce retrovirus (Robbiani et. al. 2008). Primary B cells stimulated with LPS and IL-4 were cultured as described (Robbiani et. al., 2008). After 24 hr in culture, B cells were infected with shRNA-expressing retroviral supernatants as described (Robbiani et. al., 2008) and cultured for an additional 3 days with LPS and IL-4, followed by FACS analysis for IgG1 and Spt5 protein analysis by western blotting.
Immunoprecipitation (IP)
For Flag-IPs, 2 mg of WCE (prepared as described in Supplemental Information) was incubated with 20 μl Flag Agarose resin (Sigma) for 2 hr at 4°C in IP buffer (identical to WCE preparation buffers above adjusted to 150 mM NaCl for fibroblasts assays, and 200 mM NaCl for B cells and CH12 assays). This was followed by three washes in IP buffer and elution with 0.2 mg/ml Flag peptide (Sigma) for 1 hr at 4°C. Eluates were subjected to SDS-PAGE and western blot analysis. For anti-Spt5 IPs, 2 mgs of WCE were incubated with 3 μg of anti-Spt5 (Santa Cruz Biotechnology) for 2 hrs at 4°C followed by capture of the immune complexes with 20 μl Protein A agarose (Roche) for 1 hr at 4°C. Beads were washed three times with IP buffer and bound material was extracted by boiling in 100 μl of Laemmli sample buffer. Eluted material was analyzed by SDS-PAGE and western blot. Antibodies used for probing western blots were as follows: Flag (Sigma), Spt5 (H300) (SantaCruz Biotechnology), Pol II (4H8) (Abcam) and Phospho-Ser PKC Substrate (Cell Signaling).
AID-Spt5 interaction in vitro
GST fusion proteins were expressed in E. coli and immobilized on Glutathione Sepharose 4 Fast Flow beads (GE Healthcare). Beads were incubated with 500 ng of purified DSIF (Spt5-Spt4) complex (a generous gift from Dr. Sohail Malik, The Rockefeller University) in 200 μl final volume of binding buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.1 % NP-40, 1 mM EDTA, Protease Inhibitor cocktail (Roche), 0.5 mM PMSF, 1 mM DTT, 0.5 mg/ml BSA) for 2 hours at 4°C with gentle rotation. After four washes with binding buffer, bound proteins were eluted by boiling in NuPAGE LDS loading buffer (Invitrogen). Samples were then subjected to SDS-PAGE followed by western blot analysis.
Chromatin Immunoprecipitation and sequencing (ChIP-seq)
ChIP-seq was performed exactly as described (Kuchen et al., 2010). In brief, cells were fixed with 1% paraformaldehyde at 37°C for 10 minutes followed by sonication. Chromatin fragments were then immunoprecipitated with antibodies specific for Spt5 (Santa Cruz Biotechnology (H300) and BD Biosciences (anti-DSIF)), RNA Pol II (Abcam, (4H8)) or Ser5-phosphorylated RNA Pol II (Abcam, (phospho-S5)). Immunoprecipitates were processed following Illumina’s protocol and sequenced on a Genome Analyzer. During analysis, short sequence tags were trimmed to 25 nts and aligned to the mouse genome (NCBI37/mm9) using Bowtie. Uniquely aligned reads were analyzed by SICER (Zang et al., 2009) using an expectation value E of 50 in a random background model. The requirement for unique alignment was not applied for IgSμ or IgSγ1 because of their high repetitive nature and low mappability (Figure 5A). Reads on significant islands as defined by SICER were normalized to the total number of reads on islands. Downstream analysis was carried out in R and C++.
Quantitative AID ChIP
CH12 cells were infected with shRNAs to Spt5 as above and subjected to ChIP analysis using two different anti-AID antibodies (Chaudhuri et al., 2004; McBride et al., 2006). Assays were performed as described (Vuong et. al., 2009). The ChIP’d material was analyzed by Q-PCR (described above) and raw values were normalized to the input signals for each sample (Vuong et. al., 2009). Reactions were performed in triplicate. Forward and reverse primers used for Sμ amplification were 5′ TAGTAAGCGAGGCTCTAAAAAGCAT 3′ and 5′ AGAACAGTCCAGTGTAGGCAGTAGA 3′ respectively. Forward and reverse primers used for Gas5 amplification were 5′ TATGGCTTCGGGCCTTGGA 3′ and 5′ CCTCCTAAAGTTTCCAGCTTGTGC 3′ respectively.
Calculation of the stalling index
The stalling index was calculated based on Pol II ChIP-seq reads as described (Rahl et. al., 2010; Zeitlinger et. al., 2007). Briefly, the Pol II and Spt5 stalling indices are calculated in the same way and represent the ratio of read density at the promoter to the average gene body density. The promoter was defined as a 1 kb region extending from −0.5 kb to +0.5 kb relative to the TSS, and the gene body was defined as the region from +1kb downstream of the TSS up to the transcription termination site (TTS) (Rahl et. al., 2010; Zeitlinger et. al., 2007).
Additional experimental procedures can be found in the Supplemental Information.
Supplementary Material
Figure S1. Expression of shRNA-resistant Spt5 cDNA restores CSR in CH12 cells treated with shSpt5-1. Related to Figure 2.
(A) Representative flow cytometry plots of CH12 cells infected with control vector (pMX), or full length Spt5 cDNA, or a 3′UTR truncated Spt5 cDNA (Spt5Δ) which is missing the shSpt5-1 target site. 24 hours later, cells were infected with control (pLKO), or shSpt5-1, or shAID GFP expressing lentiviruses. The numbers show the percentage of IgA expressing, GFP positive, puromycin resistant cells.
(B) Graph summarizes the data from 4–8 independent experiments. Each point represents an independent culture. The amount of CSR in the pLKO-control was assigned a value of 100%. The effect of the shRNA was calculated relative to pLKO-control. Numbers and a bar show the average. The p values are indicated.
Figure S2. Effect of Spt5 knockdown on cell proliferation. Related to Figure 2.
Duplicate experiments showing CH12 cells infected with shRNAs as described in the main text, followed by puromycin selection. After 48 hr of selection, cells were labeled with CFSE (5 μM) for 10 min at 37°C followed by CSR induction with cytokines. FACS analysis was done at 48 hr post-stimulation. Histograms show CFSE dye dilution for un-stimulated (blue) and stimulated (red) cells. Numbers indicate percentage of IgA+ cells.
Figure S3. Analysis of Spt5 knockdown in primary B cells. Related to Figure 2.
Primary B cells were retrovirally infected in duplicate with shRNAs against shSpt5-1, shSpt5-2 and LMP vector, as described in Figure 2E, in the presence of LPS and IL-4 stimulation for 4 days. WCEs were analyzed for Spt5 protein levels by SDS-PAGE and western blot. Threefold dilutions are shown. β-Actin is a loading control.
Figure S4. Gene Targeting Strategy and Characterization of Flag-AID “knock in” Mice. Related to Figure 3.
(A) Schematic representation of “knock in” strategy. A tandem Flag and HA tag (MDYKDDDDKGGYPYDVPDYA), depicted by red box and coded by ATGGACTACAAGGACGACGATGACAAGGGAGGATATCCGTATGATGTTCCTGATTATGCT was incorporated in frame to the amino terminus of AID in Exon 1. A neomycin resistance cassette flanked by FRT sites was used for positive selection. Gray arrows show the location of PCR primers, black boxes depict exons, triangles depict FRT sites. The neomycin cassette was removed by breeding to FRT mice.
(B) PCR screen for AIDF/F mice. PCR on genomic DNA with primers F- (GGACCCAACCCAGGCGGCAGCTGT) and R- (CCTCTAAGGCTTCGCTGTTATTACCAC). PCR results in 480bp band for wild type and 660 bp band for FlagHA-knockin allele after excision of the Neomycin selection cassette. Result for wild type, AIDF/+ and AIDF/F are shown.
(C) Expression of FlagHA-AID in AIDF/F B cells. Anti-AID immunoblot of lysates immunoprecipitated with indicated antibody from wild type, AID−/− or AIDF/+ splenocytes stimulated for 72 hours with IL-4 and LPS.
(D) CSR to IgG1 in AIDF/F B cells. FACS plot of IgG1 expression by wild type, AID−/− AIDF/+ or AIDF/F splenocytes stimulated for 72 hours with IL-4 and LPS.
Figure S5. AID-Spt5 association is not affected by Pol II depletion or AID phosphorylation. Related to Figure 3.
(A) WCEs from fibroblasts transfected with F-AID were prepared in the presence of Lambda phosphatase (λ) or PhosStop phosphatase inhibitor cocktail (Roche) as indicated, followed by Flag-IP as in (A). As a control for the efficacy of phosphatase and phosphatase inhibitor treatments, a region of the input blot was probed with Phospho-(Ser) PKC Substrate antibody (Cell Signaling) which detects phosphoserine residues on several endogenous proteins.
(B) F-AIDWT, F-AIDS38A and F-AIDT140A were transfected into 293T cells, and WCEs subjected to Flag-IP. Eluted material was analyzed by SDS-PAGE and blotted using antibodies against Spt5 and Flag. Threefold serial dilutions are shown.
(C) Left panel: WCEs from F-AID transfected fibroblasts were immunodepleted of Pol II using anti-Pol II or anti-HA isotype control. The flow-through was analyzed by SDS-PAGE and western blot using antibodies against Pol II, Spt5 and Flag as indicated. Twofold serial dilutions are shown. Right panel: Flag IP was performed with samples shown in (A) and analyzed by SDS-PAGE and western blot using antibodies against Pol II, Spt5 and Flag. Threefold serial dilutions are shown.
Figure S6. Summary of mutation analysis of Spt5hi and Spt5lo genes in IgκAID and AID−/− B cells as shown in Figure 6A. The data is represented as number of unique mutations/nucleotides sequenced. P values were calculated by the Student’s T test. Related to Figures 5 and 6.
Table S1. Gene Expression Analysis in Resting and Stimulated CH12 Cells. Related to Figure 1.
RNA was extracted from resting cells or cells stimulated for 48 hr with IL-4, αCD40 and TGFβ with the RNA extraction kit (Qiagen). Gene array analysis was performed on an Affymetrix platform at the Genomics Core Laboratory, Memorial Sloan-Kettering Cancer Center. Microarray data were analyzed using Affymetrix GeneSpring 7.0 software.
Table S2. List of Genes and Corresponding shRNA Hairpin Sequences in the Lentiviral Library. Related to Figure 1.
The library was prepared, arrayed and validated as described (Klein et al., 2003; Moffat et al., 2006; Root et al., 2006). Viral supernatants were supplied in 96-well plates. Each plate represented ~17 genes with ~5 shRNAs per gene along with several control shRNAs (LacZ, GFP, RFP and AID). The distribution of shRNAs was randomized in each plate to accommodate position effects.
Table S3. List of hits from the lentiviral shRNA screen with known or predictive functions in CSR. Related to Figure 1.
These genes have either been shown to be involved in CSR or are expected to be involved in CSR, based on their function and/or location in well described signaling pathways, DNA repair etc.
Table S4. ChIP-seq analysis for Spt5 and Pol II gene occupancy (−1Kb to TTS relative to TSS) from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S5. ChIP-seq analysis for Spt5 and Pol II gene occupancy at proximal promoter sequences (−1Kb to +2Kb relative to TSS) from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S6. ChIP-seq analysis for Spt5 and Pol II miRNA gene occupancy from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S7. Primer sequences used in Q-PCR and somatic mutation analysis. Related to Figures 2 and 6.
For sequencing, the amplified region encompasses the peak of Spt5 around the TSS (see Figure 5) and extends downstream for ~500–600 nucleotides.
Acknowledgments
We thank members of the Nussenzweig lab for helpful discussions, Klara Velinzon for FACS sorting, and Tom Eisenreich and David Bosque for animal management. We thank Drs Jayanta Chaudhuri and Urszula Nowak for ChIP protocols and anti-AID antibody, Dr. Sohail Malik for generously providing purified recombinant DSIF and Dr. Alan Derr for assistance with informatics. M.D.V. is a fellow of the American-Italian Cancer Foundation. R.P was a recipient of The Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute. The work was supported by NIH grant (AI037526) to M.C.N. M.C.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amir-Zilberstein L, Dikstein R. Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF) J Biol Chem. 2008;283:1317–1323. doi: 10.1074/jbc.M706767200. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec M, Akie T, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald D, Lin S, Cantor A, Orkin S, Zon L. TIF1γ Controls Erythroid Cell Fate by Regulating Transcription Elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nature Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Ganesh K, Xue K, Lu M, Rada C, Neuberger MS. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen H, Gazumyan A, Yamane A, Cho Y, Sun H, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP Promotes Chromatin Changes Critical for Immunoglobulin Class Switch Recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem. 2010;285:5317–5326. doi: 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurrence A, Yasuda T, Peng S, Hu-Li J, et al. Regulation of MicroRNA Expression and Abundance during Lymphopoesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Brand C, Lefebvre B, Ozato K. Chromosomal Integration of Retinoic Acid Response Elements Prevents Cooperative Transcriptional Activation by Retinoic Acid Receptor and Retinoid X Receptor. Mol Cell Biol. 2002;22:1446–1459. doi: 10.1128/mcb.22.5.1446-1459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K, Gazumyan A, Woo E, Schwickert T, Chait B, Nussenzweig M. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Kondo S, Sugai M, Nazarea M, Imamura S, Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Takahashi N, Kakegawa E, Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I, Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet. 2008;182:144–149. doi: 10.1016/j.cancergencyto.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo D, Santos G, Liu L, Gao Y, Adelman K. Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci U S A. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The Biochemistry of Somatic Hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Raschke EE, Albert T, Eick D. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J Immunol. 1999;163:4375–4382. [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Todd S, McGuire KL. The effect of IL-2 treatment on transcriptional attenuation in proto-oncogenes pim-1 and c-myb in human thymic blast cells. J Immunol. 1996;157:643–649. [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Shen HM, Poirier MG, Allen MJ, North J, Lal R, Widom J, Storb U. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J Exp Med. 2009;206:1057–1071. doi: 10.1084/jem.20082678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U, Shen HM, Longerich S, Ratnam S, Tanaka A, Bozek G, Pylawka S. Targeting of AID to immunoglobulin genes. Adv Exp Med Biol. 2007;596:83–91. doi: 10.1007/0-387-46530-8_8. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:4286. doi: 10.1128/mcb.11.8.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Vuong B, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight G, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207:141–153. S141–146. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999a;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999b;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, Alt FW. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression of shRNA-resistant Spt5 cDNA restores CSR in CH12 cells treated with shSpt5-1. Related to Figure 2.
(A) Representative flow cytometry plots of CH12 cells infected with control vector (pMX), or full length Spt5 cDNA, or a 3′UTR truncated Spt5 cDNA (Spt5Δ) which is missing the shSpt5-1 target site. 24 hours later, cells were infected with control (pLKO), or shSpt5-1, or shAID GFP expressing lentiviruses. The numbers show the percentage of IgA expressing, GFP positive, puromycin resistant cells.
(B) Graph summarizes the data from 4–8 independent experiments. Each point represents an independent culture. The amount of CSR in the pLKO-control was assigned a value of 100%. The effect of the shRNA was calculated relative to pLKO-control. Numbers and a bar show the average. The p values are indicated.
Figure S2. Effect of Spt5 knockdown on cell proliferation. Related to Figure 2.
Duplicate experiments showing CH12 cells infected with shRNAs as described in the main text, followed by puromycin selection. After 48 hr of selection, cells were labeled with CFSE (5 μM) for 10 min at 37°C followed by CSR induction with cytokines. FACS analysis was done at 48 hr post-stimulation. Histograms show CFSE dye dilution for un-stimulated (blue) and stimulated (red) cells. Numbers indicate percentage of IgA+ cells.
Figure S3. Analysis of Spt5 knockdown in primary B cells. Related to Figure 2.
Primary B cells were retrovirally infected in duplicate with shRNAs against shSpt5-1, shSpt5-2 and LMP vector, as described in Figure 2E, in the presence of LPS and IL-4 stimulation for 4 days. WCEs were analyzed for Spt5 protein levels by SDS-PAGE and western blot. Threefold dilutions are shown. β-Actin is a loading control.
Figure S4. Gene Targeting Strategy and Characterization of Flag-AID “knock in” Mice. Related to Figure 3.
(A) Schematic representation of “knock in” strategy. A tandem Flag and HA tag (MDYKDDDDKGGYPYDVPDYA), depicted by red box and coded by ATGGACTACAAGGACGACGATGACAAGGGAGGATATCCGTATGATGTTCCTGATTATGCT was incorporated in frame to the amino terminus of AID in Exon 1. A neomycin resistance cassette flanked by FRT sites was used for positive selection. Gray arrows show the location of PCR primers, black boxes depict exons, triangles depict FRT sites. The neomycin cassette was removed by breeding to FRT mice.
(B) PCR screen for AIDF/F mice. PCR on genomic DNA with primers F- (GGACCCAACCCAGGCGGCAGCTGT) and R- (CCTCTAAGGCTTCGCTGTTATTACCAC). PCR results in 480bp band for wild type and 660 bp band for FlagHA-knockin allele after excision of the Neomycin selection cassette. Result for wild type, AIDF/+ and AIDF/F are shown.
(C) Expression of FlagHA-AID in AIDF/F B cells. Anti-AID immunoblot of lysates immunoprecipitated with indicated antibody from wild type, AID−/− or AIDF/+ splenocytes stimulated for 72 hours with IL-4 and LPS.
(D) CSR to IgG1 in AIDF/F B cells. FACS plot of IgG1 expression by wild type, AID−/− AIDF/+ or AIDF/F splenocytes stimulated for 72 hours with IL-4 and LPS.
Figure S5. AID-Spt5 association is not affected by Pol II depletion or AID phosphorylation. Related to Figure 3.
(A) WCEs from fibroblasts transfected with F-AID were prepared in the presence of Lambda phosphatase (λ) or PhosStop phosphatase inhibitor cocktail (Roche) as indicated, followed by Flag-IP as in (A). As a control for the efficacy of phosphatase and phosphatase inhibitor treatments, a region of the input blot was probed with Phospho-(Ser) PKC Substrate antibody (Cell Signaling) which detects phosphoserine residues on several endogenous proteins.
(B) F-AIDWT, F-AIDS38A and F-AIDT140A were transfected into 293T cells, and WCEs subjected to Flag-IP. Eluted material was analyzed by SDS-PAGE and blotted using antibodies against Spt5 and Flag. Threefold serial dilutions are shown.
(C) Left panel: WCEs from F-AID transfected fibroblasts were immunodepleted of Pol II using anti-Pol II or anti-HA isotype control. The flow-through was analyzed by SDS-PAGE and western blot using antibodies against Pol II, Spt5 and Flag as indicated. Twofold serial dilutions are shown. Right panel: Flag IP was performed with samples shown in (A) and analyzed by SDS-PAGE and western blot using antibodies against Pol II, Spt5 and Flag. Threefold serial dilutions are shown.
Figure S6. Summary of mutation analysis of Spt5hi and Spt5lo genes in IgκAID and AID−/− B cells as shown in Figure 6A. The data is represented as number of unique mutations/nucleotides sequenced. P values were calculated by the Student’s T test. Related to Figures 5 and 6.
Table S1. Gene Expression Analysis in Resting and Stimulated CH12 Cells. Related to Figure 1.
RNA was extracted from resting cells or cells stimulated for 48 hr with IL-4, αCD40 and TGFβ with the RNA extraction kit (Qiagen). Gene array analysis was performed on an Affymetrix platform at the Genomics Core Laboratory, Memorial Sloan-Kettering Cancer Center. Microarray data were analyzed using Affymetrix GeneSpring 7.0 software.
Table S2. List of Genes and Corresponding shRNA Hairpin Sequences in the Lentiviral Library. Related to Figure 1.
The library was prepared, arrayed and validated as described (Klein et al., 2003; Moffat et al., 2006; Root et al., 2006). Viral supernatants were supplied in 96-well plates. Each plate represented ~17 genes with ~5 shRNAs per gene along with several control shRNAs (LacZ, GFP, RFP and AID). The distribution of shRNAs was randomized in each plate to accommodate position effects.
Table S3. List of hits from the lentiviral shRNA screen with known or predictive functions in CSR. Related to Figure 1.
These genes have either been shown to be involved in CSR or are expected to be involved in CSR, based on their function and/or location in well described signaling pathways, DNA repair etc.
Table S4. ChIP-seq analysis for Spt5 and Pol II gene occupancy (−1Kb to TTS relative to TSS) from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S5. ChIP-seq analysis for Spt5 and Pol II gene occupancy at proximal promoter sequences (−1Kb to +2Kb relative to TSS) from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S6. ChIP-seq analysis for Spt5 and Pol II miRNA gene occupancy from WT splenic B cells stimulated for 48 hrs with LPS and IL-4. Related to Figures 4–6.
Table S7. Primer sequences used in Q-PCR and somatic mutation analysis. Related to Figures 2 and 6.
For sequencing, the amplified region encompasses the peak of Spt5 around the TSS (see Figure 5) and extends downstream for ~500–600 nucleotides.