Abstract
Induced osteogenesis includes a program of microRNAs (miRs) to repress the translation of genes that act as inhibitors of bone formation. How expression of bone-related miRs is regulated remains a compelling question. Here we report that Runx2, a transcription factor essential for osteoblastogenesis, negatively regulates expression of the miR cluster 23a∼27a∼24-2. Overexpression, reporter, and chromatin immunoprecipitation assays established the presence of a functional Runx binding element that represses expression of these miRs. Consistent with this finding, exogenous expression of each of the miRs suppressed osteoblast differentiation, whereas antagomirs increased bone marker expression. The biological significance of Runx2 repression of this miR cluster is that each miR directly targets the 3′ UTR of SATB2, which is known to synergize with Runx2 to facilitate bone formation. The findings suggest Runx2-negative regulation of multiple miRs by a feed-forward mechanism to cause derepression of SATB2 to promote differentiation. We find also that miR-23a represses Runx2 in the terminally differentiated osteocyte, representing a feedback mechanism to attenuate osteoblast maturation. We provide direct evidence for an interdependent relationship among transcriptional inhibition of the miR cluster by Runx2, translational repression of Runx2 and of SATB2 by the cluster miRs during progression of osteoblast differentiation. Furthermore, miR cluster gain of function (i.e., inhibition of osteogenesis) is rescued by the exogenous expression of SATB2. Taken together, we have established a regulatory network with a central role for the miR cluster 23a∼27a∼24-2 in both progression and maintenance of the osteocyte phenotype.
Keywords: bone formation by miRs, Runx2-regulation of microRNA cluster, phenotype attenuation by miRs, feed forward-feedback miR control, SATB2 targeted by cluster miRs
Numerous regulatory pathways governing osteoblast differentiation involve transcription factors, signaling molecules, and chromatin modifiers (1–3). Recent evidence has shown that osteogenic induction and differentiation are also regulated by posttranscriptional mechanisms, most significantly by temporally expressed microRNAs (miRs) (4, 5).
miRNAs are small noncoding RNAs that significantly regulate the translation of protein coding genes in higher organisms (6, 7). These small RNAs (approximately 22 nt) are involved in almost every biological process, including early development, lineage commitment, growth and differentiation, cell death, and metabolic control (4–7). The identification of miRs that characterize cancer and genetic and metabolic abnormalities provide new approaches for treatment of diseases (8–11).
The recent discovery of miRNAs in regulating in vivo bone formation (12) and in vitro osteoblast differentiation has provided insights into the potent activity of miRs in the skeleton (4, 5, 13–15). Studies show that the osteogenic BMP2 down-regulates a cohort of miRs that inhibit bone formation (4, 16). In an introductory miR profiling study of MC3T3-E1 osteoblasts, we observed up-regulation of approximately 60 miRs during differentiation from the osteoprogenitor cell to the final osteocyte stage of tissue mineralization (5). Among this data set are included three miRs that belong to a cluster, miR-23a∼27a∼24-2.
Intergenic miR clusters are characterized by their own promoter, but there are relatively few studies regarding their roles in key biological processes. The miR-23a∼27a∼24-2 cluster was previously described in promoting cell proliferation in several cancers (17–19). Recently, a regulatory interplay was described in megakaryopoiesis between miR-27a and AML1 (Runx1), an essential regulator of hematopoiesis (20). Also, in patients, the AML1-ETO leukemia factor was found to induce expression of miR-24 in this cluster (21). These findings prompted us to examine the regulation and functional activity of the miR-23a∼27a∼24-2 cluster in osteoblasts and in relation to Runx2/AML3, which is required for osteogenesis.
Here we demonstrate that Runx2 down-regulates expression of each miR in the cluster. Significantly, each miR targets SATB2, an activator of osteogenesis. A feed-forward mechanism was characterized by Runx2 suppression of the cluster to release SATB2 from repression. Additionally, we find miR-23a directly suppresses Runx2 through 3′ UTR binding. This feedback mechanism regulates the terminal stage of osteoblast differentiation by both increasing the miR cluster expression and downregulating Runx2. Our studies have identified a regulatory circuit involving Runx2, SATB2, and the miR cluster 23a∼27a∼24-2, which has a critical central role in controlling progression and attenuation of a specific cell phenotype.
Results
Functional Activity of Runx2 and miR-23a∼27a∼24-2 Cluster Expression During Osteoblast Differentiation.
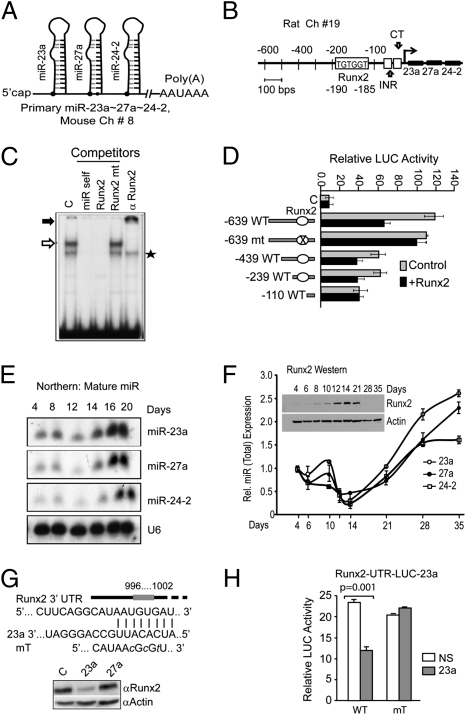
The miRNA cluster miR-23a∼27a∼24-2 RNA Pol II transcript, first identified in human chromosome 19, is very similar in structure to mouse and rat transcripts (22, 23) (Fig. 1 A and B and Fig. S1). Sequence analysis of the miR cluster promoter region identified one consensus Runx-binding site (TGTGGT) immediately upstream of the previously characterized transcription start site (Fig. 1B and Fig. S1). We postulated that Runx2 directly regulates miR expression in a cell type-specific manner. Direct binding of Runx2 to the miR-23a∼27a∼24-2 promoter was confirmed by an EMSA using nuclear proteins from MC3T3-E1 osteoblasts (Fig. 1C, lane 1). A Runx2 protein–DNA complex was confirmed by oligonucleotide competition (Fig. 1C, lanes 2–4) and antibody supershifts (Fig. 1C, arrow, lane 5). Transcriptional activity of the miR cluster promoter mediated by this Runx2 binding site was examined with a series of promoter-reporter constructs cotransfected with or without Runx2 in osteoblasts (Fig. 1D). In the presence of exogenous Runx2, promoter activity of the −639, −439, and −239 deletion fragments of the cluster is significantly down-regulated. However, no effect was found with mutants −639 LUC and −110 LUC, each lacking the Runx2-binding site. These results indicate a functional Runx2 regulatory element is present from −190 to −185 in the proximal promoter of the miR cluster 23a∼27a∼24-2.
Fig. 1.
A functional Runx2 DNA binding site regulates expression of the miR-23a∼27a∼24-2 cluster and miR-23a regulates Runx2. (A) Stem loop structure of the transcript for the murine miR cluster in chromosome 8. (B) Representation of the rat −0.639-kb miR-23a∼27a∼24-2 cluster promoter fragment. Transcription factor analysis of the proximal promoter showing the Runx binding site by TRANSFAC, TESS program (TGTGGT, −185 to −190 for rat), the INR motif (initiator, CCCCACCTCC), and the CT motif (CTCT…) sequence at −56 to −34 (23). (C) EMSA of nuclear proteins from MC3T3-E1 cells using an oligonucleotide WT and mutant probe (Table S1) for the Runx2 site in the miR promoter. Control binding (C, lane 1), self-competitor for WT (miR self, lane 2), Runx2 consensus WT (Runx2, lane 3) or mutant (Runx2 mt, lane 4), and antibody supershift (lane 5) are shown. Open arrow, Runx2 complex; solid arrow, supershifted Runx2 protein–DNA complex; star, NS band. (D) Functional activity of the Runx2 site in the −0.639 kb human miR cluster promoter and its deletion and mutant constructs with luciferase reporter. The reporter constructs were cotransfected with control vector (gray bars) or Runx2 expression construct (solid black bars) in MC3T3-E1 cells (Materials and Methods describes quantification). (E) Northern blot for expression of mature miR-23a, -27a, and -24-2 during primary ROB cell differentiation (days 4–20). U6 was used for control. (F) Expression of the miR cluster by qPCR and Runx2 protein (Inset; Western). (A) Total RNA from ROB cells was assayed for each miR (precursor and mature) normalized to U6 expression as indicated. Runx2 protein detected by a mouse monoclonal antibody (MBL International). Actin was used for control. (G) Upper: Diagram of the 3′ UTR of Runx2 mRNA illustrating miR-23a binding site and a mutation (mT). Lower: Western analysis of Runx2 protein in day 20 ROB cells transduced on day 4 with lentiviral overexpression of miR-23a and -27a. Actin was used as loading control. (H) Runx2 3′ UTR LUC assay demonstrating miR-23a regulation using MC3T3-E1 cells (Materials and Methods).
Because Runx2 directly regulates this miR cluster, we examined its expression over a time course of osteoblast differentiation (day 0–20) using primary rat osteoblasts (ROBs). A temporal expression pattern for mature miRs (Fig. 1E; Northern blot) was found with very low expression from the osteoprogenitor to the mature osteoblast up to day 12 and maximum expression during the mineralization stage, days 16 to 20, for all three cluster members (Fig. 1E). We next performed a more extensive time course to examine relation to total miRNA expression (premiR + mature miR) by quantitative PCR (qPCR) and Runx protein levels (Fig. 1F). The mature miR profile (Fig. 1E) is similar to the miR expression profile. An initial slight increase is found in proliferating osteoblasts (days 4–10) correlating with low Runx2 protein. The miRs rapidly decline as the Runx2 is induced in mature osteoblasts (days 12–21). However, Runx2 protein declines after day 21, decreasing to a negligible level in osteocytes (after mineralization days 28–35). As this occurs, expression of the miR cluster increases.
We tested whether the mechanism for attenuation of Runx2 involved the cluster miRNAs. Three different miR databases predicted mirR-23a to target the Runx2 3′ UTR (Fig. 1G). Expression of miR-23a, but not miR-27, nearly completely inhibited Runx2 protein levels in differentiated osteoblasts (Fig. 1G, Lower). Furthermore, LUC activity of the Runx2 3′ UTR reporter was suppressed 50% by miR-23a whereas no effect was observed when the binding site was mutated compared with nonspecific (NS) control miR (Fig. 1H). Taken together, the reciprocal expression pattern between Runx2 and the miR cluster and miR-23 control of Runx2, validated by specific binding site mutation analysis, suggest a regulatory loop between Runx2 and the miR cluster to regulate the osteoblast phenotype at multiple stages.
Runx2 Binding Directly Represses miR-23a∼27a∼24-2 Biosynthesis.
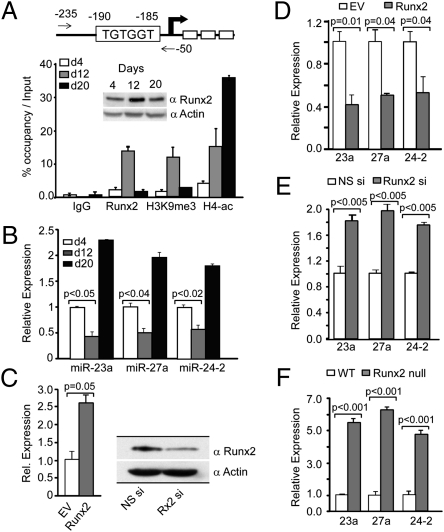
To address the timing of in vivo binding of Runx2 to the miR-23a∼27a∼24-2 promoter during differentiation, chromatin immunoprecipitation (ChIP) assays were performed using ROBs at three stages of differentiation [days 4 (proliferation), 12 (matrix maturation), and 20 (mineralization); Fig. 2A]. There is reciprocal association of Runx2 at the miR cluster promoter, with highest occupancy on day 12, correlating with the lowest expression of each precursor miR (Fig. 2B) and consistent with the highest Runx2 cellular protein levels (Fig. 2A, Inset). Increased H3K9me3 histone modification in the proximal miR cluster promoter indicates the promoter is repressed when Runx2 protein is maximally associated (day 12); whereas the lowest Runx2 binding correlates with histone H4 acetylation of the miR promoter (Fig. 2A; day 20) and highest levels of miR expression (Fig. 2B). Thus, a Runx2 regulatory element bound by Runx2 in osteoblasts negatively regulates expression of the miR-23a∼27a∼24-2 cluster for progression of osteoblast differentiation.
Fig. 2.
Mechanism of Runx2 regulation of each miR within a cellular context. (A) In vivo occupancy of Runx2 proteins on the miR promoter during ROB cell differentiation. Upper: Arrow indicates position of primers (Table S1) used in ChIP. Lower: ChIP was performed at the indicated days of ROB differentiation using H3K9me3, H4 Penta acetyl, Runx2, and control IgG antibodies. Inset, Top: Runx2 protein expression during days 4, 12, and 20 of ROB cell differentiation. (B) Expression (real-time PCR analysis) of the precursor miR cluster members normalized by U6 at the indicated time points. (C) Overexpression for 24 h (Left, qPCR) and knockdown by siRNA for 72 h (Right; Western) of Runx2 in MC3T3-E1 cells to detect Runx2 levels. (D) Consequences of overexpression of Runx2 for 24 h on precursor miR cluster RNA by qRT-PCR. (E) Precursor miRNA analysis of miR-23a, -27a, and -24-2 upon Runx2 knockdown. (F) Expression analysis of precursors of miR-23a∼27a∼24-2 cluster members by real-time qPCR in genetically deleted Runx2−/− mouse calvarial osteoblast cells.
To further establish that Runx2 functionally regulates endogenous miR-23a∼27a∼24-2 transcription in an in vivo cellular context, we examined expression of these miRs during three biological conditions: expression and knock-down in osteoblasts and in WT osteoblasts from Runx2-null mice (24). Forced expression of Runx2 in preosteoblasts (Fig. 2C, Left) down-regulates each member of the cluster by 40% to 60% (Fig. 2D). Consistent with this finding, knockdown of Runx2 (Fig. 2C, Right) increased endogenous expression of the cluster miRs 1.8 fold (Fig. 2E). Of greater significance, in Runx2-null cells isolated from calvaria of newborn mice (24), a five- to 6.5-fold increase in miR cluster expression occurred in three different preparations compared with osteoblasts from WT mice (Fig. 2F). Further, we find that the domains of Runx2 involved in negative regulation of the miR cluster are mediated through both DNA binding and the C terminus of the protein (Fig. S2). Taken together, these multiple approaches demonstrate a requirement for Runx2 to negatively regulate the miR cluster and suggest the miR cluster has a significant role related to osteoblastogenesis.
MiR-23a Is a Potent Inhibitor of Osteoblastogenesis and miR-27a Delays Osteoblast Differentiation Through Down-Regulation of SATB2.
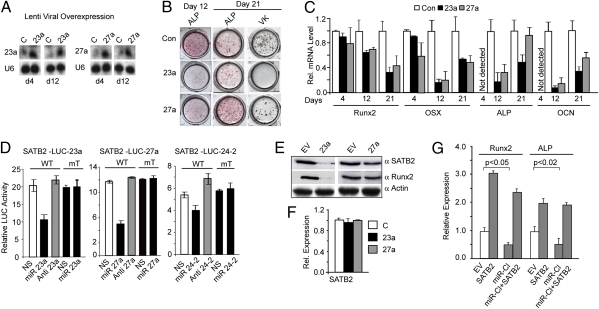
Because Runx2 down-regulates miR-23∼27a∼24-2, it is necessary to establish the functional activity of the cluster miRs on osteogenesis. We focused these studies on miR-23a and miR-27a as two representative miRs. Lentivirus expressing miR-23a and miR-27a precursors were used for transduction of primary ROBs, followed by differentiation (Fig. 3A). Histological staining for alkaline phosphatase (ALP), an early marker of bone formation, and Von Kossa staining for mineral deposition (Fig. 3B) revealed that the miRs significantly inhibited osteoblast maturation (day 12). MiR-23a reduced ALP and caused a complete block in mineralization (von Kossa stain) at day 21. However, miR-27a delayed robust ALP and reduced mineralization. These changes are supported by analysis of bone specific markers of differentiation (Fig. 3C). On day 4, expression of Runx2 and Osterix, the transcription factors required for commitment to the osteoblast phenotype, was little affected by the miRs, but down-regulation began on day 12 and decreased to 50% by day 21 for both transcription factors. The decrease in Runx2 on days 12 and 21 is the result of a block differentiation by the two miRs. Both miR-23a and miR-27a significantly decreased (approximately 80%) ALP and osteocalcin on day 12 but not on day 21, indicating strong inhibition and initial delay of ROB maturation, respectively, by the two miRs. Similar effects of miR-23a and -27a on differentiation of the mouse MC3T3-E1 cell line were observed (Fig. S3). These findings indicate that this miR cluster strongly inhibits activators essential for progression of osteoblast maturation.
Fig. 3.
Expression of miRNAs 23a and 27a inhibit primary osteoblast differentiation through targeting SATB2. (A) Northern blot showing levels of lentiviral mediated overexpressed miR-23a and -27a in primary ROB during differentiation. (B) Histochemical staining: primary ROB cells were infected with control, miR-23a, and miR-27a lentivirus at day 4 and cultured in differentiation medium for 21 d. ALP activity and von Kossa (VK) for mineral deposition at days 12 and 21 is shown. (C) mRNA expression profile of bone marker genes as indicated on days 4, 12, and 21. (D) Each miR in the cluster, as indicated, down-regulates SATB2 in LUC reporter assay, but not mutant miRs (Results). Location of miR sites are in Fig. S4. (E) Overexpression of miR-23a and -27a decrease SATB2 and Runx2 protein: Western blot after 72 h in MC3T3-E1 cells. Actin used as loading control. (F) Stability of SATB2 mRNA upon overexpression of miR-23a and -27a for 72 h (quantitative RT-PCR), normalized to U6. (G) Biological rescue of miR inhibition of osteoblast differentiation by SATAB2 in MC3T3-E1, miR cluster (miR-Cl), SATB2, or transfected together for 72 h. Total RNA was analyzed for Runx2 and ALP expression normalized to GAPDH.
To understand the mechanism by which these miRs inhibit and delay osteoblast differentiation, we sought to determine a specific target(s) of this cluster relevant to bone formation. Bioinformatics programs revealed that all three miRs have putative binding sites in the 3′ UTR of SATB2 (Fig. S4) that are evolutionarily conserved among vertebrate species. Reporter assays in MC3T3-E1 cells using SATB2 3′ UTR-LUC for each miR showed that exogenous expression of individual miRs (miR-23a, -27a, or -24-2) significantly repressed the luciferase activity (Fig. 3D). Complementary to this effect, anti-miRs slightly increased luciferase reporter activity, whereas no effect was observed when the respective miR binding sites were mutated. Additionally, SATB2 protein was repressed several fold by both premiR-23a and -27a (Fig. 3E); however, stability of the mRNA was unaffected (Fig. 3F). These studies provide proof that SATB2 expression is directly controlled through 3′ UTR regulation by each miRNA of the cluster.
We next directly examined the role of SATB2 in Runx2-miR cluster-mediated control of osteogenesis by a gain-of-function study. The effect of SATB2 expression increases osteoblast markers Runx2 and ALP (Fig. 3G). Overexpression of the miR cluster 23a∼27a∼24-2 in MC3T3-E1 cells decreased osteoblast markers by 50%. However, rescue of this inhibition occurs upon SATB2 overexpression in the presence of the miR cluster. Therefore, negative biological control of the miR cluster by Runx2 is crucial to raise SATB2 cellular levels for the progression of osteogenesis.
In Vivo Association and Regulation of SATB2 by the miR Cluster Is Linked to a Biological Network with Runx2.
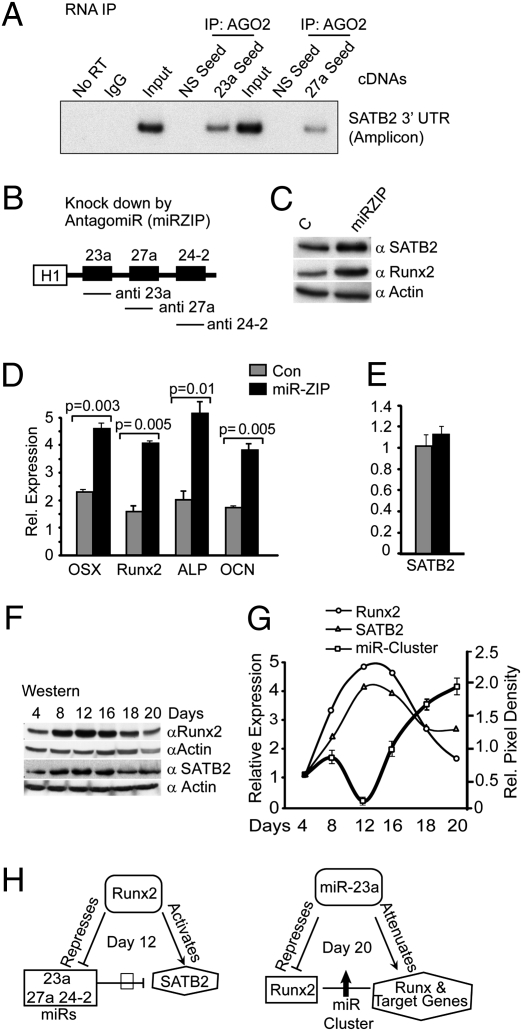
To provide direct evidence for the in vivo functional requirements for the miR cluster in regulating osteoblastogenesis through a regulatory loop involving Runx2 and SATB2, we first determined the in vivo binding of the miRs to endogenous SATB2 mRNA by ribonucleoprotein-immunoprecipitation (RNP-IP) studies (Fig. 4A). The presence of SATB2 3′ UTR fragments in miR-23a and -27a seed-specific cDNAs in the RNP-IP confirmed that SATB2 mRNA is an in vivo target in osteoblasts. Absence of SATB2-specific PCR products in IgG, No RT, and cDNA with NS seed-sequence controls ruled out the possibility of NS immunoprecipitation or DNA contamination. Thus, in osteoblasts, miR-23a and -27a directly associate with SATB2 mRNA to regulate its expression. To address direct miR regulation of SATB2, we used the miRZIP system to simultaneously knockdown all three miRs in the cluster (Fig. 4B). The anti-miRs increased SATB2 and Runx2 protein levels (Fig. 4C) and increased osteoblast differentiation markers two- to fourfold after 72 h (Fig. 4D) without affecting SATB2 mRNA levels (Fig. 4E). Thus, the inhibitory effect of the miR-23a∼27a∼24-2 cluster on osteogenesis (Fig. 3C) is contributed by the activity of all three miRs mediating translational control of SATB2 mRNA.
Fig. 4.
Feed-forward control by Runx2 and feedback regulation by miR-23a maintains the physiology of bone formation. (A) In vivo identification of miR-23a, -27a, and target SATB2 by RNP-IP. The miR-23a and 27a-SATB2 mRNA that is complexed with the AGO2 (Materials and Methods) was analyzed for the presence of SATB2 mRNA association. The 3′ UTR of SATB2 mRNA was amplified with primers specific to the binding sites (Table S1). The miR-23a and -27a seed lanes shows SATB2 3′ UTR fragment with the miR-23a and -27a binding. No RT, normal IgG and 2% input were used as negative and positive controls, respectively. (B) Anti-miR (miRZIP) construct and knockdown of miR-23a, -27a, and -24-2. H1 promoter drives the synthesis of all three miRs. (C) miRZIP specific for miR-23a, -27a, and -24-2 knockdown increases SATB2 and Runx2 protein shown by Western blot with antibodies described in Materials and Methods and actin protein as loading control. (D) Biological effect of miRZIP expression increases expression of osteoblast differentiation markers normalized to GAPDH (real-time qPCR). (E) miRZIP does not affect SATB2 mRNA. (F) Coordinated expression of Runx2 and SATB2 in primary ROB cells at indicated days of differentiation by Western blot. Actin was used as loading control. (G) Illustrated is the reciprocal relationship of SATB2 and Runx2 protein with miR-23a∼27a∼24-2 expression (qPCR average of Fig. 1F) in ROB cell differentiation. In the same experiment, Runx2 and SATB2 protein (F) were densitometrically quantified and plotted. (H) Summary of the central role of miR cluster regulation of osteoblastogenesis. Left: Model demonstrates that Runx2 controls the SATB2 functions by down-regulating miR cluster expression in a feed-forward mechanism at day 12, whereas miR-23a down-regulation of Runx2 at day 20 in a feedback loop attenuates Runx2 and target genes for terminal osteoblast differentiation.
The biological linkage among the miR cluster, Runx2, and SATB2 is further appreciated by comparing the protein levels of Runx2 and SATB2 throughout a time course of osteoblast differentiation (Fig. 4F). The Western blot reveals a similar profile of peak protein levels after the proliferation period (day 8) and continuing through the matrix mineralization stage (day 16, ALP-positive cells). In the mature cultures (representing the osteocyte end stage), both proteins are down-regulated as expression of the miR cluster increases (Fig. 4 F and G show graphic quantification). This attenuation of the proteins (days 18–20) and switch in reciprocal expression of the miR cluster continues to day 35 (Fig. 1F) and is consistent with negative regulation of SATB2 by all miRs in the cluster (Fig. 3D) and of Runx2 by miR-23a (Figs. 1G and 3E). Consistent with these findings, SATB2 and Runx2 were previously shown to form a coregulatory complex that promotes bone formation in vivo (25). Thus, the miR cluster has a central role in regulation of osteogenesis (Fig. 4H) that begins in undifferentiated cells to suppress osteoblast differentiation (Fig. 2F, Runx2 null cells, and Fig. 3B), then must be down-regulated by Runx2 at the onset of the differentiated osteoblast phenotype (day 12) to increase SATB2 to work in concert with Runx2 to promote further maturation.
Discussion
To date, Runx2 remains the earliest of the transcriptional regulators critical for bone formation. Here, we have uncovered a pathway regulating progression of the osteoblast phenotype through activity of the miR-23a∼27a∼24-2 cluster that is controlled by the bone-specific Runx2 transcription factor. Our studies show that (i) miR-23a, -27a, and -24-2 belong to a cluster whose promoter is directly and negatively regulated by Runx2; (ii) the miR cluster functionally inhibits osteogenesis and therefore requires suppression to promote differentiation; (iii) the mechanism of inhibition is that each miR member of this cluster down-regulates SATB2, also a critical regulator of osteoblast differentiation, through direct binding to its 3′ UTR; and (iv) one member of the cluster, miR-23a, reaches peak levels in mature osteoblasts and directly targets Runx2 to down-regulate its expression and facilitate maximal miR expression at terminal stages of osteoblast differentiation. This regulatory network results in attenuation of osteoblast-like activity in osteocytes in a mineralized matrix. We propose that cross-regulation between Runx2 and the miR cluster results in the activation of SATB2 (i.e., feed-forward mechanism), whereas the attenuation of Runx2 by miR-23a (i.e. feedback mechanism) fine tunes the pace of progression of the osteoblast phenotype. Our studies have identified the central role of this cluster in physiologic regulation of osteoblast maturation and maintenance of terminally differentiated bone cells.
Our results show that miRs in the cluster inhibit or delay maturation to osteocytes in a mineralized matrix. Thus, there is a requirement for negatively regulating expression of all miRs in the cluster for differentiation of osteoprogenitors to osteoblasts. The ChIP studies demonstrate direct down-regulation of the miR promoter by Runx2 through modification of histones. Significantly, a biological mechanism coupled to down-regulation of miRs is the identification of SATB2 as a direct target of all three miRs using in vitro reporter assays and demonstrating in vivo binding of miRs to SATB2 mRNA. SATB2 is a member of the family of special AT-rich binding transcription factors that interacts with nuclear matrix attachment regions and activates transcription (25). Null mouse models and human mutations of SATB2 established that the protein is involved in craniofacial development and osteoblast differentiation (25–27). SATB2 physically interacts with Runx2 and also ATF4, a transcription factor known to promote the mineralization stage of bone formation (25, 27). Thus SATB2 has multiple inputs into transcriptional control during bone formation. Therefore, the posttranscriptional regulation of SATB2 by an miRNA cluster whose expression is controlled by Runx2 brings together an integrated network of pathways that coordinate the temporal events of bone formation.
Mechanisms involving miR control of osteoblastogenesis have significant in vivo relevance for control of bone formation, as revealed by two key findings. The present studies identified a mechanism for attenuation of Runx2 protein in osteocytes in the mineralized cell layers through direct miR-23a targeting of the 3′ UTR. This mechanism is consistent with previous findings that demonstrated excessive Runx2 can cause osteopenia in mice (28), and emphasizes the importance of maintaining appropriate cellular Runx2 levels at stages of maturation. Second, deletion of the Dicer enzyme in mature osteoblasts, which results in the absence of functional miRs, stimulated bone formation and significantly increased bone mass (12). A number of miRNAs are increased during the late stages of osteoblast differentiation, indicating the importance of miR regulation for attenuating continued bone formation at the final osteocyte stage of differentiation (5, 12). Hence, the finding of feed-forward/Runx2 and feedback/miR-23a regulatory loops during osteoblast differentiation suggests a functional role for the miR cluster in maintaining a balance in the activation and repression of osteogenic/osteoblast genes during the temporal progression of the osteoblast phenotype. We postulate that this regulatory loop supports the ordered temporal progression of the osteogenic program and the stability of the terminal osteocyte phenotype. In this regard, two recent reports showed that miR-27a and -27b both down-regulated PPARγ and blocked adipogenesis (29, 30). Thus, in addition to our findings of direct involvement of miR-27 in promoting osteoblast differentiation, the targeting of PPARγ by miR-27 provides a second biological function to retain osteoblast identity during differentiation.
In summary, posttranscriptional control by miRNAs fulfills many functions related to biological processes. This study, which has identified the relationships among an miR cluster—a tissue-specific transcription factor essential for commitment and differentiation of osteoblasts—and SATB2—a key regulator of skeletal development—provides a compelling example of the very significant layer of posttranscriptional regulation controlled by miRNAs in the process of tissue formation.
Materials and Methods
Cell Culture.
Primary fetal ROBs were isolated by sequential collagenase digestion as previously described (31). These cells and murine preosteoblast cell line MC3T3-E1 clone 4 from American Type Culture Collection were maintained and differentiated for 28 d (31). Cells were harvested at indicated times for protein (total or nuclear) and mRNA or fixed in 2% paraformaldehyde for histochemical detection of ALP and mineral deposition (12).
miRNA Lentivirus Clones and Transfection.
miRNA Lenti-miR vectors (System Biosciences) were used to produce mature miRNA-23a and -27a individually or together from CMV promoter or anti-miR cluster (miRZIP, System Biosciences) for anti-miR-23a, -27, and -24-2 from H1 promoter with GFP reporter to monitor miRNA expressing cells. Virus was packaged in HEK293T (ATCC) cells (32). ROB or MC3T3-E1 cells were infected at 60% to 70% confluence for 48 h, trypsinized, and seeded at 1 × 104 cells/cm2 to assay osteogenic functions of each miR following differentiation for 28 d. MC3T3-E1 cells at 30% to 50% confluence were also transfected with miR-23a, -27a and -24-2 premiR, anti-miRNA, or NS miRNAs (Applied Biosystems/Ambion) at 50 or 100 nM using Oligofectamine (Invitrogen) following the manufacturer's instructions and harvested after 48 h for protein and mRNA analysis.
DNA Constructs and siRNA.
For functional analysis, SATB2 3′ UTR (approximately 100–150 bp) spanning the miR-23a, -27a, or -24-2 binding sites from mouse genomic DNA were cloned into the pMIR-REPORT miRNA Expression Reporter Vector (Applied Biosystems/Ambion). The miR-23a∼27a∼24-2 promoter deletion constructs (−639 LUC to −110 LUC) were the generous gift from V. Narry Kim (National University, Seoul, Korea). CMV-driven SATB2 clone was a kind gift from Rudolf Grosschedl (Max-Planck Institute of Immunobiology, Freiburg, Germany). Runx2 knockdown siRNAs were purchased from Qiagen (Table S1).
Luciferase Reporter Assay.
MC3T3-E1 cells were cotransfected with three luciferase constructs containing miR-23a, -27a, or -24-2 binding sites (SATB2-LUC-UTR and Runx2-LUC-UTR WT or mutated; Fig. S4 provides sequences) along with phRL-null (Renilla plasmid for normalization) and 100 nm of NS miR, pre-, or anti-miR of 23a, 24-2, or 27a. Luciferase assays were performed according to Promega Dual Luciferase Assay System. MiR-23a∼27a∼24-2 promoter LUC assays were performed in similar manner by using 500 ng of promoter deletion mutants cotransfected with 250 ng of control or Runx2 overexpression construct. Three independent experiments were performed for 24 h and assayed in triplicate per group. Data represent the SE for three experiments and triplicate samples. Relative luciferase activity (firefly/Renilla) was expressed in relative luminescence units and plotted.
Northern Blot and Western Analyses.
Northern blot analysis of miRNAs was done as described (4). The oligonucleotide probe sequences (complementary to mature miR) are listed in Table S1. Whole-cell lysate or nuclear extract was subjected to Western blot analysis as detailed elsewhere (31).
Electrophoretic Mobility Shift Assay (EMSA).
Detailed EMSA procedures have been described elsewhere (31). Complexes were visualized by autoradiography of a 6.5% acrylamide gel.
ChIP Assays.
The procedure for ChIP in osteoblast cells has been described (31). Primers used to analyze the bound DNA fragments are listed in Table S1. NS antibody was used as a control.
Antibodies.
The following antibodies were used for Western blot, EMSA, and ChIP studies. RUNX2 (SC-10758) and Actin (SC-1616) were purchased from Santa Cruz Biotechnology. Anti-hyperacetylated histone H4 (06–946; Penta) was purchased from Millipore. Histone H3 (tri-methyl K9) antibody ChIP-grade (ab8898) and SATB2 antibody (ab34735) were purchased from Abcam. Runx2 mouse monoclonal antibody (clone 6B4) was obtained from MBL.
RNP-IP.
Polysomal extracts from MC3T3-E1 cells were immunoprecipitated with affinity-purified silencing complex-specific argonaute antibody Ago2 (N-13; sc-32659; Santa Cruz Biotechnology) as previously described (33). The RNA isolated from RNP-IP were subjected to cDNA synthesis using primers derived from specific seed sequence for miR-23a, -27a, and miR-9 (NS). The first strand cDNA was further amplified with forward and reverse primers derived from the immediate upstream sequence of the 3′ UTR target of the SATB2 mRNA (Table S1). The detailed methodology has been described (33).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants AR039588 (to G.S.S.) and R37 DE012528 (to J.B.L).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007698107/-/DCSupplemental.
References
- 1.Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36:25–32. doi: 10.1002/biof.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soltanoff CS, Yang S, Chen W, Li YP. Signaling networks that control the lineage commitment and differentiation of bone cells. Crit Rev Eukaryot Gene Expr. 2009;19:1–46. doi: 10.1615/critreveukargeneexpr.v19.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaidi SK, et al. Mitotic bookmarking of genes: A novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 11.Li H, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur T, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340:10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luzi E, et al. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 14.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem. 2009;284:19272–19279. doi: 10.1074/jbc.M109.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oskowitz AZ, et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972–978. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 19.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci USA. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi SK, et al. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLOS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 25.Dobreva G, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Leoyklang P, et al. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat. 2007;28:732–738. doi: 10.1002/humu.20515. [DOI] [PubMed] [Google Scholar]
- 27.Britanova O, et al. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet. 2006;79:668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geoffroy V, Kneissel M, Fournier B, Boyde A, Matthias P. High bone resorption in adult aging transgenic mice overexpressing cbfa1/runx2 in cells of the osteoblastic lineage. Mol Cell Biol. 2002;22:6222–6233. doi: 10.1128/MCB.22.17.6222-6233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karbiener M, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392:323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Hassan MQ, et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 32.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 33.Hassan MQ, et al. Ribonucleoprotein immunoprecipitation (RNP-IP): A direct in vivo analysis of microRNA-targets. J Cell Biochem. 2010;110:817–822. doi: 10.1002/jcb.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.