Abstract
The oncogene cyclin D1 is highly expressed in many breast cancers and, despite its proliferation-activating properties, it has been linked to a less malignant phenotype. To clarify this observation, we focused on two key components of malignant behavior, migration and proliferation, and observed that quiescent G0/G1 cells display an increased migratory capacity compared to cycling cells. We also found that the down-regulation of cyclin D1 in actively cycling cells significantly increased migration while also decreasing proliferation. When analyzing a large set of premenopausal breast cancers, we observed an inverse proliferation-independent link between cyclin D1 and tumor size and recurrence, suggesting that this protein might abrogate infiltrative malignant behavior in vivo. Finally, gene expression analysis after cyclin D1 down-regulation by siRNA confirmed changes in processes associated with migration and enrichment of our gene set in a metastatic poor prognosis signature. This novel function of cyclin D1 illustrates the interplay between tumor proliferation and migration and may explain the attenuation of malignant behavior in breast cancers with high cyclin D1 levels.
Breast cancer represents the most frequently diagnosed disease in women in the Western world with 1 million new patients annually. Even though the five-year survival rates are as high as 86%, mortalities still total approximately 410,000 each year.1 Distant metastases from the primary tumor represent the main cause of these deaths,2 and as such, identification of new prognostic markers and targets for novel treatments of patients at risk of developing metastases is of paramount importance.
The process of metastasis represents a complex series of events, comprehensively covering many functional capabilities of a cell. These include tumor cell migration/proliferation and invasion into the tissues surrounding the primary tumor location, their subsequent intravasation into the blood or lymph vessels, extravasation back through vasculature to distal organs, and finally colonization/proliferation and angiogenesis of a new location to produce a second tumor body.3
Research in this area has focused on elucidation of the factors involved in metastasis initiation, and a plethora of candidates have been described in this setting; VEGF is reported to promote metastatic colonization and desmoplasia in breast tumors,4 gene expression analysis indicates an overexpression of DNA repair genes is associated with tumor metastasis,5 and recently, the role of the cell cycle regulatory protein cyclin D1 in the progression of metastatic events has also been addressed.6,7,8
Cyclin D1 along with its binding partners CDK4/6 partially mediate G1 to S-phase transition in the cell cycle.9 Overexpression of the protein has been reported in 25 to 60% of invasive breast carcinomas and amplification of its corresponding gene CCND1 occurs in 10 to 30% of cases.10,11,12 Previous studies by our group and others have implicated cyclin D1 with less aggressive tumor characteristics such as lower histological grade and retained pRb function.11,13 This suggests that activation of the oncogene cyclin D1 in breast cancer could be linked to less aggressive tumor properties and reduced metastatic potential–a novel hypothesis given the protein’s well-described proliferation enhancing properties. To clarify this apparently conflicting relationship between cyclin D1 and aggressiveness, we have focused on two key components in breast cancer malignancies, migration and proliferation. In this study, we demonstrate how cyclin D1 influences these properties using cell line models, in vivo studies of large collections of primary breast cancer samples linked to clinical data and gene expression array technology.
Materials and Methods
Cell Culture
The human breast cancer cell line MDA-MB-231 and the melanoma cell line MDA-MB-43514 (ATCC, Int., Manassas, VA) were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), sodium pyruvate (1 mmol/L) and 1xPEST (streptomycin 90 μg/ml, penicillin 90 IU/ml). Cells were maintained in a humidified atmosphere of 5% CO2/95% air at 37°C.
Synchronization and Flow Cytometry
For synchronization, 1 × 106 cells were seeded in 28.3-cm2 culture dishes and quiesced in 4 ml serum-free medium (SFM) for 24 hours. The SFM was then replaced with medium containing 10% FCS (SM) and the cells were grown for 4, 8, 12, 16, and 20 hours, respectively before harvest. For asynchronous experiments, 5 × 105 cells were seeded in 28.3-cm2 culture dishes and cultured for 24 hours in SM before harvest. For flow cytometric analyzes we refer to Berglund et al.15
Assessment of Ki-67 Expression
1 × 105 cells were seeded in 12.6-cm2 culture dishes and synchronized as described above. At each time point, cells were washed once with PBS and fixed for 30 minutes in PBS containing 4% paraformaldehyde. After fixation, cells were washed once and subsequently permeabilized in PBS containing 2% NP-40. After washing, cells were allowed to air-dry before incubation for 1 hour primary antibody against Ki-67 (M7240, DAKO, Glostrup, Denmark, diluted 1:25 in PBS). Cells were washed three times in PBS before incubation (1 hour) with a FITC-conjugated secondary antibody (F0313, DAKO, 1:50 in PBS). After a final wash step, cells were stained with DAPI (4′6-diamidino-2-phenylindole) and mounted with a glass coverslip. Images of the Ki-67-FITC/DAPI–stained cells were taken using a fluorescence microscope, and the analysis of Ki-67 fluorescence intensities was carried out with the Image-Pro Plus software (Media Cybernetics, L.P., MD). DAPI staining allowed cell nuclei to be defined and the intensity of green light/Ki-67 expression quantified. From the continuous light intensities measured, a cut-off for Ki-67-positivity was arbitrarily chosen based on the visually detectable limit.
Treatment with U0126
The MEK1/2 inhibitor U0126 (Sigma-Aldrich, St. Louis, MO), dissolved in dimethyl sulfoxide (DMSO), was used at a concentration of 20 μmol/L with an equal volume of DMSO used as control and both were added to SM as required. For assessment of ERK1/2 phosphorylation and migratory capacity, cells were pretreated with U0126 or DMSO for 4 hours before the migration assays. In addition, the medium in the chambers and wells was supplemented with U0126 or DMSO.
siRNA and Vector Transfections
5 × 105 cells were seeded in a 28.3-cm2 culture dish with PEST-free SM for 24 hours. The media was subsequently removed and PEST-free SFM added along with 1 ml siRNA solution (OptiMEM, Gibco, Lipofectamine 2000, Invitrogen Life Technologies, Carlsbad, CA) giving a final concentration of 40 nmol/L oligonucleotides. Five hours after transfection, SFM was replaced with SM and cells were allowed to grow for 20 hours before harvesting for migration assay, flow cytometry or Western blot. ON-TARGETplus SMARTpool siRNA reagents targeting cyclin D1 (excluding duplex J-003210-15 due to nonspecific effects, Dharmacon RNA Technologies, Lafayette, CO), CDK4 and CDK6 were included as standard experimental protocol. A nontargeting pool was used as negative control. For vector experiments, 3 μg of GFP-tagged vector with an insert of the ORF of cyclin D1, or GFP only (control) was used (OriGene Technologies Inc., Rockville, MD). Cyclin D1 transfection efficiency was 45% as determined by flow cytometry analyses of the GFP-tagged cyclin D1 construct, and total cell numbers were quantified by migration assay.
Western Blotting
Western blot was performed as described previously,15 and the following antibodies were used: α-cyclin D1 (1:500, DCS-6, DAKO, Glostrup, Denmark), α-cyclin E (1:500, HE12, Santa Cruz Biotechnology, Santa Cruz, CA), α-cyclin A2 (1:500, Santa Cruz), α-p21 (1:500, SX118, DAKO), α-phospho-pRb Ser807/811 (1:500, Cell Signaling Technology, Inc., Beverly, MA), α-pRb (1:500, IF8, Santa Cruz), α-phospho-p44/42 MAP Kinase Thr202/Tyr204 (1:500, Cell Signaling Technology, Inc.), and α-p44/42 MAP Kinase (1:500, Cell Signaling Technology, Inc.) α-CDK4 (1:500, DCS-35, Chemicon, Rockford, IL), α-CDK6 (1:1000, DCS83, Cell Signaling Technology, Inc.). Actin (1:1000 I-19, Santa Cruz) was included as loading control. Densitometric analysis of Western blot of phospho bands was performed and corrected with the values of respective total bands of ERK1/2 and actin, using ImageJ.16
Migration Assays
Boyden Chamber
Cell migration was routinely carried out in 8-μm-pore polycarbonate membrane Transwell chambers with a diameter of 6.5 mm (Corning, Inc. Corning, NY). The membranes were incubated in 150 μl serum-free RPMI 1640 for an initial equilibrium period of 1 hour. Cells were resuspended in serum-free medium (1 × 106 cells/ml), and 100,000 cells were added to each migration chamber. The chambers were placed into wells containing 600 μl 10% FCS medium, and cells were allowed to migrate for 3 hours or overnight, after siRNA or vector transfections, respectively. Cells remaining in the chamber were removed with a cotton swab, and the migrated cells situated on the lower side of membranes were fixed for 15 minutes in PBS containing 4% paraformaldehyde. Membranes were cut and mounted on glass slides for DAPI staining and counted using a fluorescent microscope (cells in three ×10 magnification fields representing the composition of the membrane were counted). Assays were repeated at least three times with at least three migration chambers for every treatment. Each migration assay was related to the mean number of migrated control siRNA treated cells in that particular experiment to avoid interexperimental variation. This is termed “relative unit” in the figures. Each dot in the figure represents one migration chamber.
Time Lapse Microscopy
For time lapse experiments cells were plated at a confluency of 2 × 104 cells/cm2 in a 24-well plate after transfection with siRNA, before being placed in an enclosed full environmental chamber surrounding a Zeiss Axiovert 200M microscope. Sample, gases, chamber, and objective lens were environmentally maintained and monitored. Visualization was undertaken with Metamorph (Molecular Devices, Downingtown, PA) and a Roper Coolsnap HQ camera, with three fields of view imaged per well. Images were taken every 5 minutes for 4 hours, and once captured data were exported to Metamorph for particle tracking analysis. Particle data were subsequently imported to Image J and analyzed the with the Ibidi chemotaxis plug-in.
Cell Sorting for Cell Cycle Analysis
MDA-MB-231 cells were resuspended at 1 × 106 cells/ml in 500 μl 10 μg/ml Hoechst 33342 (Sigma) and incubated for 45 minutes at 37°C. 7AAD (10 μl/1 × 106 cells) (BD Biosciences) was subsequently added to cells and incubated for 15 minutes at room temperature to allow exclusion of dead cells from sorting. After incubation, cells were sorted at 16PSI by FACS using a BX Influx (BD Biosciences) operated using Spigot software. FlowJo version 7.6.1 analysis software (Treestar, Inc., Ashland, OR) was used to set gates for sorting of cells into three populations G0/G1, S, and G2/M. Cells were collected in PBS and transwell migration assays performed as previously described with the exception that 50,000 cells were seeded in upper chambers of membranes and allowed to migrate for 5 hours.
Microarray Analysis
MDA-MB-231 cells were transfected with control, cyclin D1, and CDK4/6 siRNA as described previously, harvested 12 hours posttransfection and resuspended in RNeasy Lysis Buffer (QIAGEN Sciences, Germantown, MD) before RNA isolation and microarray analysis by AROS Applied Biotechnology A/S (Aarhus, Denmark). Experiments were made in triplicates. Briefly, RNA extraction was performed using TRIzol reagent (Invitrogen, Carlsbad, CA) with subsequent quantification on Nanodrop (ND-1000) Spectrophotometer at an optical density of A260 nm. RNA was then labeled using the TotalPrep RNA Labeling Kit (Ambion, Austin, TX) before hybridization to the whole genome Illumina Human WG-6 v3.0 Expression gene chip (Illumina, San Diego, CA). Raw data were examined in BeadStudio (Illumina, Inc.), with subsequent quantile normalization and identification of differentially expressed genes performed by significance analysis of microarrays (SAM)17 in R18, using the lumi19 and siggenes BioConductor packages,20 respectively. Data were then exported to the GoMiner high-throughput web-interface to determine significant gene ontology processes and to gene-set enrichment analysis software (GSEA, Broad Institute MIT) for comparison to previously published data sets and functional processes.
Analysis of Primary Tumors
The immunohistochemical analysis was conducted with a formalin-fixed paraffin-embedded tumor material consisting of 500 primary invasive stage II breast cancers from premenopausal patients. These patients were enrolled in a previously described randomized clinical trial of adjuvant tamoxifen treatment.21 The tumors were assembled in tissue microarrays (TMAs) followed by the procedure described in Berglund et al.15 Cyclin D1 intensity had been assessed previously as absent = 0, low = 1, intermediate = 2, and strong = 3,22 as had Ki-67, which was analyzed as fraction of positive nuclei where 0 = 0 to 1%, 1 = 1 to 10%, 2 = 11 to 25%, 3 = 26 to 50%, and 4 = 51 to 100%.23 For tumor growth pattern analysis, the characteristics of the tumors had been previously evaluated on whole tumor sections. Growth patterns were classified into four groups, defined by the mode of infiltration, from the most infiltrative cases with a “sieving” and diffuse tumor border (“exclusively infiltrative”) to a more pushing, clearly defined tumor margin (“exclusively pushing”). Other than these two extreme groups, two intermediary groups were also defined as either demonstrating “predominantly infiltrative” or “predominantly pushing” growth. For the present analysis, the “exclusively infiltrative” group of tumors was analyzed against the remainders of tumors, included in the group here termed “pushing feature.” For more details on the assessment of growth pattern and representative images, we refer to Berglund et al.24 Evaluation of phosphorylated pRb was performed by using an α-phospho-pRb antibody targeting phosphorylation sites Ser807/811 (1:200, cat. no #9308, Cell Signaling Technology, Inc., Beverly, MA).
Statistical Methods
To examine the statistical significance of the differences seen in the Boyden migration assays and in the distribution of cell cycle phases, a two-tailed Student’s t-test was used, assuming unequal variance. Unless stated otherwise, the SEM is shown. Further on, Spearman’s rank-order correlation coefficient (ρ) was used to test the statistical significance of the association between cyclin D1 nuclear intensity and various categorical variables. For estimation of recurrence-free survival, the Kaplan–Meier method, log-rank test and Cox regression analyses were used. Analyses were carried out using the SPSS software (version 15.0, SPSS, Chicago, IL) and a P value less than 0.05 was considered significant.
Results
Cell Cycle Specific Variations in the Protein Expression and Migratory Capacity of MDA-MB-231 Cells
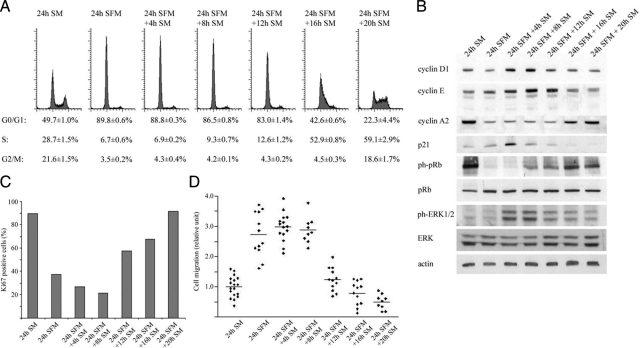
To determine the baseline migratory behavior of breast cancer cells in relation to cell cycle phase distribution, we synchronized MDA-MB-231 cells by serum starvation for 24 hours followed by serum stimulation and assessed proliferation, expression of cell cycle related proteins and migratory behavior at consecutive time-points. Cells entered a resting G0-like phase after of serum starvation, and proceed through G0/G1 and S- phases after 12 and 16 hours of serum stimulation, respectively (Figure 1A). Expression of the cell cycle regulatory proteins cyclin D1, cyclin E, and cyclin A2 peaked in consecutive order over a 24-hour period with concomitant increasing phosphorylation of pRb. The CDK inhibitor p21 exhibited an early transient peak similar to that of phospho-ERK1/2 with expression of both proteins declining as cells entered S-phase (Figure 1B). The fraction of cells positive for the proliferation marker Ki-67 decreased substantially after 24 hours of serum starvation but increased after 12 hours of serum stimulation indicating entrance into active cell cycle (Figure 1C).
Figure 1.
Effect of cell phase on MDA-MD-231 cell cycle protein expression and migration. Cells were quiescent for 24 hours before serum stimulation and monitored at the indicated times for changes in cell phase distribution, cell-cycle related protein expression, and migration. Control is nonquiescent, actively cycling cells. Blots are representative, histograms and migration plots are averaged from at least three independent experiments. A: Cell phase analysis by flow cytometry. B: Cyclin D1, cyclin E, cyclin A2, p21, pRb, phospho-pRb, ERK1/2, and phospho-ERK1/2 expression by Western blot. C: Ki-67 expression analyzed by immunocytochemistry. D: Cell migration analyzed by Boyden chamber assay. Twenty-four hour SM vs. 24 hours SFM + 8 hours SM, P < 0.001 two-tailed Student’s t-test. SM, serum medium; SFM, serum-free medium.
We next used a Boyden chamber assay for examination of cell migratory capacity. Cells serum stimulated for up to 8 hours exhibited an average of 2.9 ± 0.15-fold increase in migration relative to actively cycling control cells. After 12 and 16 hours, however, this had fallen to 1.2 ± 0.11 and 0.80 ± 0.11-fold of control, respectively (Figure 1D). These results suggest that resting G0/early G1 cells have greater migratory abilities compared with both late G1/S-phase and G2/M cells as well as unsynchronized, actively cycling cells.
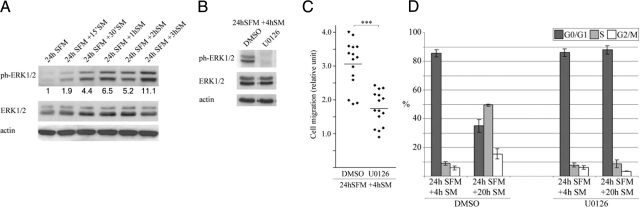
MDA-MB-231 Cell Cycle Progression and Migration Are Partially Modulated through ERK1/2 Phosphorylation
As ERK1/2 phosphorylation has been established as participating in cell migratory and cycling activities,25,26,27 we decided to assess the significance of the observed phosphorylation of the protein after 4 and 8 hours of serum stimulation. For this, the specific inhibitor of the ERK1/2 phosphorylation protein MEK, U0126, was used with subsequent examination of MDA-MB-231 cell phase distribution and migration. Initial investigations revealed a detectable band of ERK1/2 phosphorylation 15 minutes after serum stimulation that increased and maintained intensity up to a 3 hours time point (Figure 2A). This band was absent after the inclusion of U0126 in the serum-containing media (Figure 2B). In addition, treatment with the inhibitor reduced the cells migratory ability by ∼40% (Figure 2C) and prevented cell cycle progression to S-phase on serum stimulation (Figure 2D). Cyclin D1 protein levels were not affected by MEK inhibition (data not shown). Taken together, these results suggest that the migratory capacity of G0/early G1 cells is strongly influenced by, but not solely dependent on, ERK1/2 activity.
Figure 2.
Effect of MEK1/2 inhibitor U0126 on MDA-MB-231 cell phase distribution and migration. Cells were quiescent for 24 hours before serum stimulation in the absence or presence of 20 μmol/L U0126 and monitored at the indicated times for changes in phospho-ERK1/2 expression, cell phase distribution, and migration. Control is DMSO-treated cells. Blots are representative, and histograms and migration plots are averaged from at least three independent experiments. A: Short-term phospho-ERK1/2 expression by Western blot. Densitometric analysis of phospho-ERK1/2 bands was corrected with the values of respective total ERK1/2 bands and actin. B: Phospho-ERK1/2 expression after inhibitor treatment by Western blot. C: Cell migration analyzed by Boyden chamber assay. ***P < 0.001, two-tailed Student’s t-test. D: Cell phase distribution analyzed by flow cytometry. SM, serum medium; SFM, serum-free medium.
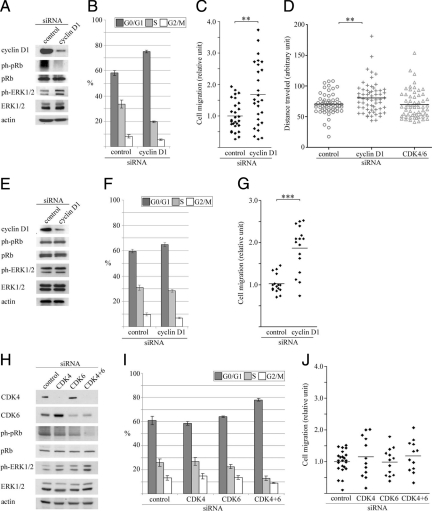
Silencing of the Cyclin D1 Gene Increases MDA-MB-231 and MDA-MB-435 Cell Migration in a CDK4/6-Independent Manner
After the observed link between proliferation and migration in serum starved and partially synchronized cells we examined the role of the main G1-cyclin, cyclin D1, in a migratory and proliferative capacity with actively cycling MDA-MB-231 and -435 cell lines. Cyclin D1 protein was efficiently down-regulated using siRNA with subsequent diminished pRb phosphorylation and proliferation in MDA-MB-231 but not MDA-MB-435 cells (Figure 3, A, B and E, F). In parallel with the decrease in cyclin D1 expression, a substantial increase in ERK1/2 phosphorylation in MDA-MB-231 but not MDA-MB-435 cells was found. Interestingly, both cells lines displayed significantly increased migration after cyclin D1 silencing (1.7 ± 0.19 and 1.8 ± 0.14-fold, respectively) relative to control transfected cells in Boyden chamber assay (Figure 3, C and G) and by time-lapse microscopy in MDA-MB-231 cells (Figure 3D). A comparable result was found when using an alternative siRNA sequence targeting cyclin D1 in MDA-MB-231 cells by Boyden chamber assay (see Supplemental Figure 1, A and B at http://ajp.amjpathol.org).
Figure 3.
Effect of cell cycle–related siRNA treatment on MDA-MB-231, -435 cell-cycle, and migratory properties. Unquiescent MDA-MB-231 cells were monitored 20 hours posttransfection with the indicated siRNA (cyclin D1 or CDK 4/6) for changes in cell cycle–related protein expression, cell phase distribution, and migration by Boyden chamber assay and time-lapse microscopy. Blots are representative, and histograms and migration plots are averaged from at least three independent experiments. MDA-MB-231 cells: A: Western blot; B: cell phase distribution. G0/G1 control versus cyclin D1 knockdown, P = 0.001, two-tailed Student’s t-test. C: Cell migration by Boyden chamber assay. **P = 0.003, two-tailed Student’s t-test. D: Cell migration by time-lapse microscopy, **P = 0.006, two-tailed Student’s t-test. MDA-MB-435 cells: E: Western blot; F: cell phase distribution. G0/G1 control versus cyclin D1 knockdown, P = 0.056, two-tailed Student’s t-test. G: Cell migration. ***P < 0.001, two-tailed Student’s t-test. MDA-MB-231 cells: H: Western blot; I: cell phase distribution. G0/G1 control versus CDK4/6 knockdown, P < 0.001, two-tailed Student’s t-test. J: Cell migration. Control siRNA versus CDK4/6 siRNA, P = 0.30, two-tailed Student’s t-test. CDK, cyclin-dependent kinase.
Simultaneous silencing of the cyclin D1 associated kinases CDK4 and CDK6 in actively cycling MDA-MB-231 cells resulted in a similar reduction in pRb phosphorylation and proliferation with concurrent increase in ERK1/2 phosphorylation (Figure 3, H and I). Notably, cell migration was unaffected relative to control after CDK4/6 silencing (Figure 3J). These results indicate that increased migration in both MDA-MB-231 and MDA-MB-435 cells after cyclin D1 silencing is ERK1/2 and CDK4/6 independent.
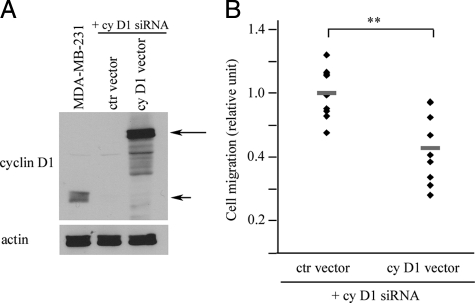
To further demonstrate that the migration difference observed after cyclin D1 silencing was not due to unspecific siRNA effects, we reintroduced cyclin D1 to cyclin D1 siRNA-silenced MDA-MB-231 cells. As illustrated in Figure 4, A and B, overexpression of the protein in cyclin D1 silenced cells decreased migration significantly compared to a control vector, again confirming the importance of cyclin D1 in breast cancer cell migratory behavior. To investigate whether the cyclin D1–mediated increase in migration was specific for a cell cycle phase we sorted siRNA transfected cells using the DNA dye Hoechst 33342. As illustrated in Figure 5A, the most pronounced increase in migration (relative to control) after cyclin D1 silencing occurred in the S- and G2/M phases of the cell cycle, with limited effects in G0/G1 phase. An increase was also noted in migratory capacity of control siRNA treated G0/G1 cells compared to S and G2/M cells, confirming our initial observations in Figure 1A.
Figure 4.
Reintroduction of cyclin D1 into cyclin D1–silenced MDA-MB-231 cells. A: Western blot of MDA-MB-231 cells. First lane shows untreated cells as positive control for endogenous cyclin D1 (short arrow). Long arrow indicates reintroduced GFP-tagged cyclin D1. B: Cell migration. **P = 0.002, two-tailed Student’s t-test.
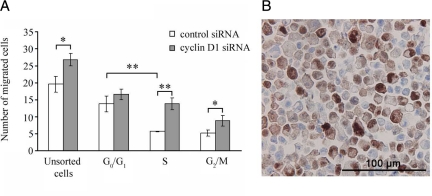
Figure 5.
The effect of cyclin D1 silencing on migration capacity of cells in different cell cycle phases. A: MDA-MB-231 cells were sorted according to cell cycle phase after siRNA treatment and migration was assessed. Unsorted cells: control versus cyclin D1 siRNA, *P = 0.018; S-phase: control versus cyclin D1 siRNA, **P = 0.001; G2/M-phase: control versus cyclin D1 siRNA, *P = 0.04. G0/G1 phase versus S-phase, control siRNA, **P = 0.004. Two-tailed Student’s t-test. B: Expression of cyclin D1 in actively cycling MDA-MB-231 cells viewed by immunohistochemistry at ×40 magnification.
Differential nuclear and cytoplasmic expression of cyclin D1 was observed in a large proportion of actively cycling MDA-MB-231 cells (Figure 5B) revealing cyclin D1 expression in multiple cell cycle phases.
Low Levels of Cyclin D1 Are Associated with an Exclusively Infiltrative Tumor Growth Pattern and Decreased Recurrence-Free Survival
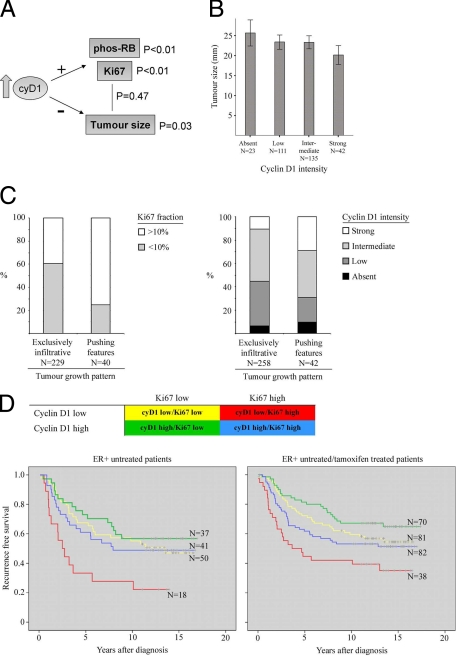
Experimental findings were validated through examination of cyclin D1 protein expression in a primary breast cancer material consisting of 500 tumors from premenopausal patients. We focused on ER-positive tumors to obtain a more homogenous group of breast cancer with intact pRb machinery. As anticipated, strong cyclin D1 intensity correlated with both pRb phosphorylation and high Ki-67 expression (Figure 6A), but not with tumor grade or node status (Table 1). Notably, cyclin D1 intensity was inversely correlated to tumor size (Table 1 and Figure 6B) but no significant link was found between proliferation and tumor size (Table 1, summarized in Figure 6A).
Figure 6.
Correlation of cyclin D1 and Ki-67 expression to tumor size and growth pattern in vivo. Five hundred tumors from ER+ premenopausal breast cancer patients were evaluated for cyclin D1, Ki-67, and phos-pRb protein expression. A: Schematic representation of the relationship between cyclin D1, Ki-67, phospho-pRb, and tumor size. B: Relationship between tumor size and cyclin D1 (Spearman’s rho = −0.123, P = 0.03). C: Distribution of Ki-67 and cyclin D1 in tumors subgrouped based on exclusively infiltrative versus pushing features. Highly proliferative tumors were significantly overrepresented among tumors with pushing features (P < 0.001, χ2 test) as was strong cyclin D1 intensity (P = 0.006, χ2 test). D: Kaplan–Meier curves illustrating the prognostic information of combining cyclin D1 and Ki-67 expression in untreated patients (log rank test, P = 0.011) and in untreated and tamoxifen patients combined (log rank test, P = 0.004).
Table 1.
Cyclin D1 Expression and Clinicopathological Parameters in ER+ Premenopausal Breast Cancer Patients
| Variable | Grade | Node status | Tumor size | Ki67* | Statistics |
|---|---|---|---|---|---|
| Cyclin D1† | 0.059 | 0.038 | −0.123 | 0.263 | rho‡ |
| 0.303 | 0.504 | 0.03 | <0.001 | P value | |
| 310 | 312 | 311 | 272 | N | |
| Grade§ | −0.056 | 0.151 | 0.450 | rho | |
| — | 0.317 | 0.007 | <0.001 | P value | |
| 319 | 318 | 277 | N | ||
| Node status¶ | −0.367 | 0.046 | rho | ||
| — | <0.001 | 0.439 | P value | ||
| 323 | 280 | N | |||
| Tumor size∥ | 0.044 | rho | |||
| — | 0.469 | P value | |||
| 279 | N |
Nuclear fraction (0–1%, 2–10%, 11–25%, 26–50%, 51–100%).
Nuclear intensity (absent, low, intermediate, strong).
Spearman’s rho.
Nottingham histological grade.
Lymph node metastasis: negative versus positive.
Size in mm.
Tumor growth pattern in relation to cyclin D1 expression was studied by defining tumors with an exclusively infiltrative growth pattern (86% of total ER+ tumors) and those with “pushing features” (14% of total ER+ tumors). There was a significant overrepresentation of exclusively infiltrative behaviors in cyclin D1 low and Ki-67 low tumors (Figure 6C) and contrastingly, pushing behaviors were linked to high cyclin D1 expression.
A further overrepresentation of absent or low cyclin D1 expression was observed in the highly infiltrative subgroup of lobular breast cancer (see Supplemental Table 1 at http://ajp.amjpathol.org). Our findings of a negative link between cyclin D1 and tumor size as well as infiltrative behaviors in cyclin D1 low tumors supports the experimental findings that cyclin D1 down-regulation is linked to increased tumor migratory properties.
We next examined the relationship between cyclin D1, tumor size, and Ki-67 in relation to recurrence-free survival (RFS) in the untreated ER+ cohort. Interestingly, cyclin D1 was positively and Ki-67 negatively associated with RFS in a multivariate analysis independent of tumor grade and lymph node status (Table 2), despite a positive link between cyclin D1 and proliferation (Table 1). We next combined cyclin D1 and Ki-67 expression, constructing four subgroups: cyD1low/Ki-67low, cyD1low/Ki-67high, cyD1high/Ki-67low, and cyD1high/Ki-67high. The best and worst clinical outcomes were represented by the cyD1high/Ki-67low and cyD1low/Ki-67high, respectively (Figure 6D, left panel). Similar results were found when tamoxifen-treated patients were included in the analysis (Figure 6D, right panel). These results indicate that absent or low levels of cyclin D1 are associated with malignant properties irrespective of proliferative status, further validating our experimental findings.
Table 2.
Hazard Ratios for Recurrence-Free Survival in ER+ Patients in the Untreated Control Cohort Calculated with Cox Uni- and Multivariate Analyses (N = 173)
| Variable | Univariate
|
Multivariate
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Cyclin D1* | 0.818 | 0.625–1.070 | 0.143 | 0.711 | 0.527−0.959 | 0.026 |
| Grade† | 1.905 | 1.354–2.680 | <0.001 | 1.449 | 0.931–2.256 | 0.101 |
| Node status‡ | 1.411 | 0.832–2.393 | 0.202 | 1.670 | 0.927–3.009 | 0.088 |
| Tumor size§ | 1.107 | 0.884–1.452 | 0.461 | 1.014 | 0.681–1.511 | 0.944 |
| Ki67¶ | 1.442 | 1.151–1.808 | 0.001 | 1.431 | 1.102–1.859 | 0.007 |
Nuclear intensity (absent, low, intermediate, strong).
Nottingham histological grade.
Lymph node metastasis: negative versus positive.
Size in mm.
Nuclear fraction (0–1%, 2–10%, 11–25%, 26–50%, 51–100%).
Silencing of the Cyclin D1 and CDK4/6 Genes in MDA-MB-231 Cells Impacts on Migration-Related Cellular Processes
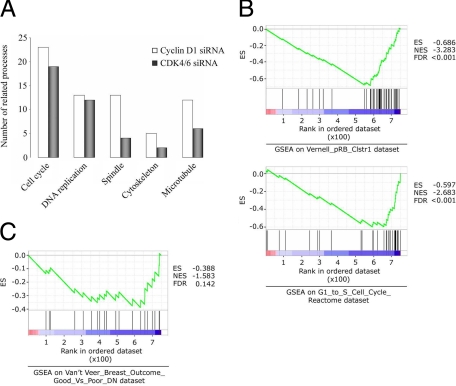
We next examined the response to cyclin D1 and CDK 4/6 silencing using a gene expression microarray approach, identifying a number of significantly altered transcripts (see Supplemental Tables 2–5 at http://ajp.amjpathol.org) and used the GoMiner program28 to interpret the cell processes affected. As expected, numerous proliferation-associated processes were altered and notably, a number of processes traditionally associated with migration (including spindle, cytoskeleton and microtubule changes) were increased in cyclin D1 siRNA, relative to CDK4/6 siRNA-treated cells, when both were compared to control (Figure 7A).
Figure 7.
Effect of cell-cycle related siRNA treatment on MDA-MB-231 cell processes and gene set enrichment. Gene expression data were submitted to GO miner and GSEA programs for analysis of gene ontology terms and data set enrichment, respectively. A: GoMiner processes traditionally associated with migration (spindle, cytoskeleton, and microtubule) were more frequently changed in cyclin D1 siRNA-treated cells over CDK4/6 siRNA treatment, when both were compared to control transfected cells. P < 0.001 for all processes. B: Comparison of genes changed after cyclin D1 siRNA treatment of cells to published data sets and functional pathways (P < 0.001). C: Comparison to published poor-outcome gene expression set (P = 0.04). ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate-adjusted q value.
Through direct comparison of the cyclin D1 and CDK4/6 silenced gene lists we identified a set of gene products that were only changed by cyclin D1 and not CDK4/6 silencing (see Supplemental Table 6 at http://ajp.amjpathol.org), potentially harboring candidate genes mediating the cyclin D1 linked migration effect.
Finally, we compared our differentially expressed cyclin D1 silenced genes (relative to control silenced) to published gene sets and known functional gene sets using gene-set enrichment analysis (GSEA) software. Of 472 sets of genes whose products are involved in ‘metabolic and signaling pathways’ and 50 previously published sets containing genes coregulated in response to ‘genetic and chemical perturbations,’29 two of the most significant negatively enriched gene sets corresponded to the ‘Vernell pRb cluster 1 study’30 (cluster 1 genes are down-regulated by pRb) and the ‘G1 to S cell cycle reactome’ (Figure 7B). As cyclin D1 is central in both processes these results indicate the validity of using the GSEA software to compare our gene set to published data. Notably, our gene set was also negatively enriched in genes (eg, FBXO5, MAD2L1, EZH2) correlated to poor prognosis in breast cancer patients in the previously published van’t Veer et al study (Figure 7C).31 These results further implicate loss of cyclin D1 in breast cancer cell migration and malignant tumor properties.
Discussion
Tumor cell migratory and proliferative processes are central to the initiation and progression of tumor metastatic events, and previous reports have proposed a contrasting relationship between proliferative and infiltrative behaviors in cancer.15,32,33 Although it is not generally applicable to all malignancies, this may reflect relevant biological characteristics of certain tumors and be of potential clinical significance. Hence, we have delineated how tumor cells combine proliferation with the process of migration in both in vitro and in vivo settings.
Our initial investigations demonstrated that when quiescent and subsequently serum-stimulated MDA-MB-231 breast cancer cells progress through cell cycle, they have a peak migratory capacity during G0/G1 phases, which declines as they progress into S-phase. Serum stimulation of quiescent cells most likely evokes a multitude of signaling events, making it challenging to derive simple causal connections between proliferative and migratory responses. Despite this, the overall impression from the experiments clearly suggests that these activities are to some extent coordinated. The decreased migratory potential concomitant with S-phase entrance is most likely influenced by increased cell cycle activity. In addition, the starved G0-population displayed a significantly increased migratory capacity compared to asynchronously growing populations containing few resting G0 cells. This suggests that cells in G0 have a higher migratory potential compared to actively cycling cells and are more prone to migrate toward chemo-attractant gradients. Moreover, actively cycling cells that continuously cycle between a modified G1-phase and M-phase, might have difficulties in combining a high level of proliferation with efficient migration. This interpretation is supported by our previous work where cyclin E overexpression resulted in an increased S-phase population as well as decreased cell motility and invasiveness.15
As the mitogen-activated protein (MAP)-kinase pathway has been shown to regulate many cellular processes, including proliferation and migration,26,27 we assessed the role of one of its members, ERK1/2, in influencing these properties of MDA-MB-231 cells. Our findings indicate that its activity is not only necessary for cell cycle progression but is also instrumental in migration. Moreover, silencing of cyclin D1 and CDK4/6 in these cells elevated ERK1/2 phosphorylation, indicating a novel mechanism whereby cyclin D1-CDK4/6 complexes are able to regulate ERK1/2 activity. Initially, the increased migratory capacity of cyclin D1 siRNA-silenced cells appeared to be related to this elevation in ERK1/2 activity, however two results challenged this hypothesis. Silencing of CDK4/6 resulted in an elevated ERK1/2 activity without affecting migration and cyclin D1 knock-down in MDA-MB-435 cells increased the migratory capacity without affecting ERK1/2 activity. This implies an ERK1/2-independent regulation of migration by cyclin D1.
These observations were further reflected in cyclin D1–silenced MDA-MB-231 and MDA-MB-435 cells, where despite differences in the size of the G0/G1 populations and phopsho-ERK1/2 expression between these cell types, migratory capacity was equivalently increased. Moreover, even though silencing of the cyclin D1 binding partners CDK4/6 increased the G0/G1 population, it did not enhance the migratory capacity of the MDA-MB-231 cells. This suggests cyclin D1 is able to modulate tumor cell migration by mechanisms that are both CDK independent and unrelated to the role of cyclin D1 in regulating proliferation. This assessment is supported by previous reports showing that cyclin D1 exerts CDK independent functions through binding and regulation of several transcription factors such as the estrogen and androgen receptors, DMP1, STAT3, SP1, and C/EBPβ.34,35 In addition, several cyclin D1-bound genes have recently been identified in developing mouse retinas and gene expression profiles of wild-type and cyclin D1 knock-out mouse retinas suggesting that cyclin D1 can both activate and down-regulate gene transcription, independently of CDKs.36
The finding that high cyclin D1 levels would confer a less migratory phenotype is somewhat contradicted by the rapid up-regulation of cyclin D1 during the synchronized cell phase experiments, where the migratory capacity peaks. Our data from flow cytometry sorted siRNA-treated cells in different cell cycle phases nevertheless suggests that with regard to migratory potential, it is mainly S and G2/M cells that are affected by cyclin D1 silencing (Figure 5A). In contrast, synchronized cells having high migratory capacity with concurrent high cyclin D1 levels were predominantly in G0/G1 phase. This tends toward a hypothesis that high cyclin D1 can be tolerated in G0/G1 migratory cells, whereas S and G2/M cells have a migratory advantage with reduced cyclin D1. Moreover it suggests that cyclin D1 has distinct functions in different cell cycle phases. This may also account for the modest, yet significant, increase in migration of cyclin D1 silenced actively cycling cells consisting of a mixed population of cell cycle phases.
Previous studies have attempted to address the role of cyclin D1 in breast cancer cell migration and Pestell et al demonstrated cyclin D1−/− mouse embryo fibroblasts (MEFs) and mouse mammary epithelial cells (MECs) cells have a reduced capacity to migrate.7,37 Ectopic cyclin D1 expression in these cells resulted in an increased movement velocity and this effect was dependent on the ability of cyclin D1 to interact with CDK4 and p27. In our model, the increased migration induced by silencing of cyclin D1 occurred in the absence of p27 and could not be mimicked by CDK knock-down. This implies that the link between cyclin D1 and migration reported here is not necessarily comparable with earlier reports. Future studies will address whether silencing of cyclin D1 in varying subtypes of breast cancer cells increases their migratory capacity, however it is more likely that a genetic background of abnormalities influences how cyclin D1 affects cell migration machinery.
To validate our experimental findings we analyzed cyclin D1 in a large cohort of ER+ premenopausal breast cancer patients. We focused on ER+ breast tumors, as ER− tumors are often cyclin D1 negative, pRb-inactivated, and highly proliferative, thus representing a separate entity of breast cancer.38,39 Translating our experimental results to the in vivo situation would imply that increased cyclin D1 levels should hamper infiltrative behavior, leading to overall reduced tumor aggressiveness. As expected increased cyclin D1 expression correlated to higher proliferation, however cyclin D1 was inversely correlated with tumor size, which in turn, was not linked to proliferation. Tumor size is a complex parameter that reflects many clinical aspects, including both tumor cell proliferation and infiltrative properties. As the size of a tumor is defined as the largest diameter including tumor cells marked and measured on a hematoxylin-eosin stained slide, a tumor comprising highly invasive but slowly proliferating cells may generally be of larger size when compared to one which is highly proliferative and less infiltrative. In this regard, our observation that tumors expressing low levels of cyclin D1 were, in general, of larger size, is in line with our in vitro findings.
We have previously analyzed the relationship between cyclin E and tumor growth pattern and found cyclin E high tumors to present with a less infiltrative, pushing growth pattern.15,24 Despite the limitation of a small number of low infiltrative ER+ tumors in this cohort, a significant association was found between high levels of proliferation and the presence of a pushing growth pattern, and tumors with the highest cyclin D1 expression were overrepresented among tumors with pushing features relative to the exclusively infiltrative cases. Together with the observation of an inverse association between cyclin D1 and tumor size, this could indicate that increased cyclin D1 expression impedes infiltrative behavior in vivo.
In the multivariate Cox analysis of untreated ER+ patients, cyclin D1 was an independent prognostic factor, inversely linked to recurrence-free survival (Table 2). Moreover, adding Ki-67 to our analysis successfully defined subgroups of patients with significantly different outcome and implied that cyclin D1 is linked to favorable tumor properties that affect disease outcome independently of both grade and node status.
It is pertinent to mention that our choice of cell lines (MDA-MB-231, -435) does not directly match the properties exhibited by tumors in our cohort, and are ER-, PR-,40 and E-cadherin–41,42 negative. However, these cell lines were chosen for their highly migratory and invasive qualities.43 They are also vimentin-positive43 and represent mesenchymal-like cells in the epithelial-mesenchymal transition (EMT) system. The exact link between proliferation and migration in relation to EMT needs to be further clarified in future studies, because EMT is relevant regarding both malignant and migratory properties in breast cancer.44,45
Finally, to identify gene candidates that may be increasing migration in response to decreased cyclin D1 expression we performed a gene expression microarray experiment. Transcripts associated with cyclin D1 as well as CDK4/6 silencing were identified separately, and there was an overrepresentation of changes in processes associated with migration in the cyclin D1 siRNA–treated cells compared to CDK4/6 siRNA–treated cells (Figure 7A). These results further support the observation from Boyden chamber and time lapse experiments that cyclin D1 has the capacity to influence breast cancer cell migration. As we only observed migratory differences in cyclin D1 and not CDK4/6-silenced cells, we compared the microarray results for the two treatments to identify proteins potentially mediating this effect, while excluding pure cell cycle–dependent changes. Transcripts up- or down-regulated in the cyclin D1 siRNA treated cells, but unaffected or moderately changed by the CDK4/6 siRNA (see Supplemental Table 6 at http://ajp.amjpathol.org) include YAP1,46 CNN2,47 PTP4A1,48 ID1,49 IRS2,50 and MAPK651 which have been linked to migration related processes and are potentially important mediators in proliferation and migration responses in tumors. However, these candidate genes require significant experimental analysis before any roles in proliferation and migration with regard to cyclin D1 can be validated. Transcripts associated with cyclin D1 silencing showed negative enrichment of genes proposed by van’t Veer et al to predict poor prognosis and a short interval to distant metastasis31 in premenopausal breast cancer, again linking cyclin D1 with a migratory phenotype in patient samples.
Our results indicate several key processes in migration may be influenced by the cyclin D1–specific effect (microtubule linked processes, actin binding, cytoskeleton, focal adhesion), and it is likely that the mechanism by which cyclin D1 executes functions is highly complex and not mediated by a single pathway or gene product.
In summary, our findings establish a clear function for cyclin D1 in tumor cell migration as well as proliferation and further support an important role for the protein in defining malignant behaviors in premenopausal breast cancer.
Acknowledgments
We thank Elise Nilsson for technical assistance. We also thank the South Swedish and South-East Swedish Breast Cancer Groups.
Footnotes
Address reprint requests to Prof. Göran Landberg, Breakthrough Breast Cancer Research Unit, School of Cancer, Enabling Sciences and Technology, University of Manchester, Manchester Academic Health Science Centre, Paterson Institute for Cancer Research, The Christie NHS Foundation Trust, Wilmslow Road, Manchester, M20 4BX, UK. E-mail: glandberg@picr.man.ac.uk.
Supported by grants from the Swedish Cancer Society, Malmö University Hospital Research Cancer Funds and by Breakthrough Breast Cancer.
S.L. and N.P.T. contributed equally to this study.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Qu Z, Van Ginkel S, Roy AM, Westbrook L, Nasrin M, Maxuitenko Y, Frost AR, Carey D, Wang W, Li R, Grizzle WE, Thottassery JV, Kern FG. Vascular endothelial growth factor reduces tamoxifen efficacy and promotes metastatic colonization and desmoplasia in breast tumors. Cancer Res. 2008;68:6232–6240. doi: 10.1158/0008-5472.CAN-07-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659:49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Caldon CE, Swarbrick A, Lee CS, Sutherland RL, Musgrove EA. The helix-loop-helix protein Id1 requires cyclin D1 to promote the proliferation of mammary epithelial cell acini. Cancer Res. 2008;68:3026–3036. doi: 10.1158/0008-5472.CAN-07-3079. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, Lisanti MP, Katiyar S, Pestell RG. Cyclin D1 induction of cellular migration requires p27(KIP1). Cancer Res. 2006;66:9986–9994. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- Revillion F, Lhotellier V, Hornez L, Leroy A, Baranzelli MC, Giard S, Bonneterre J, Peyrat JP. Real-time reverse-transcription PCR to quantify a panel of 19 genes in breast cancer: relationships with sentinel lymph node invasion. Int J Biol Markers. 2008;23:10–17. [PubMed] [Google Scholar]
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- Courjal F, Louason G, Speiser P, Katsaros D, Zeillinger R, Theillet C. Cyclin gene amplification and overexpression in breast and ovarian cancers: evidence for the selection of cyclin D1 in breast and cyclin E in ovarian tumors. Int J Cancer. 1996;69:247–253. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Gillett C, Smith P, Gregory W, Richards M, Millis R, Peters G, Barnes D. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- McIntosh GG, Anderson JJ, Milton I, Steward M, Parr AH, Thomas MD, Henry JA, Angus B, Lennard TW, Horne CH. Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene. 1995;11:885–891. [PubMed] [Google Scholar]
- Loden M, Stighall M, Nielsen NH, Roos G, Emdin SO, Ostlund H, Landberg G. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002;21:4680–4690. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- Berglund P, Stighall M, Jirstrom K, Borgquist S, Sjolander A, Hedenfalk I, Landberg G. Cyclin E overexpression obstructs infiltrative behavior in breast cancer: a novel role reflected in the growth pattern of medullary breast cancers. Cancer Res. 2005;65:9727–9734. doi: 10.1158/0008-5472.CAN-04-3984. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Computat Graph Stat. 1996;5:299–314. [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden L, Jonsson PE, Chebil G, Dufmats M, Ferno M, Jirstrom K, Kallstrom AC, Landberg G, Stal O, Thorstenson S, Nordenskjold B. Two years of adjuvant tamoxifen in premenopausal patients with breast cancer: a randomised, controlled trial with long-term follow-up. Eur J Cancer. 2005;41:256–264. doi: 10.1016/j.ejca.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Jirstrom K, Stendahl M, Ryden L, Kronblad A, Bendahl PO, Stal O, Landberg G. Adverse effect of adjuvant tamoxifen in premenopausal breast cancer with cyclin D1 gene amplification. Cancer Res. 2005;65:8009–8016. doi: 10.1158/0008-5472.CAN-05-0746. [DOI] [PubMed] [Google Scholar]
- Jirstrom K, Ryden L, Anagnostaki L, Nordenskjold B, Stal O, Thorstenson S, Chebil G, Jonsson PE, Ferno M, Landberg G. Pathology parameters and adjuvant tamoxifen response in a randomised premenopausal breast cancer trial. J Clin Pathol. 2005;58:1135–1142. doi: 10.1136/jcp.2005.027185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund P, Stighall M, Jirstrom K, Ryden L, Ferno M, Nordenskjold B, Landberg G. Cyclin E confers a prognostic value in premenopausal breast cancer patients with tumours exhibiting an infiltrative growth pattern. J Clin Pathol. 2008;61:184–191. doi: 10.1136/jcp.2007.047688. [DOI] [PubMed] [Google Scholar]
- Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. Regulation of cell motility by mitogen-activated protein kinase. J Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Svensson S, Nilsson K, Ringberg A, Landberg G. Invade or proliferate? Two contrasting events in malignant behavior governed by p16(INK4a) and an intact Rb pathway illustrated by a model system of basal cell carcinoma. Cancer Res. 2003;63:1737–1742. [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, Condeelis JS. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, Zahnow CA, Patterson N, Golub TR, Ewen ME. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, Frampton GM, Cole MF, Odom DT, Odajima J, Geng Y, Zagozdzon A, Jecrois M, Young RA, Liu XS, Cepko CL, Gygi SP, Sicinski P. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, Dye C, Yang J, Dai M, Ju X, Zhang X, Li A, Burbelo P, Stanley ER, Pestell RG. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26:4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen NH, Emdin SO, Cajander J, Landberg G. Deregulation of cyclin E and D1 in breast cancer is associated with inactivation of the retinoblastoma protein. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- Jares P, Rey MJ, Fernandez PL, Campo E, Nadal A, Munoz M, Mallofre C, Muntane J, Nayach I, Estape J, Cardesa A. Cyclin D1 and retinoblastoma gene expression in human breast carcinoma: correlation with tumour proliferation and oestrogen receptor status. J Pathol. 1997;182:160–166. doi: 10.1002/(SICI)1096-9896(199706)182:2<160::AID-PATH814>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Tong D, Czerwenka K, Sedlak J, Schneeberger C, Schiebel I, Concin N, Leodolter S, Zeillinger R. Association of in vitro invasiveness and gene expression of estrogen receptor, progesterone receptor, pS2 and plasminogen activator inhibitor-1 in human breast cancer cell lines. Breast Cancer Res Treat. 1999;56:91–97. doi: 10.1023/a:1006262501062. [DOI] [PubMed] [Google Scholar]
- Hiraguri S, Godfrey T, Nakamura H, Graff J, Collins C, Shayesteh L, Doggett N, Johnson K, Wheelock M, Herman J, Baylin S, Pinkel D, Gray J. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res. 1998;58:1972–1977. [PubMed] [Google Scholar]
- Sarrio D, Palacios J, Hergueta-Redondo M, Gomez-Lopez G, Cano A, Moreno-Bueno G. Functional characterization of E- and P-cadherin in invasive breast cancer cells. BMC Cancer. 2009;9:74. doi: 10.1186/1471-2407-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150:534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, Thompson EW. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, Pardo O, Crook T, Mein CA, Lemoine NR, Jones LJ, Basu S. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15:1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- Huang QQ, Hossain MM, Wu K, Parai K, Pope RM, Jin JP. Role of H2-calponin in regulating macrophage motility and phagocytosis. J Biol Chem. 2008;283:25887–25899. doi: 10.1074/jbc.M801163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V, Pallen CJ, Manser E, Hong W. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63:2716–2722. [PubMed] [Google Scholar]
- Jang KS, Han HX, Paik SS, Brown PH, Kong G. Id-1 overexpression in invasive ductal carcinoma cells is significantly associated with intratumoral microvessel density in ER-negative/node-positive breast cancer. Cancer Lett. 2006;244:203–210. doi: 10.1016/j.canlet.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Cui X, Kim HJ, Kuiatse I, Kim H, Brown PH, Lee AV. Epidermal growth factor induces insulin receptor substrate-2 in breast cancer cells via c-Jun NH(2)-terminal kinase/activator protein-1 signaling to regulate cell migration. Cancer Res. 2006;66:5304–5313. doi: 10.1158/0008-5472.CAN-05-2858. [DOI] [PubMed] [Google Scholar]
- Crowe DL. Induction of p97MAPK expression regulates collagen mediated inhibition of proliferation and migration in human squamous cell carcinoma lines. Int J Oncol. 2004;24:1159–1163. [PubMed] [Google Scholar]