Abstract
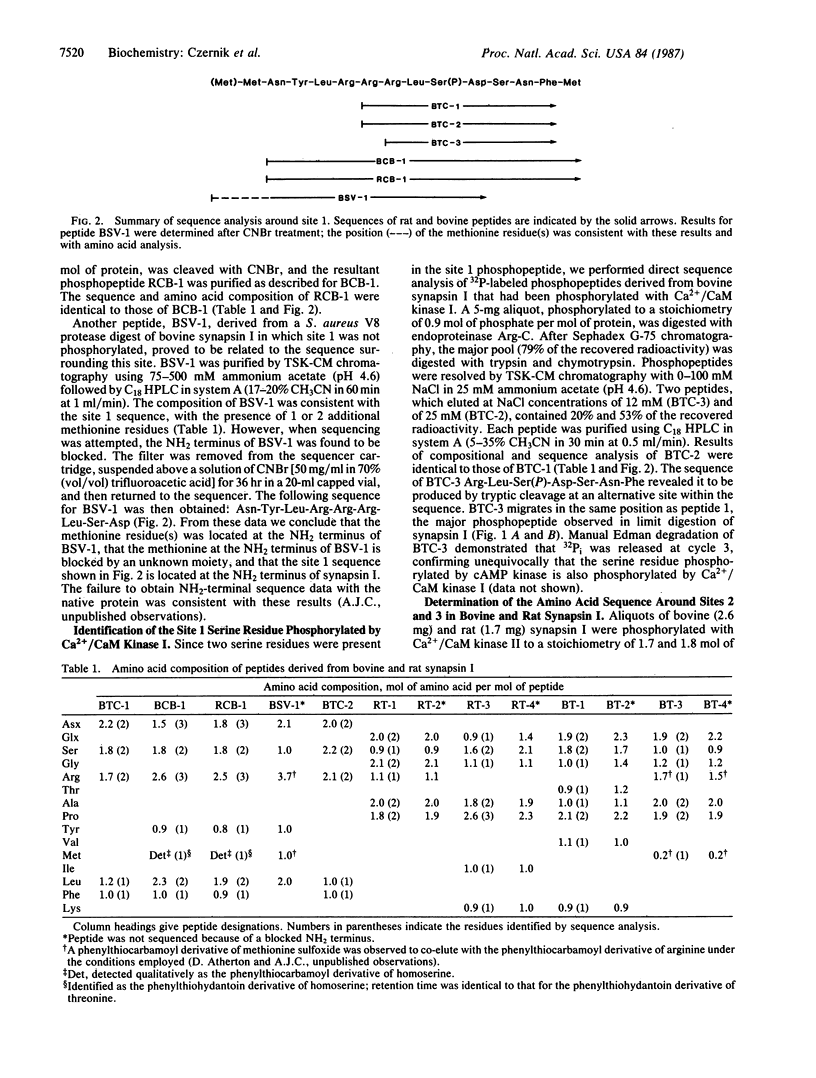
The amino acid sequences surrounding three major phosphorylation sites in rat and bovine synapsin I have been determined by employing automated gas-phase sequencing and manual Edman degradation of purified phosphopeptide fragments. Site 1 is a serine residue phosphorylated by cAMP-dependent protein kinase and by calcium/calmodulin-dependent protein kinase I. The sequence around site 1 was derived from tryptic/chymotryptic phosphopeptides and overlapping cyanogen bromide cleavage fragments. This sequence, identical in rat and bovine synapsin I, is Asn-Tyr-Leu-Arg-Arg-Arg-Leu-Ser(P)-Asp-Ser-Asn-Phe-Met. Site 1 is located at the NH2 terminus of the protein, within the collagenase-resistant head region. Sites 2 and 3 are serine residues phosphorylated by calcium/calmodulin-dependent protein kinase II. The sequences surrounding bovine site 2 and site 3 were derived from tryptic phosphopeptides and overlapping fragments generated by cleavage with chymotrypsin, collagenase, and endoproteinase Lys-C. The sequence around bovine site 2 is Thr-Arg-Gln-Thr-Ser(P)-Val-Ser-Gly-Gln-Ala-Pro-Pro-Lys, and the sequence around bovine site 3 is Thr-Arg-Gln-Ala-Ser(P)-Gln-Ala-Gly-Pro-Met-Pro-Arg. Sites 2 and 3 are located within the COOH-terminal, collagenase-sensitive tail region of the molecule, separated by 36 amino acids. The sequences surrounding rat site 2 and site 3 were derived from tryptic phosphopeptides. The sequence around rat site 2 is Gln-Ala-Ser(P)-Ile-Ser-Gly-Pro-Ala-Pro-Pro-Lys, and the sequence around rat site 3 is Gln-Ala-Ser(P)-Gln-Ala-Gly-Pro-Gly-Pro-Arg. Thus, the sequences surrounding the four sites that are phosphorylated by calcium/calmodulin-dependent protein kinase II, namely sites 2 and 3 in rat and bovine synapsin I, exhibit a high degree of homology.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Bähler M., Greengard P. Synapsin I bundles F-actin in a phosphorylation-dependent manner. Nature. 1987 Apr 16;326(6114):704–707. doi: 10.1038/326704a0. [DOI] [PubMed] [Google Scholar]
- De Camilli P., Greengard P. Synapsin I: a synaptic vesicle-associated neuronal phosphoprotein. Biochem Pharmacol. 1986 Dec 15;35(24):4349–4357. doi: 10.1016/0006-2952(86)90747-1. [DOI] [PubMed] [Google Scholar]
- De Camilli P., Harris S. M., Jr, Huttner W. B., Greengard P. Synapsin I (Protein I), a nerve terminal-specific phosphoprotein. II. Its specific association with synaptic vesicles demonstrated by immunocytochemistry in agarose-embedded synaptosomes. J Cell Biol. 1983 May;96(5):1355–1373. doi: 10.1083/jcb.96.5.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- Huttner W. B., DeGennaro L. J., Greengard P. Differential phosphorylation of multiple sites in purified protein I by cyclic AMP-dependent and calcium-dependent protein kinases. J Biol Chem. 1981 Feb 10;256(3):1482–1488. [PubMed] [Google Scholar]
- Huttner W. B., Greengard P. Multiple phosphorylation sites in protein I and their differential regulation by cyclic AMP and calcium. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5402–5406. doi: 10.1073/pnas.76.10.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. E., Graves D. J., Benjamini E., Krebs E. G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977 Jul 25;252(14):4888–4894. [PubMed] [Google Scholar]
- Kennedy M. B., Greengard P. Two calcium/calmodulin-dependent protein kinases, which are highly concentrated in brain, phosphorylate protein I at distinct sites. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1293–1297. doi: 10.1073/pnas.78.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. B., McGuinness T., Greengard P. A calcium/calmodulin-dependent protein kinase from mammalian brain that phosphorylates Synapsin I: partial purification and characterization. J Neurosci. 1983 Apr;3(4):818–831. doi: 10.1523/JNEUROSCI.03-04-00818.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinás R., McGuinness T. L., Leonard C. S., Sugimori M., Greengard P. Intraterminal injection of synapsin I or calcium/calmodulin-dependent protein kinase II alters neurotransmitter release at the squid giant synapse. Proc Natl Acad Sci U S A. 1985 May;82(9):3035–3039. doi: 10.1073/pnas.82.9.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery C. A., DeGennaro L. J. Determination and analysis of the primary structure of the nerve terminal specific phosphoprotein, synapsin I. EMBO J. 1986 Dec 1;5(12):3167–3173. doi: 10.1002/j.1460-2075.1986.tb04625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness T. L., Lai Y., Greengard P., Woodgett J. R., Cohen P. A multifunctional calmodulin-dependent protein kinase. Similarities between skeletal muscle glycogen synthase kinase and a brain synapsin I kinase. FEBS Lett. 1983 Nov 14;163(2):329–334. doi: 10.1016/0014-5793(83)80846-1. [DOI] [PubMed] [Google Scholar]
- Nairn A. C., Greengard P. Purification and characterization of Ca2+/calmodulin-dependent protein kinase I from bovine brain. J Biol Chem. 1987 May 25;262(15):7273–7281. [PubMed] [Google Scholar]
- Nairn A. C., Hemmings H. C., Jr, Greengard P. Protein kinases in the brain. Annu Rev Biochem. 1985;54:931–976. doi: 10.1146/annurev.bi.54.070185.004435. [DOI] [PubMed] [Google Scholar]
- Navone F., Greengard P., De Camilli P. Synapsin I in nerve terminals: selective association with small synaptic vesicles. Science. 1984 Dec 7;226(4679):1209–1211. doi: 10.1126/science.6438799. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Woodgett J. R., Cohen P., Kemp B. E. Substrate specificity of a multifunctional calmodulin-dependent protein kinase. J Biol Chem. 1985 Nov 25;260(27):14471–14476. [PubMed] [Google Scholar]
- Podell D. N., Abraham G. N. A technique for the removal of pyroglutamic acid from the amino terminus of proteins using calf liver pyroglutamate amino peptidase. Biochem Biophys Res Commun. 1978 Mar 15;81(1):176–185. doi: 10.1016/0006-291x(78)91646-7. [DOI] [PubMed] [Google Scholar]
- Schiebler W., Jahn R., Doucet J. P., Rothlein J., Greengard P. Characterization of synapsin I binding to small synaptic vesicles. J Biol Chem. 1986 Jun 25;261(18):8383–8390. [PubMed] [Google Scholar]
- Steiner J. P., Ling E., Bennett V. Nearest neighbor analysis for brain synapsin I. Evidence from in vitro reassociation assays for association with membrane protein(s) and the Mr = 68,000 neurofilament subunit. J Biol Chem. 1987 Jan 15;262(2):905–914. [PubMed] [Google Scholar]
- Tarr G. E. A general procedure for the manual sequencing of small quantities of peptides. Anal Biochem. 1975 Feb;63(2):361–370. doi: 10.1016/0003-2697(75)90358-9. [DOI] [PubMed] [Google Scholar]
- Ueda T., Greengard P. Adenosine 3':5'-monophosphate-regulated phosphoprotein system of neuronal membranes. I. Solubilization, purification, and some properties of an endogenous phosphoprotein. J Biol Chem. 1977 Jul 25;252(14):5155–5163. [PubMed] [Google Scholar]