Abstract
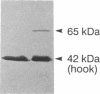
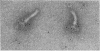
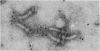
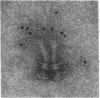
The M ring is a substructure of the flagellar basal body of bacteria, which lies in the cytoplasmic membrane and is therefore close to the site where the energy of the transmembrane proton potential is converted into mechanical work of rotation of the motor. The protein from which this ring is constructed has not been identified. Flagellar hook-basal body complexes from Salmonella typhimurium were used as the immunogen for the preparation of monoclonal antibodies. An antibody obtained was directed against a major basal-body component, a 65-kDa protein that from mutant studies has been assigned as the product of the flaAII.1 gene. By immunoelectron microscopy, the antibody was observed to bind the innermost feature of the basal body: the cytoplasmic-facing surface of the M ring. We conclude that the 65-kDa protein is a component--probably the main component--of this important substructure of the flagellar motor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aizawa S. I., Dean G. E., Jones C. J., Macnab R. M., Yamaguchi S. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol. 1985 Mar;161(3):836–849. doi: 10.1128/jb.161.3.836-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad D., Koshland D. E., Jr Isolation, characterization and complementation of Salmonella typhimurium chemotaxis mutants. J Mol Biol. 1975 Sep 15;97(2):225–235. doi: 10.1016/s0022-2836(75)80036-2. [DOI] [PubMed] [Google Scholar]
- Bartlett D. H., Matsumura P. Identification of Escherichia coli region III flagellar gene products and description of two new flagellar genes. J Bacteriol. 1984 Nov;160(2):577–585. doi: 10.1128/jb.160.2.577-585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C. Dynamic properties of bacterial flagellar motors. Nature. 1974 May 3;249(452):77–79. doi: 10.1038/249077a0. [DOI] [PubMed] [Google Scholar]
- Block S. M., Berg H. C. Successive incorporation of force-generating units in the bacterial rotary motor. 1984 May 31-Jun 6Nature. 309(5967):470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- Clegg D. O., Koshland D. E., Jr Identification of a bacterial sensing protein and effects of its elevated expression. J Bacteriol. 1985 Apr;162(1):398–405. doi: 10.1128/jb.162.1.398-405.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cyoplasmic membrane. J Bacteriol. 1971 Jan;105(1):396–407. doi: 10.1128/jb.105.1.396-407.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard J. Y., Hoffman T. Enzyme-linked immunosorbent assay for screening monoclonal antibody production using enzyme-labeled second antibody. Methods Enzymol. 1983;92:168–174. doi: 10.1016/0076-6879(83)92016-5. [DOI] [PubMed] [Google Scholar]
- Glagolev A. N., Skulachev V. P. The proton pump is a molecular engine of motile bacteria. Nature. 1978 Mar 16;272(5650):280–282. doi: 10.1038/272280a0. [DOI] [PubMed] [Google Scholar]
- Homma M., Iino T. Excretion of unassembled hook-associated proteins by Salmonella typhimurium. J Bacteriol. 1985 Dec;164(3):1370–1372. doi: 10.1128/jb.164.3.1370-1372.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Iino T. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J Bacteriol. 1985 Apr;162(1):183–189. doi: 10.1128/jb.162.1.183-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T., Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987 Apr;169(4):1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985 Aug;163(2):464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T., Yamaguchi S. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium. J Bacteriol. 1984 Jan;157(1):100–108. doi: 10.1128/jb.157.1.100-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Ohnishi K., Iino T., Macnab R. M. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII and FlaFVIII) in Salmonella typhimurium. J Bacteriol. 1987 Aug;169(8):3617–3624. doi: 10.1128/jb.169.8.3617-3624.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Homma M., Iino T., Asakura S., Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987 Mar;169(3):1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol. 1987 Apr;169(4):1489–1492. doi: 10.1128/jb.169.4.1489-1492.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Berg H. C. Isotope and thermal effects in chemiosmotic coupling to the flagellar motor of Streptococcus. Cell. 1983 Mar;32(3):913–919. doi: 10.1016/0092-8674(83)90076-4. [DOI] [PubMed] [Google Scholar]
- Komeda Y., Kutsukake K., Iino T. Definition of additional flagellar genes in Escherichia coli K12. Genetics. 1980 Feb;94(2):277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Silverman M., Matsumura P., Simon M. Genes for the hook-basal body proteins of the flagellar apparatus in Escherichia coli. J Bacteriol. 1978 May;134(2):655–667. doi: 10.1128/jb.134.2.655-667.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S. H., Reader R. W., Kort E. N., Tso W. W., Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974 May 3;249(452):74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Läuger P. Ion transport and rotation of bacterial flagella. Nature. 1977 Jul 28;268(5618):360–362. doi: 10.1038/268360a0. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Aizawa S. Bacterial motility and the bacterial flagellar motor. Annu Rev Biophys Bioeng. 1984;13:51–83. doi: 10.1146/annurev.bb.13.060184.000411. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Tedesco P., Berg H. C., Harold F. M., Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A. 1977 Jul;74(7):3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Matsumura P., Simon M. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974 May 3;249(452):73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Taira T., Kutsukake K., Homma M., Iino T. Genetic analysis of three additional fla genes in Salmonella typhimurium. J Gen Microbiol. 1984 Dec;130(12):3339–3342. doi: 10.1099/00221287-130-12-3339. [DOI] [PubMed] [Google Scholar]