Abstract
Environmental transcriptomics (metatranscriptomics) for a specific lineage of eukaryotic microbes (e.g., Dinoflagellata) would be instrumental for unraveling the genetic mechanisms by which these microbes respond to the natural environment, but it has not been exploited because of technical difficulties. Using the recently discovered dinoflagellate mRNA-specific spliced leader as a selective primer, we constructed cDNA libraries (e-cDNAs) from one marine and two freshwater plankton assemblages. Small-scale sequencing of the e-cDNAs revealed functionally diverse transcriptomes proven to be of dinoflagellate origin. A set of dinoflagellate common genes and transcripts of dominant dinoflagellate species were identified. Further analyses of the dataset prompted us to delve into the existing, largely unannotated dinoflagellate EST datasets (DinoEST). Consequently, all four nucleosome core histones, two histone modification proteins, and a nucleosome assembly protein were detected, clearly indicating the presence of nucleosome-like machinery long thought not to exist in dinoflagellates. The isolation of rhodopsin from taxonomically and ecotypically diverse dinoflagellates and its structural similarity and phylogenetic affinity to xanthorhodopsin suggest a common genetic potential in dinoflagellates to use solar energy nonphotosynthetically. Furthermore, we found 55 cytoplasmic ribosomal proteins (RPs) from the e-cDNAs and 24 more from DinoEST, showing that the dinoflagellate phylum possesses all 79 eukaryotic RPs. Our results suggest that a sophisticated eukaryotic molecular machine operates in dinoflagellates that likely encodes many more unsuspected physiological capabilities and, meanwhile, demonstrate that unique spliced leaders are useful for profiling lineage-specific microbial transcriptomes in situ.
Keywords: metatranscriptome, eukaryote, gene expression, alveolate
Although environmental community genomic analysis (metagenomics) has proved to be a powerful tool for illuminating the genetic potentials of an aquatic microbial assemblage, corresponding gene expression profiling (metatranscriptomics) is crucial for further understanding how these molecular machines function to regulate microbial physiological processes in natural environments. Such in situ transcriptomic analysis is particularly important because only a small fraction of microorganisms are currently amenable to culturing, and laboratory cultures may not truly mimic natural populations. Recent metatranscriptomic studies on prokaryotes, by microarray (e.g., ref. 1) or cDNA sequencing (e.g., ref. 2) analyses, have benefited from the availability of rapidly growing genome sequence data, which allow the expressed genes to be mapped to a particular organism. Thus, these studies have been able to shed light on how genomic machinery operates in a dominant prokaryotic species (Prochlorococcus and Synechococcus) in the investigated natural environment (2). Similar studies of natural eukaryotic microbial assemblages have not been possible because eukaryotic microbial genome data are limited. However, transcriptomic studies for particular eukaryotic microbe lineages would be possible if there was a selective tool to separate gene transcripts of these lineages. Dinoflagellata is an excellent microbial lineage for this type of studies.
Dinoflagellates, present in both marine and freshwater ecosystems, are one of the most important primary producers and contributors of algal toxins in the marine ecosystem. In addition, the symbiotic relationship of the genus Symbiodinium with corals is indispensible for reef growth. Dinoflagellates possess enormous and highly dynamic genomes and exhibit many genomic and cytological features atypical for eukaryotes, such as a lack of nucleosome and canonical histones (as is currently believed), DNA rich in modified nucleotides (5-hydroxymethyluracil and 5-methylcytosine), chromosomes permanently condensed and closed mitosis, highly reduced plastid and mitochondrial genomes with transcripts undergoing extensive editing, and limited cis-splicing but widespread spliced-leader trans-splicing (for reviews, see refs. 3–9). Recent studies suggest that dinoflagellate genomes have been extensively remodeled by rampant organelle-to-nucleus gene transfer (10), extensive gene or genome duplication (11), and integration of reversely transcribed mRNA (12). The large genome sizes (∼3–250 gigabases) have prevented whole-genome sequencing for dinoflagellates; therefore, our knowledge of their molecular machinery has stemmed from single-gene studies or varying scales of EST analyses and is thus fragmentary. What molecular machines operate in dinoflagellate genomes to enable the organisms to thrive in their diverse habitats remains obscure.
The recently discovered unique spliced leader at the 5′ end of the nuclear-encoded mRNAs in dinoflagellates (DinoSL) (3, 4) offers a selective tool to separate dinoflagellate transcripts from a plankton assemblage. To explore the utility of DinoSL for the investigation of gene expression in natural dinoflagellate assemblages, we constructed DinoSL-based cDNA libraries from one marine and two freshwater plankton assemblages and sequenced ∼1,000 cDNA clones for each library. Analysis of the dataset, along with existing dinoflagellate EST and cDNA datasets (DinoEST), revealed a set of genes common to dinoflagellates, a typical eukaryotic set of ribosomal protein (RP) genes, and some genomic features previously unrecognized in dinoflagellates.
Results
Dinoflagellate Diversities in Several Plankton Assemblages.
Samples were collected at Mirror Lake in Storrs, CT, on June 1 (ML1) and 28 (ML2), 2007, and in Long Island Sound at Avery Point (AP) on May 31, 2007. Microscopic analysis showed that Ceratium hirundinella (dominant) and some other dinoflagellates in ML1 accounted for 51% of the total phytoplankton (3,200 cells mL−1) that also contained diatoms and green algae. ML2 (625 cells mL−1) was dominated by Oscillatoria and other small algae, with C. hirundinella (∼10%) as the only recognized dinoflagellate. In the AP sample, the plankton assemblage (2,177 cells mL−1) primarily consisted of dinoflagellates (∼57%; mainly Heterocapsa, Dinophysis, Glenodinium-like, Gymnodinium, and Polarella-like), and diatoms.
Sequencing of dinoflagellate 18S rDNA (∼1.6 kb) clone libraries yielded sequences that matched dinoflagellates (90–100% identical) from various lineages (Fig. S1A). Rarefaction curves indicated that approximately 12, 2, and 9 dinoflagellate species occurred in ML1, ML2 and AP, respectively (Fig. S1B). C. hirundinella and related taxa dominated the ML libraries (Fig. S1A). Heterocapsa triquetra (54%) and Dinophysis acuminata (31%) dominated the AP library, agreeing with microscopic observation.
Specificity of DinoSL and Dinoflagellate Origin of the Retrieved Environmental cDNAs.
A BLASTN search against the GenBank and Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA) databases showed that the 21-nt conserved region of the 22-nt DinoSL (5′ variable nucleotide excluded) was specific to dinoflagellates. Among the 5,000 top hits from GenBank, those showing substantial (>76%) identity to DinoSL were dinoflagellate spliced-leader RNA or DinoSL-containing mRNAs. Most of the 232 significant hits from the CAMERA genomic dataset were unknown sequences; 27 hits had matches in GenBank, 26 belonging to dinoflagellate spliced-leader RNA, and one DinoSL-containing RPS30. In many cases, spliced-leader RNA was in conjunction with 5S, as reported for dinoflagellates previously (4, 9), and, unexpectedly, with 18S or 28S rDNA genes (Table S1).
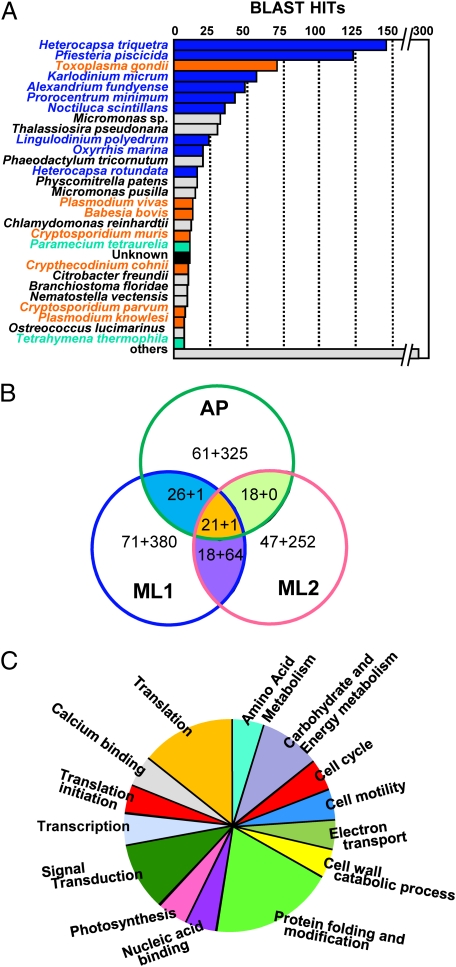
The sequencing of ≈1,000 clones for each of the field-derived DinoSL-based cDNA libraries (e-cDNAs) yielded 822, 647, and 824 good quality cDNAs for ML1, ML2, and AP, which formed 735, 563, and 753 unigene clusters, respectively. These unigene cDNAs had GC contents of 55.66–56.25% (Table S2). Of the unigenes, 47.6%, 46.3%, and 39.7%, respectively, in ML1, ML2, and AP had no match in the GenBank database, whereas 19.0%, 18.1%, and 18.1% hit unknown proteins in dinoflagellates or other alveolates. The remaining clones hit diverse genes (Fig. S2A), predominantly of dinoflagellates and other alveolates (Fig. 1A). Consistent with the above-mentioned observation that H. triquetra was the most abundant dinoflagellate in the AP sample, the largest fraction (27%) of the AP dinoflagellate e-cDNAs hit H. triquetra in DinoEST (Fig. 1A); 33 of them were confirmed to be of this species with strict BLASTN analysis (>95% nucleotide identity with >90% query coverage), including RPS30, α-tubulin, class II fructose bisphosphate aldolase, calreticulin, low-temperature/low-salt protein, calmodulin, and different unknown proteins. Overall, the matches to reported dinoflagellate/alveolate sequences were found with high levels of amino acid identity (typically 70–100%), whereas the small number to nonalveolate sequences were found with weak identity (mostly <50%), indicative of mismatches resulting from lack of corresponding alveolate gene sequences in GenBank. The no-match cDNAs had similar GC contents as those matching dinoflagellate/alveolate sequences (Table S2).
Fig. 1.
Taxonomic and functional distribution of the field-retrieved transcripts (e-cDNAs). (A) Taxonomic distribution of the e-cDNAs. The majority of the sequences either had no matches (others) or hit dinoflagellates (blue) or their alveolate relatives, apicomplexans (orange) and ciliates (green), and a small fraction hit unknown (black) or nondinoflagellate organisms (gray) with low similarities. (B) Number of common and unigene clusters in the three libraries. Numbers indicate annotated genes plus unknown genes. (C) Functional distribution of the 21 common annotated unigene clusters.
Common and Highly Expressed Genes.
In e-cDNAs, most of the unigenes were represented by 1–2 cDNAs, and only several were represented by >10 cDNAs (Fig. S2B). As shown in Fig. 1B, ML1 and ML2 shared 82 common gene clusters, whereas the marine and freshwater samples shared many fewer genes in common (≤27). The two ML libraries exhibited higher expression of ubiquitin (>26 clones) and cell-surface protein p43 (>10), whereas the AP library had a higher expression of major basic nuclear protein (44), centrin/caltractin (24), calmodulin (19), and 14-3-3 (15). Twenty-two gene clusters were common among all three libraries (Table S3), 21 of which were involved in 14 main cellular processes (Fig. 1C), and the other was of unknown function. Major basic nuclear protein, ubiquitin, centrin/caltractin, calmodulin, and 14-3-3 were the most highly expressed among the common genes (Table S3). BLAST analysis showed that these genes were also common in DinoEST (Fig. 2A). In the phylogenetic trees, these highly expressed common genes were clustered with homologs isolated from dinoflagellate cultures (Fig. S3).
Fig. 2.
Comparative analyses of common and highly expressed genes between e-cDNAs and reported datasets. (A) Taxonomic distribution (y axis) among the previously studied dinoflagellates and relative expression levels (x axis; percentage of the transcript in total number of transcripts examined for the species) of the five most highly expressed common genes in e-cDNAs. (B) Dinoflagellate RP set (Dino, purple box) identified from e-cDNAs (green box) and existing DinoEST, which is four proteins more than that in Plasmodium falciparum (Plafa, blue box), one (the Y chromosome–associated RPS4) less than that in humans (Homsa, yellow box), and one (RPLP3) less than that in plants. Asterisks depict that RPL37 is shown not to exist in P. falciparum in the RP database, but we found it in GenBank (Table S4).
RPs.
Fifty-five typical cytoplasmic RPs were identified, 26 belonging to the 40S subunit and 29 to the 60S subunit (Fig. 2B). Two more were found to fuse with ubiquitin (Table S4). All these RPs matched sequences in DinoEST with highly significant E values in BLAST and strong bootstrap support in the phylogenetic trees (examples shown in Fig. S4). Compared with the 79 RPs common in other eukaryotes (http://ribosome.med.miyazaki-u.ac.jp/), 24 were missing from e-cDNAs. By using human RPs as query (Table S4), homologs of all of the 24 RPs were identified in DinoEST, whose identities as RPs were further verified by BLASTX against the GenBank database and phylogenetic analyses.
Histones and Other Nucleosome-Associated Proteins.
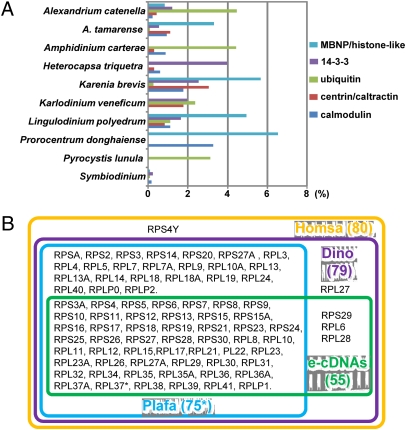
A cDNA in the ML2 library was highly similar to histone H2A.X in Alexandrium tamarense (Fig. 3A and Table 1). BLAST analysis of this sequence against DinoEST hit an EST from Crypthecodinium cohnii (EB086331). A comparison of H2A.X sequences between the three dinoflagellates and other organisms revealed conserved binding sites, the H2A.X signature motif SQEF (generally SQ[D/E]Φ, where Φ is a hydrophobic residue) (Fig. S5A), and the close phylogenetic affiliation of the dinoflagellate sequences (Fig. 3A). Histone H4 was also found in the AP library; it was highly similar to counterparts in other organisms, sharing all of the key conserved binding sites (Fig. S5B). In the phylogenetic tree, this protein fell within the alveolate group (Fig. 3B). Because nucleosomes in “typical” eukaryotes contain four core histones, we further searched in DinoEST and our ongoing Amphidinium carterae and Karlodinium veneficum cDNA datasets and found the other two, H2B and H3 (Table 1, Fig. 3C, and Fig. S5C). Also recovered were transcripts of a histone modification protein (dpy-30 like) and nucleosome assembly protein 1 (NAP1) from the e-cDNAs and histone deacetylase from the above-mentioned ongoing cDNA dataset (Table 1).
Fig. 3.
Comparison of histones H2A.X, H4, and H2B in dinoflagellates with those in other organisms. (A and B) Phylogenetic trees inferred from amino acid sequences of H2A.X (A) and H4 (B) using neighbor joining (NJ), maximum likelihood (ML), and Mr. Bayes (MB) algorithms. Bootstrap support values (NJ and ML) and posterior probabilities (MB) are shown as NJ/ML/MB at nodes; only values higher than 50% or 0.5 are shown, with “-” denoting weak support. Taxa are abbreviated species names followed by GenBank accession numbers; in bold type are e-cDNA sequences. (C) Multialignment of H2B amino acid sequences. Ampca, Amphidinium carterae (GenBank accession number HM245439); Karve, Karlodinium veneficum (HM245441); Perma, Perkinsus marinus (XP_002782087); Toxgo, Toxoplasma gondii (XP_002369740); Plafa, Plasmodium falciparum (XP_001349046); Thaps, Thalassiosira pseudonana (XP_002296171); Homsa, Homo sapiens (CAA41051).
Table 1.
Dinoflagellate nucleosome-related genes retrieved from current e-cDNAs and representatives from existing DinoEST
| Dinoflagellate species | Accession no. | Hit protein | Hit accession number | E value | Similarity, % |
| Uncultured (e-cDNA) | GU555462 | Histone H2A.X | AAX49407 | 5e−50 | 82 |
| Crypthecodinium cohnii | EB086331* | Histone H2A.X | AAX49407 | 3e−29 | 80 |
| Amphidinium carterae | HM245439 | Histone H2B | XP_002782087 | 7e−17 | 71 |
| Karlodinium veneficum | HM245441 | Histone H2B | ACF75502 | 1e−21 | 68 |
| P. lunula | AY151192* | Histone H3 | AAA20819 | 1e−17 | 57 |
| K. veneficum | EC1592402* | Histone H3 | AY151192 | 2e−50 | 70 |
| Symbiodinium sp. | FE864064* | Histone H3 | AY151192 | 9e−08 | 56 |
| Uncultured (e-cDNA) | GU554466 | Histone H4 | XP_001175663 | 9e−32 | 91 |
| A. carterae | HM245435 | Histone deacetylase | XP_002504096 | 7e−62 | 61 |
| K. veneficum | HM245436 | Histone deacetylase | NP_578547 | 2e−30 | 52 |
| Uncultured (e-cDNA) | GU554524 | dpy-30 like | XP_002499412 | 7e−13 | 74 |
| Uncultured (e-cDNA) | GU554907 | NAP1 | XP_002764795 | 1e−27 | 58 |
*Existing DinoEST.
Rhodopsin.
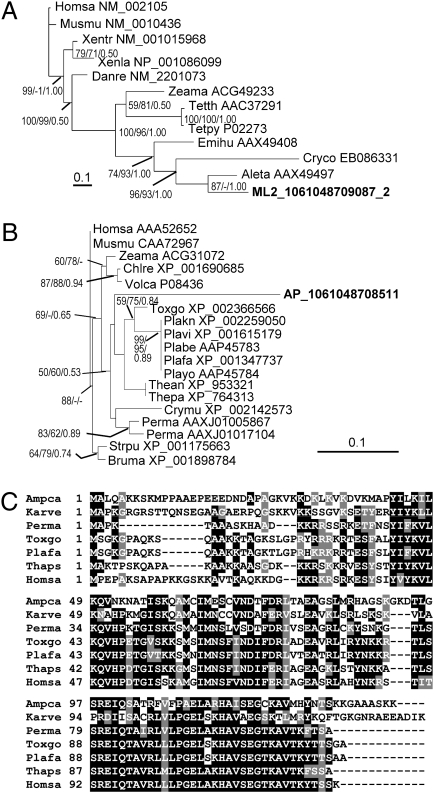
Two cDNAs from the AP library belonged to rhodopsin, a transmembrane retinal-binding protein involved in non-chlorophyll-dependent phototrophy in many bacteria. Their deduced amino acid sequences showed highest (>84%) similarity to the homologs from dinoflagellates Alexandrium catenella, Oxyrrhis marina, and Pyrocystis lunula and was next most similar (66%) to xanthorhodopsin from bacteria among many other rhodopsin sequences. Although these dinoflagellate species represent a phylogenetic spectrum from ancient (O. marina) to late-diverging (A. catenella) lineages, to broaden ecotypic coverage, we analyzed a DinoSL-based cDNA library of the polar species P. glacialis and found a homolog (HM231308). Most of the 22 aa residues in the retinal-binding pocket were conserved between dinoflagellates and the bacteria, and the conserved proton donor residue (glutamic acid) was found in dinoflagellates (Fig. S6). In the phylogenetic tree that included representatives of all different types of rhodopsin, the AP sequences formed a distinct monophyletic clade with the homologs from cultured dinoflagellates, which, although sister to that in an α-proteobacterium Octadecabacter antarcticus, was nested in the larger group of proton-pumping rhodopsin (xanthorhodopsin) from various bacteria (Fig. 4).
Fig. 4.
Phylogenetic affiliation of dinoflagellate rhodopsin with proton-pumping type proteorhodopsin. The gray arc denotes a clade of proton-pumping rhodopsin (xanthorhodopsin), and the dark arc indicates the dinoflagellate subclade, in which sequences from e-cDNAs are in gray shade. Bootstrap support of nodes leading to dinoflagellate-containing clades (arrow) was from maximum likelihood analysis. Taxa are shown by abbreviated species names followed by GenBank accession numbers. GU553957 for AP-cDNA3; GU554267 for AP_1061048708262.
Discussion
Utility of Spliced Leaders for Studying Lineage-Specific Transcriptomes.
All of the results achieved in this study were consistent in indicating that the DinoSL-based approach of cDNA library construction was highly specific for dinoflagellate transcriptome. No DinoSL-containing sequences from databanks were found to be of nondinoflagellate origin. Additional evidence includes high sequence identities of e-cDNAs to dinoflagellate or alveolate cDNAs, well-supported affiliation of e-cDNAs with cultured dinoflagellates in phylogenetic trees, and similar GC content of e-cDNAs to documented dinoflagellate cDNAs (e.g., refs. 13 and 10). Besides, the high diversity of the e-cDNAs is consistent with our earlier proposition that DinoSL is likely ubiquitous in dinoflagellate transcriptomes (3). These findings in concert demonstrate that DinoSL can be a useful tool for investigating dinoflagellate transcriptomes in the natural environment, where contamination by other organisms readily occurs otherwise (Fig. S4C). Furthermore, this approach should be adaptable to the in situ transcriptomic studies of other organisms whose mRNAs also contain unique spliced leaders, such as euglenozoids, cnidarians, rotifers, ascidians, and others (ref. 3 and references therein). Recently, kinetoplastids (euglenozoid) and other parasites were suspected of being responsible for mass mortality of deep fauna at a hydrothermal vent (14). The spliced leader–based approach could be used to investigate what kinetoplastid genes might underlie these mass deaths.
The application of this approach in the current study, albeit on a small scale, yielded interesting results. Our protocol ensured that samples would be fixed in RNA buffer within 5 min of sampling, resulting in nearly a “real-time” in situ dinoflagellate genome expression profile. With DinoEST, a large fraction of the AP e-cDNAs was mapped to H. triquetra, consistent to its being the most dominant dinoflagellate in the sample. As the DinoEST dataset grows and the metatranscriptome sequencing scale increases in the future, an in-depth understanding of how an individual dinoflagellate species’ genome is expressed in situ is achievable. Because C. hirundinella dominated the dinoflagellate ML assemblages, our ML e-cDNAs most likely belong to this species, which is not available in cultures, thus filling the taxonomic gap of dinoflagellate transcriptomic data. More importantly, the results from analyzing the e-cDNAs and further mining DinoEST have helped uncover previously unrecognized genomic features in dinoflagellates, and prompted many new questions worthy of addressing in future studies.
Transcriptional Regulation in Dinoflagellates.
Our observation that the majority of unigenes were represented by 1–2 cDNAs, i.e., most genes were expressed at a uniform background level, is no surprise because previous laboratory studies have similarly shown the general lack of transcriptional gene regulation in dinoflagellates (e.g., ref. 13). The few genes that were highly represented in the e-cDNAs are of interest. Most of them are the common genes found across systems (see Common Genes in Dinoflagellates) and associated with essential cellular processes. Although not noted in the same context, small-numbered highly expressed genes can also be found in DinoEST. For instance, in a K. brevis cDNA library (13), 4,399 of the total of 5,280 cDNA clusters contained single ESTs, 881 contained 2–31 ESTs, and the 24 most highly expressed gene clusters were each represented by 10–50 ESTs. In an A. catenella cDNA library (15), the sequenced 9,847 ESTs contained 6,496 unigene clusters, with the majority (5,475) being represented only once, and 17 genes predominated the library. More systematic investigation is needed to understand whether different genes are more highly expressed in different dinoflagellate species or under different growth conditions.
Common Genes in Dinoflagellates.
The 21 common genes identified in the e-cDNAs are indicative of the essential life activities in the natural assemblages of dinoflagellates: photosynthesis, carbohydrate and energy metabolism, amino acid metabolism, signal transduction, translation, protein folding and modification, cell-wall dynamics, cell motility, and cell cycle (Fig. 1C). Some of these common genes have been reported as highly expressed genes in cultured dinoflagellates, such as fucoxanthin chlorophyll a/c-binding protein (most highly expressed), calmodulin, and cytochrome c in K. brevis (13); fumarate reductase and ubiquitin in A. catenella (15); and fucoxanthin chlorophyll a/c-binding protein, ATP synthase, and major basic nuclear protein in A. tamarense (10). These are clearly the common and actively functional genes in all dinoflagellates. In addition, the cell-surface protein p43 found in the C. hirundinella-dominated lake samples was originally reported in the marine dinoflagellate Lingulodinium polyedrum (16), a gonyaulacoid relative of C. hirundinella. We also found it in another gonyaulacoid species A. catenella (EX462246) and the CAMERA dataset (WesternChannelOMM_READ_06137557). Thus, it is likely that this protein is common in gonyaulacoid if not in all dinoflagellates.
Eukaryotic Set of RPs in Dinoflagellates.
Eukaryotes generally have 32 small-subunit (40S) and 47 large-subunit (60S) cytoplasmic RPs (excluding the plant-specific RPLP3 and human Y chromosome–specific isoform of RPS4) with few exceptions. Only several RPs have previously been documented in dinoflagellates (e.g., refs. 10 and 13). From the e-cDNAs, we obtained 55 RPs; blast analysis matched them to various unannotated sequences in DinoEST, verifying that they were of dinoflagellate origin, meanwhile resulting in the annotation of these DinoEST sequences (Table S4). Further search using human homologs of other eukaryotic RPs against DinoEST yielded another 24 RPs, indicating that the dinoflagellate lineage possesses all of the 79 RPs of a typical eukaryote, 4 more than in Plasmodium, a close relative of dinoflagellates. Our e-cDNAs also contained nop10 (GU554874), a protein required for generation of 18S rRNA. Thus, it is most likely that the structure and function of ribosomes in dinoflagellates are similar to those in typical eukaryotes despite the many noneukaryotic features in dinoflagellates.
Histones and Nucleosome-Like Machinery in Dinoflagellates.
Nucleosome is an octamer containing two units of H2A, H2B, H3, and H4 with 150 bp of DNA wrapped around it (17). Dinoflagellates are thought to have no histones and nucleosome (18), and chromatin is thought to be organized by major basic nuclear proteins (e.g., ref. 19). The canonical H2A in some metazoan nucleosomes is replaced by H2A.X (20), which in most eukaryotes is packaged in nucleosomes during DNA replication and is important in recognizing and repairing DNA double-strand breaks (21). H2A.X has been reported in A. tamarense cDNA (10), but further characterization became possible only now that we have identified more dinoflagellate homologs. Interestingly, the signature motif in all three dinoflagellates is SQEF, identical to the two H2A.X isoforms in Xenopus laevis, in which phosphorylation of this (S135) and another (S127) serine residue regulates the early developmental cell cycle independent of DNA damage repair (20). It will be of interest to find out if H2A.X in dinoflagellates is involved in regulating the cell cycle of their reproductive cysts in which multiples of two daughter cells are produced before release (22), analogous to the rapid cell division in metazoan early embryos.
Dinoflagellate histone H3 has only previously been reported in P. lunula (23). In this study, two full-length homologs were identified for K. veneficum, and a partial cDNA was identified for Symbiodinium species in DinoEST (Table 1). These dinoflagellate H3 sequences, phylogenetically close to each other (Fig. S5C), contained most of the critical amino acid residues (e.g., R2 and K14; ref. 24). The two other core histones, H2B and H4 were also obtained in this study (Table 1 and Fig. S5). In addition, we found a histone modification transcript from e-cDNAs and histone deacetylase in K. veneficum and A. carterae (Table 1). NAP1, which is an integral component in establishing, maintaining, and modulating eukaryotic chromatin and regulating gene transcription (25), was found in e-cDNAs and K. brevis ESTs (CO062076, EX957422-23, and EX957973-74). All these results in concert suggest that dinoflagellates currently possess operative histone-based nucleosome machinery. Previous studies failed to detect histones and other nucleosome-associated proteins probably because they are expressed at low levels or only in some stage(s) of the life cycle in dinoflagellates. If not associated in any way with the organization of chromatins in dinoflagellates, then these histones and other nucleosomal proteins may be involved in the regulation of gene expression (e.g., in response to stress; ref. 26) besides DNA repair (H2A.X).
Nonphotosynthetic Photoenergy Utilization.
The conventional view that biological utilization of solar energy is solely through photosynthesis has become obsolete since the discovery of rhodopsin and the recognition of its function in marine microbes to harvest light energy nonphotosynthetically. First detected in haloarchaea, rhodopsin is now known to occur in various bacteria and eukaryotes such as fungi and algae, with various functions ranging from light sensing and chloride pumping to proton pumping (refs. 27–30 and references therein). In the latter case, the proton would be pumped across the cell membrane to generate power for cell use (through ATP synthesis) or to drive nutrient transport or flagellar motor rotation. Rhodopsin, in its role as a proton pump in a wide range of bacteria, is believed to contribute substantially to light energy entry into the marine ecosystem (30). Rhodopsin has been unknown in dinoflagellates until its recent detection in P. lunula EST, in which this gene was observed to be expressed more actively at early light phase than at early dark phase, suggesting its involvement in circadian photoreception and phase shifting (31). However, it had remained unclear as to whether this gene was common in dinoflagellates and what functions it might have. Now with the isolation of this gene from e-cDNAs and cultured P. glacialis in this study, and from A. catenella and O. marina recently (Fig. 4), it appears that rhodopsin is ubiquitous in dinoflagellates. Further, the dinoflagellate rhodopsin is phylogenetically closely related to xanthorhodopsin, a unique rhodopsin that harvests light energy with carotenoid molecules as the antenna (32). Additionally, the dinoflagellate rhodopsin sequences contain the characteristic proton donor site. All these facts suggest that dinoflagellate rhodopsin is a proton-pumping, instead of a sensory, rhodopsin. It remains to be determined which of the above-mentioned physiological processes benefits from the rhodopsin-generated energy in dinoflagellates. But it is noteworthy that light promotes heterotrophic growth of mixotrophic dinoflagellates such as K. veneficum (33), where rhodopsin-generated energy might be used to enhance prey ingestion and digestion.
Materials and Methods
In Silico Analysis of DinoSL Specificity.
DinoSL had been shown to be specific to dinoflagellate nuclear-encoded mRNAs (3). To further verify the specificity, the 21-nt highly conserved DinoSL RNA (excluding the variable first nucleotide) was used to query against the GenBank database and the total environmental DNA sequence reads in the CAMERA database (43,240,119 entries, 16,900,401,306 bp) using an E value of 0 as the cutoff. Hit sequences were retrieved and BLAST-searched against the GenBank database.
Sample Collection and Initial Processing.
At Mirror Lake in Storrs, CT (a freshwater pond, 41°48’36”N, 72°15’36”W), surface-water samples were collected at 2:00 PM on June 1 (ML1) and 28 (ML2), 2007. A marine sample was collected at the northern shore of Long Island Sound, i.e., at Avery Point (41°18’55”N, 72°03’48.6”'W) at 11:30 AM on May 31, 2007, with the use of a plankton net. At the time of sampling, the water temperature was at 29 °C (ML1), 33 °C (ML2), and 20 °C (AP), respectively. Salinity was 0‰ for the ML samples and 32‰ for the AP sample. A subsample (100 mL for ML1, 250 mL for ML2, and 500 mL for AP) was filtered on-site immediately onto a 5-μm Nuclepore filter membrane. The cell-retaining filter was then immersed in 1 mL TRIzol RNA buffer in a cooler. To capture in situ gene expression, water collection to sample preservation in RNA buffer was accomplished within 3–5 min. To analyze dinoflagellate species composition, a subsample was filtered in the same way, but the filter was immersed in 0.5 mL of DNA buffer (34), and another was fixed with Utermöhl's solution for subsequent microscopic examination.
DNA Extraction and Dinoflagellate 18S rDNA Analysis.
DNA was extracted and PCR was run with dinoflagellate-oriented 18S rDNA primers (34). A 1.6-kb amplicon was cloned, and more than 40 clones were randomly picked and sequenced. The taxonomic affiliations of the sequences were determined by BLAST against the GenBank database and phylogenetic analysis. The sequences were aligned with ClustalX, and the alignment was manually inspected and corrected. Phylogenetic analyses were performed according to our previous reported methods (34, 35).
RNA Extraction, cDNA Library Construction, and Sequencing.
Total RNA was extracted following the methods in ref. 3 and further purified with the Qiagen RNeasy Mini Kit. First-strand cDNA was synthesized and purified and then used as the template in PCR with primer set DinoSL-RACER3 to enrich the full-length dinoflagellate-specific cDNAs (3); the cDNAs were then cloned and sequenced.
cDNA Sequence Data Analysis.
The cDNA sequences were clustered with CAP3 at 99% identity cutoff to yield unigenes in each library. Codon usage and nucleotide frequencies were analyzed with the General Codon Usage Analysis (GCUA) package (http://bioinf.may.ie/GCUA/index.html). Annotation was performed using BLAST2GO under the BLASTX algorithm (http://www.blast2go.org/). Gene function was determined based on the function of hit genes from blastx and gene ontology mapping. Gene products were then described in terms of their associated biological pathway, molecular function, and cellular components.
Comparative Analyses for e-cDNAs and DinoEST Datasets.
Characterized genes common to two or all three libraries were identified based on their functional identities. Common unknown genes were identified by 80% identity-cutoff clustering. The relative expression levels of the genes were assessed by examining the total number of cDNA clones contributing to each unigene cluster. To find gene homologs in cultured dinoflagellates, e-cDNA sequences were BLAST searched against the largely unannotated DinoEST sequences downloaded from GenBank (∼130,000 entries as of June 2009). Hit sequences with significant E values (less than −10 overall, mostly much lower) were BLAST-searched against the GenBank annotated database to verify their functional identities.
Phylogenetic Analyses to Verify Dinoflagellate Origin and Investigate Potential Functions of the cDNAs.
Deduced amino acid sequences of selected cDNAs were aligned with homologs from dinoflagellates and other organisms using ClustalX. Neighbor-joining phylogenetic trees were generated (http://www.ddbj.nig.ac.jp) to indicate the relationship of the retrieved sequences to documented genes. Representative genes were further investigated by using maximum likelihood analysis in PhyML3.0 (http://www.atgc-montpellier.fr/phyml) based on the best-fit evolutionary model selected by ProtTest (http://darwin.uvigo.es/software/prottest.html) and Bayesian analysis in Mr. Bayes3.1.2 (http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=mrbayes).
Supplementary Material
Acknowledgments
We thank Dr. Sheng Liu for assistance in the microscopic analysis of the samples, Dr. Cheong Xin Chan for advice on initial data analysis, and Dr. Edward Monahan for English language review. This work was supported by National Science Foundation Grants EF-0626678 and OCE-0854719.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. GU553924–GU555965 (cDNA) and GU647163–GU647195 (18S rDNA)].
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007246107/-/DCSupplemental.
References
- 1.Parro V, Moreno-Paz M, González-Toril E. Analysis of environmental transcriptomes by DNA microarrays. Environ Microbiol. 2007;9:453–464. doi: 10.1111/j.1462-2920.2006.01162.x. [DOI] [PubMed] [Google Scholar]
- 2.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. Microbial community gene expression in ocean surface waters. Proc Acad Natl Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, et al. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci USA. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lidie KB, van Dolah FM. Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis. J Eukaryot Microbiol. 2007;54:427–435. doi: 10.1111/j.1550-7408.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 5.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: A remarkable evolutionary experiment. Am J Bot. 2004;91:1523–1534. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 6.Howe CJ, Nisbet RER, Barbrook AC. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 2008;59:1035–1045. doi: 10.1093/jxb/erm292. [DOI] [PubMed] [Google Scholar]
- 7.Waller RF, Jackson CJ. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays. 2009;31:237–245. doi: 10.1002/bies.200800164. [DOI] [PubMed] [Google Scholar]
- 8.Lin S, Zhang H, Gray MW. RNA editing in dinoflagellates and its implications for the evolutionary history of the editing machinery. In: Smith HC, editor. RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems. Hoboken, NJ: John Wiley & Sons, Inc.; 2008. [Google Scholar]
- 9.Zhang H, Campbell DA, Sturm NR, Lin S. Dinoflagellate spliced leader RNA genes display a variety of sequences and genomic arrangements. Mol Biol Evol. 2009;26:1757–1771. doi: 10.1093/molbev/msp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackett JD, et al. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics. 2005;6:80. doi: 10.1186/1471-2164-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PLoS ONE. 2009;4:e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slamovits CH, Keeling PJ. Widespread recycling of processed cDNAs in dinoflagellates. Curr Biol. 2008;18:R550–R552. doi: 10.1016/j.cub.2008.04.054. [DOI] [PubMed] [Google Scholar]
- 13.Lidie KB, Ryan JC, Barbier M, Van Dolah FM. Gene expression in Florida red tide dinoflagellate Karenia brevis: analysis of an expressed sequence tag library and development of DNA microarray. Mar Biotechnol (NY) 2005;7:481–493. doi: 10.1007/s10126-004-4110-6. [DOI] [PubMed] [Google Scholar]
- 14.Moreira D, López-García P. Are hydrothermal vents oases for parasitic protists? Trends Parasitol. 2003;19:556–558. doi: 10.1016/j.pt.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Uribe P, et al. Preparation and analysis of an expressed sequence tag library from the toxic dinoflagellate Alexandrium catenella. Mar Biotechnol (NY) 2008;10:692–700. doi: 10.1007/s10126-008-9107-8. [DOI] [PubMed] [Google Scholar]
- 16.Bertomeu T, Hastings JW, Morse D. Vectorial labeling of dinoflagellate cell surface proteins. J Phycol. 2003;39:1254–1260. [Google Scholar]
- 17.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 18.Rizzo PJ. Those amazing dinoflagellate chromosomes. Cell Res. 2003;13:215–217. doi: 10.1038/sj.cr.7290166. [DOI] [PubMed] [Google Scholar]
- 19.Wong JT, New DC, Wong JC, Hung VK. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot Cell. 2003;2:646–650. doi: 10.1128/EC.2.3.646-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shechter D, et al. A distinct H2A.X isoform is enriched in Xenopus laevis eggs and early embryos and is phosphorylated in the absence of a checkpoint. Proc Natl Acad Sci USA. 2009;106:749–754. doi: 10.1073/pnas.0812207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litaker RW, et al. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida. J Phycol. 2002;38:442–463. [Google Scholar]
- 23.Okamoto OK, Hastings JW. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene. 2003;321:73–81. doi: 10.1016/j.gene.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeld JA, et al. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park Y-J, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 28.Waschuk SA, Bezerra AG, Jr., Shi L, Brown LS. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung K-H. The distinct signaling mechanisms of microbial sensory rhodopsins in Archaea, Eubacteria, and Eukarya. Photochem Photobiol. 2007;83:63–69. doi: 10.1562/2006-03-20-IR-853. [DOI] [PubMed] [Google Scholar]
- 30.Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: An array of physiological roles? Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto OK, Hastings JW. Novel dinoflagellate clock-related genes identified through microarray analysis. J Phycol. 2003;39:519–526. [Google Scholar]
- 32.Balashov SP, Lanyi JK. Xanthorhodopsin: Proton pump with a carotenoid antenna. Cell Mol Life Sci. 2007;64:2323–2328. doi: 10.1007/s00018-007-7167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li A, Stoecker DK, Coats DW. Mixotrophy in Gymnodinium galatheanum (Dinophyceae): grazing responses to light intensity, and inorganic nutrients. J Phycol. 2000;36:33–45. [Google Scholar]
- 34.Lin S, Zhang H, Hou Y, Miranda L, Bhattacharya D. Development of a dinoflagellate-oriented PCR primer set leads to detection of picoplanktonic dinoflagellates from Long Island Sound. Appl Environ Microbiol. 2006;72:5626–5630. doi: 10.1128/AEM.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin S, Zhang H, Hou Y, Zhuang Y, Miranda L. High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob genes for dinoflagellate DNA barcoding. Appl Environ Microbiol. 2009;75:1279–1290. doi: 10.1128/AEM.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.