Protein phosphatase 1 (PP1) accumulates within the nucleolus, which plays a central role in regulation of cell growth and proliferation. Using a powerful combination of imaging and quantitative proteomics, we identify RRP1B as a nucleolar PP1 targeting subunit that sits at the hub of the pre-60S ribosomal subunit processing complex.
Abstract
A pool of protein phosphatase 1 (PP1) accumulates within nucleoli and accounts for a large fraction of the serine/threonine protein phosphatase activity in this subnuclear structure. Using a combination of fluorescence imaging with quantitative proteomics, we mapped the subnuclear localization of the three mammalian PP1 isoforms stably expressed as GFP-fusions in live cells and identified RRP1B as a novel nucleolar targeting subunit that shows a specificity for PP1β and PP1γ. RRP1B, one of two mammalian orthologues of the yeast Rrp1p protein, shows an RNAse-dependent localization to the granular component of the nucleolus and distributes in a similar manner throughout the cell cycle to proteins involved in later steps of rRNA processing. Quantitative proteomic analysis of complexes containing both RRP1B and PP1γ revealed enrichment of an overlapping subset of large (60S) ribosomal subunit proteins and pre-60S nonribosomal proteins involved in mid-late processing. Targeting of PP1 to this complex by RRP1B in mammalian cells is likely to contribute to modulation of ribosome biogenesis by mechanisms involving reversible phosphorylation events, thus playing a role in the rapid transduction of cellular signals that call for regulation of ribosome production in response to cellular stress and/or changes in growth conditions.
INTRODUCTION
The primary role of the nucleolus, a nonmembrane-bound organelle that forms around tandem repeats of rDNA in the nucleus, is to ensure that the cell receives the essential supply of ribosomes required for protein synthesis (for review see Boisvert et al., 2007). Because it must respond to dynamic changes in cell growth rate and metabolic activity with either an increase or decrease in ribosome subunit biogenesis, tight control of nucleolar pathways of rDNA transcription and ribosome subunit processing and export is critical. Several key cellular kinases and phosphatases have been linked to regulation of nucleolar events throughout the cell cycle, and our work has shown that the serine/threonine protein phosphatase 1 (PP1) accounts for ∼80% of Ser/Thr phosphatase activity within this structure (Trinkle-Mulcahy et al., 2003).
The intracellular localization and substrate specificity of the core catalytic subunit of PP1 is regulated through its association with a spectrum of interacting proteins, termed targeting subunits (for review see Cohen, 2002). Most of these targeting subunits contain conserved motifs that mediate direct binding to PP1, including the “RVxF” motif with its consensus Arg/Lys-Val/Ile-Xaa-Phe/Trp (Egloff et al., 1997; Wakula et al., 2003). Over the years biochemical, bioinformatic, and proteomic approaches have been used to identify and characterize a wide range of PP1 targeting subunits (Tran et al., 2004; Meiselbach et al., 2006; Trinkle-Mulcahy et al., 2006; Roadcap et al., 2007; Moorhead et al., 2008; Hendrickx et al., 2009), however the current list still cannot account for the large number of regulatory pathways in which PP1 is known to play a critical role.
Of the three closely-related mammalian isoforms, PP1α, PP1β, and PP1γ (Barker et al., 1993; Shima et al., 1993; Barker et al., 1994), only the β and γ isoforms show significant accumulations within nucleoli (Trinkle-Mulcahy et al., 2001; Trinkle-Mulcahy et al., 2003; Trinkle-Mulcahy et al., 2006; Lesage et al., 2005; Andreassen et al., 1998). This difference has been attributed to a specific N-terminal Arginine residue (Arg19 in PP1β and Arg20 in PP1γ) that is not present in PP1α (Lesage et al., 2005). Having validated the use of FP-PP1 fusion proteins as markers for endogenous pools of PP1 (Trinkle-Mulcahy et al., 2001), we went on to demonstrate that isoform-specific binding partners detected via affinity purification of the respective tagged proteins reflect the isoform-specific localization patterns observed by fluorescence imaging (Trinkle-Mulcahy et al., 2006). Although our initial comparison of nuclear PP1 complexes focused on the α and γ isoforms, we have now established a cell line stably expressing low levels of GFP-tagged PP1β. In addition, we have further optimized a SILAC (stable isotope labeling of amino acids in cell culture)-based quantitative immunoprecipitation approach for both increased sensitivity and reliability (Trinkle-Mulcahy et al., 2008) and developed an efficient method for extracting multiprotein complexes from purified nucleoli for interactome analyses (Chamousset et al., 2010). As presented here, nucleolar interactome screens of GFP-PP1 define a wide range of multiprotein complexes to which the phosphatase is targeted, including complexes involved in ribosome subunit biogenesis.
The pathway of ribosome subunit biogenesis is initiated in the nucleolus by RNA Pol I–mediated transcription of pre-rRNA (Russell and Zomerdijk, 2005). This transcript is further processed into 18S transcripts (which form the 40S subunit) and 5.8S and 28S transcripts (which combine with RNA Pol III-transcribed 5S rRNA to form the 60S subunit). The ribosomal proteins assemble on these complexes (RPLs with 60S, RPSs with 40S), where they are joined by a large number of nonribosomal processing proteins. In yeast, >300 proteins, many of which show transient associations, have been characterized or predicted to play roles in the maturation and export of pre-ribosomal particles (Nissan et al., 2002). Affinity purification-based analysis of mutant strains that lead to accumulations of pre-rRNAs characteristic of early, intermediate, or late steps, in combination with protein–protein interactions deposited from 32 individual publications in the BioGrid database, was recently used to build up a network of physical interactions consistent with previous work (Lebreton et al., 2008).
Importantly, structure and function are intrinsically linked in the nucleolus, and thus localization to a particular compartment can provide clues to protein function. The transcription machinery is found in the fibrillar center (FC) and rRNA transcription occurs at the border of this region with the dense fibrillar component (DFC), while additional processing occurs within the granular component (GC). A further remarkable feature of the mammalian cell nucleolus is its coordinated mitotic disassembly and post-mitotic reassembly (Dundr et al., 2000; Leung et al., 2004). Disassembly at prophase is initiated by inhibition of transcription, and reassembly in early G1 is driven by resumption of transcriptional activity. In most cases proteins with related roles remain associated throughout this process.
Using biochemical, fluorescence imaging, and quantitative proteomic approaches, we define here a specific pool of PP1 in the granular component of mammalian cell nucleoli that is associated with pre-60S ribosomal subunit processing complexes.
MATERIALS AND METHODS
Plasmids and Antibodies
All FP-PP1 constructs were described previously (Trinkle-Mulcahy et al., 2001; Trinkle-Mulcahy et al., 2003). PP1β-EGFP was prepared by subcloning the PP1β cDNA into the EGFP-N3 vector. The EYFP-NLS plasmid (nuclear localization signal cloned into EYFP-C1), which accumulates in nucleoli in addition to its cytoplasmic and nuclear localization, was a generous gift from Dr. Archa Fox (Western Australia Institute for Medical Research, Australia). RRP1B was cloned from an expressed sequence tag using oligonucleotide primers and inserted into EGFP/mCherry/pET vectors. The Val and Phe residues of the putative PP1 binding motif (residues 684 and 686) were changed to Ala using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Nol1, RPL5, and RPS23 were cloned and inserted into the EGFP-C1 vector, while EGFP/EYFP-tagged RPA39, Fibrillarin, PWP1, Gar1, B23, Pescadillo, and RPL27 were obtained as previously described (Leung et al., 2004). YFP-RRN3 and GFP-NNP-1/Nop52 were generous gifts from Drs. Joost Zomerdijk (University of Dundee, UK) and Daniela Hernandez-Verdun (Institut Jacques Monod, France). Recombinant 6His-tagged RRP1B was expressed in bacteria, purified using Ni2+-NTA beads, and injected into rabbits for the generation of the polyclonal antibodies used in this study.
Far Western Blotting
Recombinant purified wild-type and KATA mutant RRP1B (200 ng each) were electrophoresed and transferred to nitrocellulose membrane, along with control lanes containing 10 μg of total protein from rat nuclear extracts. The membranes were incubated with 10% milk to block nonspecific binding sites and then overlaid with either DIG-labeled recombinant PP1α or PP1γ. Anti-DIG-HRP antibodies (Pierce; Rockford, IL) were used to detect the binding profiles of the PP1 isoforms.
Cell Culture and Transfection Assays
HeLa and U2OS cells were obtained from ATCC (Manassas, VA). Cells were grown in Dulbecco's modified Eagles' medium supplemented with 10% fetal calf serum and 100 U/ml penicillin and streptomycin (Wisent). For immunofluorescence assays, cells were grown on coverslips and transfected (if required) using Effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturers' instructions. For live cell imaging, cells were cultured in glass-bottomed dishes (WILLCO; Ted Pella, Redding, CA). For fixed cell imaging, cells were washed in PBS, fixed for 5 min in 3.7% (wt/vol) paraformaldehyde in CSK buffer (10 mM PIPES pH 6.8, 10 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 2 mM EDTA) at room temperature, and permeabilized with 1% Triton X-100 in PBS for 10 min.
Stable Cell Lines
HeLaEGFP-PP1α and HeLaEGFP-PP1γ stable cell lines were described previously (Trinkle-Mulcahy et al., 2001; Trinkle-Mulcahy et al., 2006). The U2OSEGFP-PP1α, U2OSEGFP-PP1γ, U2OSPP1β-EGFP, U2OSEGFP-RRP1B, and U2OSEGFP-RPL27 stable cell lines were established and validated in a similar manner.
Live Cell Imaging
Time-lapse imaging and FRAP experiments were carried out as described previously (Trinkle-Mulcahy et al., 2006; Trinkle-Mulcahy et al., 2007), using a wide-field fluorescence microscope (DeltaVision CORE; Applied Precision, Issaquah, WA) equipped with a three-dimensional motorized stage, temperature- and gas-controlled environmental chamber, and 488-nm diode laser (for photobleaching EGFP). Images were collected using a 60× NA 1.4 Plan-Apochromat objective and recorded with a CoolSNAP coupled-charge device (CCD) camera (Roper Scientific, Trenton, NJ). DIC imaging was obtained with the approporiate prism insert (Olympus, Center Valley, PA). The microscope was controlled by SoftWorX acquisition and deconvolution software (Applied Precision). DNA was stained by incubating the cells for 30 min in medium containing 0.25 μg/ml Hoechst No 33342 (Sigma-Aldrich, St. Louis, MO). For FRAP experiments, a single section was imaged before photobleaching, a region of interest was then bleached to ∼50% of its original intensity using the 488-nm laser, and a rapid series of images was acquired after the photobleaching period. Recovery curves were plotted and the mobile fraction and half time of recovery were determined using SoftWorX and Excel (Microsoft, Redmond, WA).
RNAse and DNAse Treatments
Cells were grown on glass coverslips, rinsed once with PBS and once with ASE buffer (20 mM Tris pH 7.5, 5 mM MgCl2, 0.5 mM EGTA), and permeabilized by incubating for 5 min at room temperature in ASE buffer plus 0.1% Triton X-100. Cells were then treated for 20 min at room temperature with either PBS (mock treatment), RNAse (100 μg/ml; Worthington, Lakewood, NJ), or DNAse (1000 U/ml; Worthington). After treatment the cells were rinsed with PBS and fixed with 3.7% PFA in CSK buffer for 5 min. After 10 min permeabilization with 1% Triton X-100 in PBS, cells were stained sequentially with the rabbit polyclonal anti-RRP1B primary antibody, DyLight488 anti-rabbit secondary antibody (Pierce), Hoechst 33342 (2.5 μg/ml for 2 min), and Pyronin Y (33 mM for 5 s). Coverslips were mounted on glass slides using FluorSave mounting media (Merck, Whitehouse Station, NJ).
Preparation of Whole Cell Lysates and Immunoblotting
Cells were washed twice with ice-cold PBS and lysed in 0.5 ml of ice-cold 50 mM Tris-HCl pH 7.5; 0.5 M NaCl; 1% (vol/vol) Nonidet P-40; 1% (wt/vol) sodium deoxycholate; 0.1% (wt/vol) SDS; 2 mM EDTA, and protease inhibitor cocktail (Roche, Indianapolis, IN). The lysate was passed through a Qiashredder column (Qiagen) to shear DNA and cleared by centrifugation at 14,000 g for 10 min at 4°C. Lysates were separated on 4–12% Novex Nu-PAGE bis-Tris polyacrylamine gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes for immunoblotting. Primary antibodies used were anti-GFP mouse monoclonal (Roche), isoform-specific goat anti-PP1 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Nol1 (Proteintech Europe, Manchester, UK), anti-B23 (Sigma-Aldrich), anti-RPL7 (Abcam, Cambridge, MA), anti–α-tubulin (Sigma), anti-Lamin A/C (Santa Cruz Biotechnology), and anti-A190 (generous gift from Dr. Joost Zomerdijk).
Cellular Fractionation
Preparation of cytoplasmic, nucleoplasmic, and nucleolar extracts was carried out as previously described (Chamousset et al., 2010). The final buffer for each extract was RIPA (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and protease inhibitors), and all extracts were cleared by centrifuging at 2800 g for 10 min at 4°C. Nuclear extracts were prepared from purified nuclei as previously described (Trinkle-Mulcahy et al., 2006). Total protein concentrations were measured using a Qubit Fluorometer (Invitrogen).
Quantitative Immunoaffinity Purification
For the quantitative SILAC-based proteomic experiments, cells were encoded with the required isotopic amino acids as described previously (Trinkle-Mulcahy et al., 2008). Double encoding experiments (control IP vs. IP of tagged or endogenous protein) used L-arginine 13C and L-lysine 4,4,5,5-D4 (Cambridge Isotope Laboratories, Andover, MA) as the “heavy” isotopes. Triple encoding experiments designated these isotopes as “medium” and added a “heavy” condition that used L-arginine 13C/15N and L-lysine 13C/15N. All control or “light” media contained L-arginine and L-lysine (Sigma-Aldrich). GFP-tagged proteins were affinity purified by incubation for 1 h at 4°C with the GFP-Trap-A reagent (Chromotek, Martinsried, Germany). For affinity purification of endogenous RRP1B, the affinity purified rabbit polyclonal antibody was covalently conjugated to protein G Dynabeads (Invitrogen) at a concentration of 1 mg/ml. Covalently-conjugated affinity purified rabbit IgG was used for the control IP. Control and experimental IPs were carried out separately by incubating the beads with equivalent total protein amounts of cellular, nuclear or nucleolar extracts. After a 1 h incubation at 4°C, the extracts were removed and the beads washed once with ice-cold RIPA buffer. The control and experimental beads were then carefully combined and washed an additional three times with ice-cold RIPA buffer. Proteins were eluted, separated by 1D SDS-PAGE, and trypsin-digested for MS analysis as described previously (Trinkle-Mulcahy et al., 2008).
Mass Spectrometry and Data Analysis
An aliquot of the tryptic digest (prepared in 5% acetonitrile/0.1% trifluoroacetic acid in water) was analyzed by LC-MS on an LTQ-Orbitrap mass spectrometer system (ThermoElectron, Rockford, IL) as described previously (Trinkle-Mulcahy et al., 2008). Database searching and quantitation were performed using the Mascot search engine v.2.2 (Matrix Science, Boston, MA) and MaxQuant software (Cox and Mann 2008; http://www.maxquant.org).
siRNA Knockdown of RRP1B
HeLa and U2OS cells were transfected in 24-well plates with 1 μg of small interfering RNA (siRNA) duplexes (ON-TARGET plus SMART pool #L-031402-01; Dharmacon, Lafayette, CO) using DharmaFECT transfection reagent (Dharmacon). The control/scrambled siRNA duplex was also obtained from Dharmacon (5′DY647-CAG UCG CGU UUG CGA CUG G dTdT 3′) and incorporated a Cy5 tag to enable us to assess transfection efficiency. Cells were either harvested or passaged 24 h after transfection.
Sucrose Gradient Fractionation
Linear sucrose gradients (15–45% wt/wt in 25 mM Tris-HCl pH 7.5, 25 mM NaCl, 5 mM MgCl2) were prepared in ultracentrifuge tubes using a gradient maker (Hoefer Scientific Instruments, Holliston, MA) and stored at 4°C before use. Two confluent 10-cm dishes of cells were sufficient for preparation of cytoplasmic extracts, while nuclear extracts required three confluent 15-cm dishes of cells as a starting point to ensure that sufficient material was obtained. For siRNA experiments, cells were transfected in multiple 24-well plates and passaged for preparation of extracts 48 h afterward. To prepare cytoplasmic extracts, cells were washed with ice-cold PBS and harvested in low-salt lysis buffer containing 20 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 1 mM RNAsin, 1 mM DTT, 0.3% Triton X-100, and 0.05 M sucrose. Nuclei and the majority of mitochondria were sedimented by centrifugation at 10,000g at 4°C for 10 min. NaCl and MgCl2 concentrations in the supernatant (cytoplasmic extract) were adjusted to 170 mM and 13 mM, respectively. An aliquot was retained as the input sample, and the remaining extract layered onto the sucrose gradient. Preparation of nuclear extracts used a variation of our standard nuclear purification technique (Trinkle-Mulcahy et al., 2008), with the final nuclear pellet resuspended in 0.5 ml of sonication buffer (25 mM Tris pH 7.5, 100 mM KCl, 2 mM EDTA, 0.05% NP-40), transferred to a 1.5 ml microcentrifuge tube, and sonicated on ice using a microtip and 10 × 1 s pulses (with 15 s pauses in between). The resulting extract was centrifuged at 14,000g for 15 min at 4°C and the supernatant transferred to a new tube. An aliquot was retained as the input sample and the remaining extract layered onto the sucrose gradient.
Extracts were centrifuged at 36,000 rpm for 2 h at 4°C in an ultracentrifuge with an SW40 swing-out rotor (Beckman Coulter, Brea, CA). A typical profile of UV absorbance was obtained upon fractionation of the extracts of a sucrose gradient, with continuous monitoring at 254 nm using a UA-6 UV detector (ISCO, Lincoln, NE) and UNICORN 5.01 software (GE Healthcare). One-milliliter fractions were collected, with 0.9 ml of each used for protein extraction by TCA precipitation, and the remaining 0.1 ml for RNA extraction using Trizol (Invitrogen).
Reverse Transcription and 28S PCR
cDNA for each RNA sample was synthesized using the AMV Reverse Transcriptase kit (Promega, Madison, WI), as per the manufacturer's instructions. PCR was carried out using GoTaq Flexi DNA polymerase kit (Promega) and a MasterCycler ProPCR machine (Eppendorf, Hamburg, Germany). The following primers were used to amplify 28S rRNA: GTTCACCCACTAATAGGGAACG for 28S rRNA FWD and GGATTCTGACTTAGAGGGCGTT for 28S rRNA REV (Granneman and Baserga, 2004). For each sample, 10 μl of PCR product was separated on a 1% agarose gel and stained with SYBR Safe DNA gel stain (Invitrogen). Gels were imaged using a Fuji LAS 4000 Mini Chemiluminescent Imager.
In Vivo Transcription Assays
U2OS cells were grown on coverslips, incubated at 37°C for 20–40 min with 1 mM 5-fluorouridine (Sigma) and fixed for 5 min with PFA-CSK as described above. After permeabilization with 0.5% Triton X100 for 10 min, the incorporated FU was detected using anti-BrdU (Sigma) primary and DyLight 549 anti-mouse (Thermo Scientific) secondary antibodies. All coverslips were stained with Hoechst 33342 and mounted in FluorSave mounting media (Calbiochem) for imaging.
Northern Blots
Total cellular RNA was extracted from Hela cells using the RNeasy Mini Kit (Qiagen). RNA was separated for 3 h at 110 Volts (NorthernMax Kit, Ambion), transferred to BrightStar-Plus Positively Charged Nylon Membrane (Ambion, Austin, TX) for 2 h using a TurboBlotter (Whatman), and biotinylated probes detected by streptavidin reagent (BrightStar BioDetect Kit, Ambion). BrightStar Biotinylated RNA Millennium Markers (Ambion) were included on each blot. The sequences of the 5′ biotinylated probes (Dharmacon) used to detect pre-rRNA species are as follows: human pre-rRNA probe hITS1 (1076-1091) 5′biotin-AGGTCGATTTGGCGAG and human pre-rRNA probe hITS1 (869-884) 5′biotin-GACACCACCCCACAGG.
RESULTS
RRP1B Is a Nucleolar PP1 Targeting Subunit
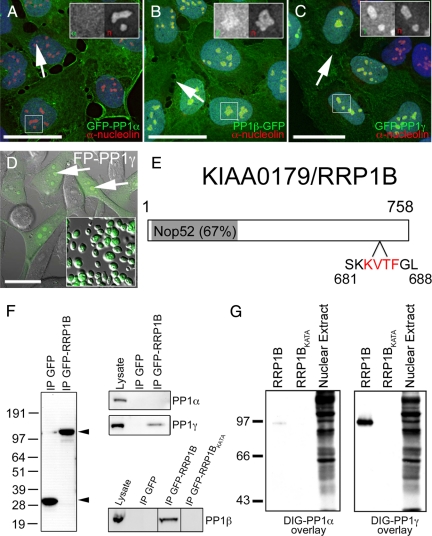
As our previous work indicated the importance of PP1 in nucleolar processes (Trinkle-Mulcahy et al., 2003), we set out to define the nucleolar PP1 interactome using a quantitative proteomics-based approach. HeLa and U2OS cell lines stably expressing the three FP-tagged PP1 isoforms at low levels were established and characterized (Trinkle-Mulcahy et al., 2001; Trinkle-Mulcahy et al., 2003; Andreassen et al., 1998). Fluorescence imaging of both fixed and live cells confirmed that the fusion proteins maintain the distinct subcellular localization patterns of their endogenous counterparts, including the accumulation of PP1β-GFP and GFP-PP1γ in nucleoli (Figure 1, B and C). PP1α distributes equally throughout the nucleoplasm and nucleoli (Figure 1A). Nucleoli are readily purified from these cell lines in large quantities and with high purity, as shown here for HeLaEGFP-PP1γ (Figure 1D, inset), and the nucleolar pool of GFP-PP1 is retained throughout the purification protocol.
Figure 1.
Identification of RRP1B as a nucleolar targeting subunit that specifically binds PP1β and PP1γ. (A) When stably expressed at low levels in U2OS cells as a GFP fusion protein, PP1α (green) is found distributed throughout the cell. (B) Stably expressed GFP-tagged PP1β (green) is also found throughout the cell but shows an additional accumulation in nucleoli (arrows), as does stably expressed GFP-PP1γ (C, green). DNA stained with Hoechst 33342 is shown in blue and anti-nucleolin, a nucleolar marker protein, in red. For each panel there is an inset showing an expanded version of the boxed region with the GFP-PP1 signal on the left and the anti-nucleolin signal on the right. (D) GFP-PP1γ (green), shown here superimposed on a differential interference contrast (DIC) image in HeLaGFP-PP1γ cells, accumulates in nucleoli (arrows) and retains its nucleolar association in purified nucleoli (inset). Scale bars are 15 μm. (E) RRP1B is a 758-aa protein that contains a Nop52 domain (67% identical to yeast NOP52) and a putative PP1 binding RVxF motif (aa 683-686). (F) GFP-Trap pulls down similar amounts of free GFP and GFP-RRP1B from U2OSGFP and U2OSGFP-RRP1B cells, respectively. Endogenous PP1γ, but not PP1α, copurifies specifically with GFP-RRP1B. Endogenous PP1β also copurifies with GFP-RRP1B, but not with the GFP-RRP1BKATA mutant in which the PP1 binding domain has been disrupted. (G) Far Western blot demonstrating that RRP1B preferentially binds PP1γ over PP1α and that the KATA mutation greatly weaken its ability to bind PP1. Equal amounts of purified recombinant DIG-labeled PP1α and PP1γ were overlayed on 200 ng of purified recombinant wild type and KATA mutant RRP1B. The control lane (10 μg rat liver nuclear extract) illustrates that DIG-PP1α binds other target proteins with high affinity.
We initially immunoprecipitated GFP-PP1γ and nucleolar-targeted YFP (as a negative control) from nucleolar extracts derived from HeLaEGFP-PP1γ and HeLaEYFP-NLS cells (according to Trinkle-Mulcahy et al., 2008). We quantified all of the proteins identified and found KIAA0179/RRP1B to be the most abundant putative PP1 interaction partner (Figure 1E). After our optimization of nucleolar protein extraction (Chamousset et al., 2010), we repeated this experiment with similar results (Figure 6D) and also identified RRP1B as an interaction partner for nucleolar GFP-tagged PP1β (data not shown). In addition, RRP1B was identified as a PP1 interactor in HeLa nuclear extracts using a peptide displacement chromatography method (Moorhead et al., 2008).
Figure 6.
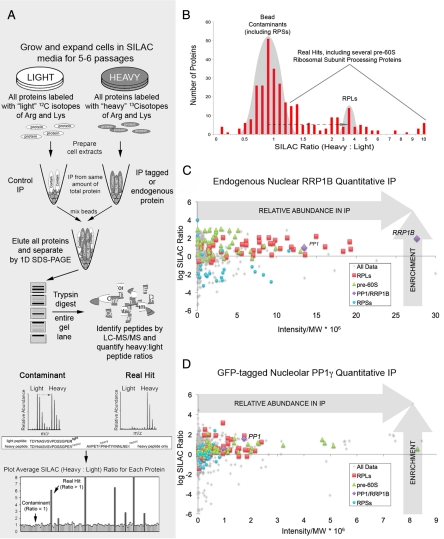
Quantitative immunoprecipitation of RRP1B selectively enriches molecular complexes containing 60S (RPL) proteins and pre-60S ribosomal subunit processing proteins. (A) Design of a typical quantitative SILAC IP experiment. (B) Real hits are distinguished by their high SILAC ratios (>1), which shift them from the bell curve distribution of contaminants (ratio of ∼1). Plotting log SILAC ratio versus relative peptide intensity highlights the RPLs and pre-60S processing proteins purified with both endogenous nuclear RRP1B (C) and GFP-tagged nucleolar PP1γ (D).
RRP1B contains a “Nucleolar Protein of 52 kDa” (NOP52) homology domain and the canonical “RVxF” motif (Figure 1E) found in most PP1 targeting subunits (Wakula et al., 2003). In immunoprecipitation experiments, endogenous PP1γ and PP1β, but not PP1α, copurified specifically with GFP-RRP1B (Figure 1F). This interaction was disrupted when the hydrophobic Val and Phe residues in the RVxF motif were mutated to Ala (RRP1B-KATA, Figure 1F). The specificity of binding to PP1γ over PP1α was also confirmed by far Western blot analysis. When overlaid on nuclear extracts, recombinant DIG-labeled PP1α and PP1γ both detect a large number of protein bands, however PP1γ interacts more strongly with recombinant RRP1B (Figure 1G). The interaction between RRP1B and PP1 is disrupted when the RVxF motif is mutated to KATA, confirming this is the binding site (Figure 1, F and G).
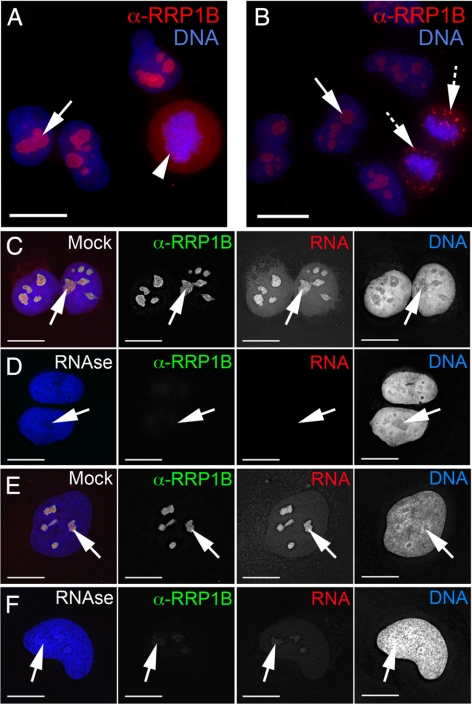
Antibodies raised against endogenous RRP1B (see Materials and Methods) recognize a single band at ∼84 kDa in cell lysates. Specificity of the antibody was confirmed by loss of this band on Western blots after siRNA-mediated knockdown of RRP1B, and by loss of the cell staining pattern under these same conditions (Supplemental Figure 4). Immunofluorescence microscopy analysis using our anti-RRP1B antibodies confirmed that the RRP1B protein is predominantly nucleolar (Figure 2, A and B, arrows). Additional accumulations were observed in the perichromatin region in metaphase cells (Figure 2A, arrowhead) and in cytoplasmic foci in late telophase cells (Figure 2B, hashed arrows).
Figure 2.
Localization of endogenous RRP1B is predominantly nucleolar and sensitive to RNAse treatment. (A) In interphase U2OS cells, endogenous RRP1B (red) is predominantly nucleolar (arrow). The metaphase cell in this field shows perichromatin accumulations of RRP1B (arrowhead). (B) The telophase cell in this field shows that RRP1B is found initially in bright foci that likely represent prenucleolar bodies (PNBs; hashed arrows) at the end of mitosis, and later in mature nucleoli (arrows). (C) Endogenous RRP1B stained with anti-RRP1B antibodies (green) in U2OS cells colocalizes with the nucleolar RNA signal (arrow) visualized by Pyronin Y staining (red). (D) Treatment of live cells with RNAse before fixation results in a near complete loss of both the RNA and anti-RRP1B, but not the DNA signal. (E) HeLa cells show a similar colocalization in nucleoli (arrows) of RRP1B (green) and Pyronin Y-stained RNA (red) and also demonstrate the sensitivity of RRP1B to RNAse treatment (F). Scale bars are 15 μm.
Nucleolar RRP1B Localization Is RNA-Dependent
The RRP1B antibody signal shows significant colocalization with the Pyronin Y–labeled RNA signal in nucleoli in both U2OS (Figure 2C) and HeLa (Figure 2E) cells. Treatment of cells with RNAse results in a near total loss of the RNA and nucleolar RRP1B antibody signals in both cell lines (Figure 2, D and F). Because the cells are lightly permeabilized to permit access of the RNAse, any RRP1B protein displaced by digestion of RNA would be lost. In contrast, neither mock treatment nor DNAse treatment affects the localization of endogenous RRP1B (Supplemental Figure 1).
GFP-RRP1B Shows a Similar Localization to the Endogenous Protein and Can Recruit Excess PP1 to the Nucleolus when Overexpressed
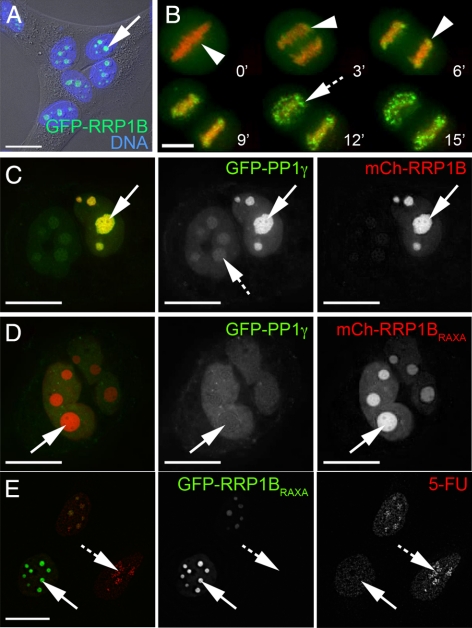
We established a U2OS cell line stably expressing GFP-RRP1B and compared localization of the fusion protein with that of the endogenous protein. Both are predominantly nucleolar (Figure 3A, arrow) in interphase cells and also show a similar distribution throughout the cell cycle, first associating with the perichromatin region during metaphase and early anaphase (Figure 3B, arrowheads) and later appearing in small foci during telophase (Figure 3B, hashed arrow), before reaccumulation in nucleoli during G2.
Figure 3.
GFP-tagged RRP1B shows a similar localization to the endogenous protein, and overexpression of exogenous RRP1B alters nucleolar levels of PP1. (A) GFP-RRP1B stably expressed in U2OS cells shows a similar nucleolar accumulation (arrow) to the endogenous protein, as shown here by a fluorescent signal superimposed on a differential interference contrast (DIC) image of the cells. DNA stained with Hoechst 33342 is shown in blue. (B) The fusion protein GFP-RRP1B also shows similar perichromatin accumulations during metaphase and anaphase (B, arrowheads) and likely prenucleolar body (PNB) accumulations in telophase (hashed arrow). DNA stained with Hoechst 33342 is shown in red. (C) Overexpression of mCherry-RRP1B (red) in U2OSGFP-PP1γ cells leads to an increased level of GFP-PP1γ (green) in the nucleolus (arrow) compared with the nucleoplasm. (D) Overexpression of the mCherry-RRP1BKATA mutant in U2OSGFP-PP1γ cells leads to a decreased level of GFP-PP1γ (green) in the nucleolus (arrow) relative to the nucleoplasm. (E) High levels of mCherry-RRP1BKATA overexpression induce changes in nucleolar morphology, specifically a “rounding up” (arrow), and these nucleoli show reduced levels of 5-fluorouridine incorporation compared with nucleoli in nontransfected cells (hashed arrows). All experiments were repeated three times in two different cell lines. Scale bars are 15 μm.
Previous studies have shown that overexpression of a PP1 targeting subunit can cause redistribution of the PP1 catalytic subunit to the cellular localization of its interaction partner (Trinkle-Mulcahy et al., 2001; Trinkle-Mulcahy et al., 2006). Similarly, overexpression of mCh-RRP1B in HeLaGFP-PP1γ cells increases the nucleolar pool of GFP-PP1γ (Figure 3C). Overexpression of mCh-RRP1B in HeLaGFP-PP1α cells does not recruit excess PP1α to nucleoli (Supplemental Figure 4A), again confirming the selective interaction of RRP1B with PP1β and PP1γ. Conversely, overexpression of the nonPP1 binding GFP-RRP1BKATA mutant decreases the nucleolar pool of GFP-PP1γ (Figure 3D). High levels of GFP-RRP1BKATA expression also induced a “rounding-up” of nucleoli (Figure 3E, arrows). This “rounding up” is accompanied by a clear reduction of nucleolar 5-fluorouridine incorporation (Figure 3E, arrow) when compared with cells expressing little or no mutant protein (Figure 3E, hashed arrow).
RRP1B Is a Predominantly GC-Associated Nucleolar Protein
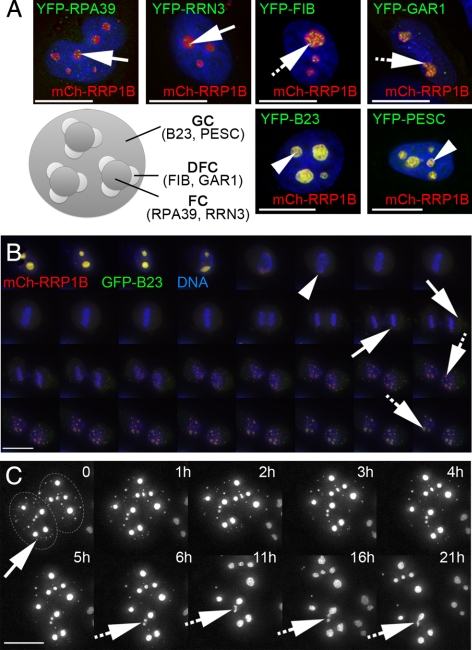
To determine the predominant subnucleolar localization of RRP1B, YFP/GFP-tagged protein markers for the fibrillar centre (FC), dense fibrillar component (DFC), and granular component (GC), respectively, were coexpressed with mCherry-RRP1B (Figure 4A). The RRP1B signal is distinct from that of the FC markers RPA39 and RRN3 and the DFC markers Fibrillarin and Gar1 but colocalizes with the GC markers B23 and Pescadillo.
Figure 4.
RRP1B is a GC protein that colocalizes with B23 throughout the cell cycle. (A) Coexpression of mCh-RRP1B (red) with markers (green) for the three subnucleolar compartments, the fibrillar centre (FC), dense fibrillar component (DFC), and granular component (GC), reveals that the subnucleolar distribution of RRP1B is spatially distinct from both the RPA39 subunit of RNA Pol I and its associated RRN3 factor (FC; arrows) and from Fibrillarin and Gar1 (DFC; hashed arrows). RRP1B colocalizes with B23 and Pescadillo, suggesting that it is predominantly a GC protein (arrowheads). (B) Time-lapse imaging of mCh-RRP1B (red) coexpressed with GFP-B23 (green) in U2OS cells stained with Hoechst 33342 to visualize DNA (blue) reveals that these proteins colocalize throughout the cell cycle. When nucleoli break down in prometaphase they both accumulate in the perichromatin region (arrowhead) and later appear in prenucleolar bodies (PNBs; arrows), eventually relocalizing to reformed mature nucleoli (hashed arrows). Images are 2D projections of 3D datasets captured every 10 min. (C) Time-lapse imaging of GFP-RRP1B in U2OSGFP-RRP1B cells shows the transfer of RRP1B from PNBs to nucleoli in late telophase. For most PNBs (arrow) the RRP1B signal disappears and reappears in nucleoli, although fusion of small foci was also observed (hashed arrows). Images are 2D projections of 3D datasets and time (in hours) is indicated. The two adjacent nuclei are outlined in the first panel for clarity. Scale bars are 15 μm.
Time-lapse triple-wavelength imaging of mCh-RRP1B, GFP-B23 and Hoechst 33342-stained DNA revealed that these two granular component proteins also exhibit similar localization patterns throughout mitosis (Figure 4B). Specifically, when nucleoli break down at the onset of prometaphase, both proteins become predominantly diffuse, with additional accumulations observed in the perichromatin region of the condensed chromosomes (Figure 4B, arrowhead). During late telophase, accumulations are observed in prenucleolar bodies (PNBs; Figure 4B, arrows). The contents of these PNBs later appear in newly-formed nucleoli (Figure 4B, hashed arrows). Although many of the PNBs disappear over time as their contents transfer to nucleoli, fusion of small bodies into larger nucleoli over time was also observed (Figure 4C; hashed arrows).
RRP1B Localization and Mobility Changes upon Induction of Nucleolar Reorganization by Drug Treatment
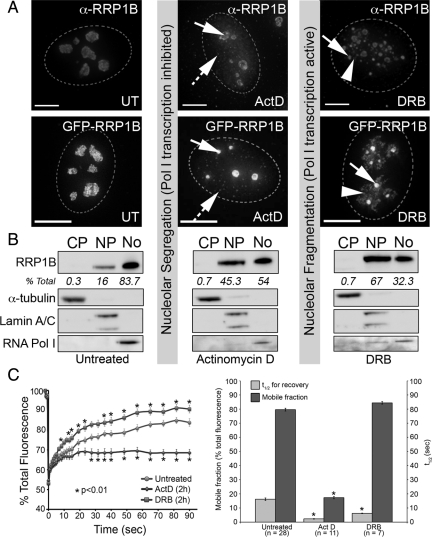
Overexpression of a nonPP1 binding RRP1B variant induces morphological changes in nucleoli and leads to a reduction in 5-fluorouridine incorporation (Figure 3E). Moreover, RRP1B colocalizes with B23 and Pescadillo (Figure 4A). Taken together these observations suggest a role for PP1-RRP1B in rRNA metabolism. To investigate this further, we treated cells with actinomycin D (ActD) at low levels (0.5 μg/ml) to inhibit RNA Pol I and at high levels (2.5 μg/ml) to inhibit RNA Pol I and II. Both endogenous (Figure 5A, top panels) and GFP-tagged RRP1B (Figure 5A, bottom panels) relocalize to the nucleoplasm in response to ActD and accumulate in small nucleoplasmic foci of unknown function (Figure 5A, hashed arrows). This relocalization occurs more quickly in response to higher levels of ActD. A pool of nucleolar RRP1B is retained in the remnant central body (Figure 5A, arrows) and perinucleolar region. This perinucleolar accumulation does not overlap markers for the well-characterized fibrillarin and coilin caps (Supplemental Figure 2E) (Shav-Tal et al., 2005).
Figure 5.
Relocalization and altered dynamics of RRP1B in response to drugs that induce nucleolar segregation. (A) Treatment with actinomycin D (ActD) at a concentration that inhibits both RNA Pol I and II (2.5 μg/ml for 2 h) leads to a redistribution of a pool of nucleolar RRP1B to the nucleoplasm. The nucleoplasmic pool of RRP1B includes small foci distributed throughout the nucleus (hashed arrows). The remaining nucleolar RRP1B is retained in both the central body (arrow) and at the nucleolar periphery. GFP-RRP1B shows a similar relocalization in response to ActD treatment. Treatment with DRB (25 μg/ml for 2 h) leads to a breakdown in nucleolar structure and redistribution of RRP1B to small granules (arrows) and additional masses surrounding these granules (arrowheads). GFP-RRP1B shows a similar response. Scale bars are 15 μm. (B) Untreated, ActD-treated, and DRB-treated U2OS cells were fractionated into cytoplasm, nucleoplasm, and nucleoli, and cell-equivalent volumes of each separated by SDS-PAGE and transferred to nitrocellulose membrane for Western blot analysis. Antibodies raised against markers for the cytoplasm (α-tubulin), nucleus (lamin A/C), and nucleolus (RNA Pol I subunit A190) demonstrate the purity of the subcellular fractions. Staining with the antibodies raised against endogenous RRP1B revealed that it is predominantly nucleolar (83.7%) in untreated cells, with 16% found in the nucleoplasm and 0.3% in the cytoplasm. This distribution changes after both ActD and DRB treatment, with a large increase in the nucleoplasmic fraction (45.3 and 67%, respectively) and decrease in the nucleolar fraction (54 and 32%, respectively). RRP1B is not found in appreciable amounts in the cytoplasm under any of these conditions. (C) Photobleaching analysis of nucleolar GFP-RRP1B in U2OSGFP-RRP1B cells reveals that ActD treatment leads to a significant reduction in the mobile fraction. DRB treatment does not alter the mobile fraction, however there is a significant decrease in the t1/2 of recovery. For each condition, datasets from 10 different cells collected in two separate experiments were averaged and SEs calculated. Asterisks (*) indicate time points at which there is a significant (p < 0.01) alteration from the untreated recovery curve, as calculated by a two-tailed paired Student's t test.
The nucleoplasmic redistribution of RRP1B was confirmed by cell fractionation and Western blot analysis (Figure 5B). In untreated cells, RRP1B is predominantly nucleolar (∼80% of total protein found in this structure). The remaining signal is nucleoplasmic, and little or no RRP1B is found in the cytoplasm. On inhibition of transcription with ActD treatment, the nucleolar signal falls to ∼50% total protein as a pool of RRP1B relocalizes to the nucleoplasm. There is little or no change in the cytoplasmic fraction.
Clearly, RRP1B localization responds to chemical treatment influencing RNA Pol I functionality. To discriminate between a potential role for RRP1B in rRNA transcription or rRNA processing, we exploited the unique properties of the nucleoside analog 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB). DRB inhibits certain protein kinases, including casein kinase II (CK2), and indirectly inhibits RNA Pol II transcription. DRB reversibly dissociates rRNA transcription from later processing steps, causing segregation of nucleolar components into transcriptional “beads” and separate processing bodies (Scheer et al., 1984). Despite the breakdown in nucleolar structure, RNA Pol I transcription continues, with rRNA transcripts originating in the transcriptional beads and diffusing to the neighboring processing bodies. On treatment of U2OS cells with DRB, both endogenous and GFP-RRP1B relocalize to smaller granular bodies (Figure 5A, arrows) surrounded by dispersed masses (Figure 5A, arrowheads). The relocation of core nucleolar RRP1B to the nucleoplasm upon DRB treatment was also demonstrated by Western blot analysis (Figure 5B). On removal of DRB, nucleoli reform and RRP1B and GFP-RRP1B resume their normal GC localization pattern (Supplemental Figure 3A). Coexpression of RRP1B and markers for either transcription/early processing (fibrillarin, Supplemental Figure 2F) or later rRNA processing (RPL27, Supplemental Figure 2D) confirmed that the RRP1B granular bodies are indeed processing bodies, distinct from the transcriptional “beads.”
The association of RRP1B with rRNA transcripts is supported by the observation that the protein remains associated with the major nucleolar RNA signal after both ActD and DRB treatment. As in untreated cells, this localization is sensitive to RNAse treatment (Supplemental Figure 1, C and D). While ActD and DRB treatment led to relocalization of RRP1B-containing processing complexes within nuclei, we surmise that this is caused by different signal transduction events because the primary targets of both chemicals are distinct biomolecular complexes. Using a Fluorescence Recovery After Photobleaching (FRAP) approach, we measured the dynamic turnover of nucleolar GFP-RRP1B in untreated cells compared with cells treated with either ActD or DRB. The kinetics of recovery of a photobleached GFP-tagged protein can reflect the degree and affinity of its association with other proteins and/or nucleic acids. While both drug treatments induce reorganization of the nucleolus, ActD induces a significant (p < 0.01 in a two-tailed Student's t test) decrease in the mobile fraction of nucleolar GFP-RRP1B (17.3 ± 0.82% for Act D vs. 79.6 ± 1.0% for untreated; Figure 5C). Conversely, the nucleolar pool of GFP-RRP1B after DRB treatment has a similar mobile fraction to that observed in untreated cells, but the recovery rate is increased when compared with untreated cells (Figure 5C). These differences in the dynamic turnover rate of GFP-RRP1B between untreated, ActD-, and DRB-treated cells reveals distinct underlying interaction profiles. Their precise nature awaits investigation.
RRP1B Coprecipitates 60S Ribosomal Subunit Processing Complexes
To identify the molecular complexes with which RRP1B associates, we first carried out a quantitative SILAC-based immunoprecipitation (Figure 6) of GFP-RRP1B from U2OSGFP-RRP1B–derived nuclear extracts. The most highly enriched interaction partner was Nol1, a known pre-60S (large) ribosomal subunit processing protein, followed by several other 60S processing proteins. We also noted a specific enrichment of the ribosomal proteins associated with the large/60S ribosomal subunit proteins (RPLs) over the small/40S ribosomal subunit proteins (RPSs) (full dataset presented in Supplemental Table 1). When coexpressed in cells, GFP-RRP1B and mCh-Nol1 colocalize throughout the entire cell cycle, supporting their presence in overlapping molecular complexes (Supplemental Figure 2A). A quantitative immunoprecipitation of the nonPP1 binding mutant GFP-RRP1BKATA revealed a similar binding profile to that of GFP-RRP1B, with the obvious lack of PP1 (data not shown).
Although the GFP-RRP1B immunoprecipitation experiment identified RRP1B as a factor present in pre-60S ribosome subunit processing complexes, a caveat of overexpressed exogenous proteins is that they may not behave identically to the endogenous protein. Thus, we used our RRP1B antibody for immunodepletion of the endogenous protein and carried out a quantitative proteomic screen of endogenous nuclear RRP1B. As shown in Figure 6C, endogenous RRP1B, like GFP-RRP1B, specifically enriches a large pool of RPLs (36/40 of the total) and an even more comprehensive array of known pre-60S processing proteins. Significant overlap was found between the interactomes of endogenous RRP1B and GFP-Nol1 (quantitatively immunoprecipitated from U2OSGFP-Nol1-derived nuclear extracts; data not shown), confirming that these proteins are found in similar complexes. When we highlighted pre-60S ribosomal subunit-related proteins in our nucleolar GFP-PP1γ interactome, we confirmed both the enrichment of RRP1B and of these particular factors (Figure 6D), indicating that pulldown of nucleolar PP1 enriches a subset of pre-60S ribosomal subunit processing complexes to which it is targeted by RRP1B. Consistent with the plurifunctional nature of this enzyme, the nucleolar PP1 interactome also contains additional phosphatase complexes not involved in pre-60S ribosome biogenesis.
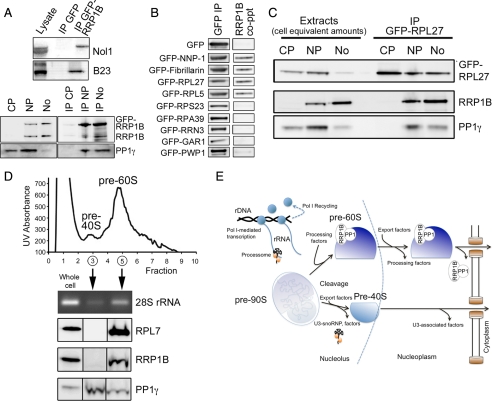
The quantitative aspect of these experiments, including built-in negative controls, combined with stringent analyses and the application of “bead proteomes” to flag potential false positives (see Materials & Methods) (Trinkle-Mulcahy et al., 2008), provides a high level of confidence in the protein interaction partners identified. Nevertheless, we felt it important to confirm the interaction of RRP1B with a representative subset of proteins identified in our proteomic screen, including PP1β and PP1γ (Figure 1), Nol1 and B23 (Figure 7A), two RPLs (Figure 7B) and the pre-60S processing factor fibrillarin (Figure 7B). We also confirmed that RRP1B coprecipitates NNP-1/Nop52, indicating that both of these NOP52 domain-containing proteins are likely present in pre-60S processing complexes. This is intriguing as both are putative mammalian orthologues of yeast Rrp1p.
Figure 7.
RRP1B associates with 60S ribosomal processing proteins and is found with pre-60S ribosomal subunits in both the nucleolus and nucleoplasm. (A) The top panel shows validation of the copurification of endogenous Nol1 and B23 with GFP-RRP1B and not GFP alone. In the bottom panel, IP of GFP-RRP1B copurifies PP1γ from nucleoplasmic and nucleolar but not cytoplasmic cellular fractions. Endogenous RRP1B also coprecipitates with GFP-RRP1B, indicating that more than one molecule may be incorporated into pre-60S processing complexes. (B) Validation of the copurification of endogenous RRP1B with GFP-tagged RRP1/NNP-1/Nop52, Fibrillarin, RPL27, and RPL5, but not GFP alone. Other nucleolar proteins not found in the RRP1B interactome, including RPS23, RPA39, RRN3, GAR1, and PWP1, do not copurify endogenous RRP1B. (C) GFP-RPL27, a marker for 60S ribosomal subunits, is found in cytoplasmic, nucleoplasmic, and nucleolar extracts, as is PP1γ. RRP1B is limited to the nucleoplasm and nucleoli. IP of GFP-RPL27 copurified both RRP1B and PP1γ, but only from the nucleoplasmic and nucleolar fractions. (D) Sucrose gradient fractionation of nuclear extracts into pre-40S (peak fraction 3) and pre-60S (peak fractions 4 and 5) ribosomal subunits confirms the specific association of RRP1B with pre-60S subunits. Protein and RNA were extracted from the fractions, and both Western blot analysis of the 60S marker protein RPL7 and RT-PCR of the 60S marker RNA species 28S confirmed the fractionation. While RRP1B is found only in the pre-60S fraction, PP1γ is found with both pre-40S and pre-60S subunits. (E) Model depicting the interaction of RRP1B with (and targeting of PP1 to) pre-60s ribosomal subunits in the nucleolus and nucleoplasm, and loss of this association before export of the mature subunits to the cytoplasm.
We further present a set of negative controls (i.e., proteins which, based upon their lack of enrichment in our RRP1B proteomic screens, should not be part of the RRP1B interactome). Indeed, Western blot analyses show that the small (40S) ribosomal subunit protein RPS23 and the H/ACA snoRNP protein Gar 1 do not coprecipitate RRP1B (Figure 7B). Furthermore, the RNA Pol I-related proteins RPA39 and RRN3, absent from our RRP1B interactome studies, do not coprecipitate RRP1B, again confirming the predicted role for RRP1B in later rRNA processing steps. Lastly, the 60S processing protein PWP1, which we found enriched with PP1 but not RRP1B, coprecipitated little to no endogenous RRP1B (Figure 7B). It should be noted that the small amount of RRP1B could be due to the difference in approach, as antibodies can be more sensitive than the mass spectrometer, and trace amounts of PWP1 may indeed be present in the RRP1B interactome.
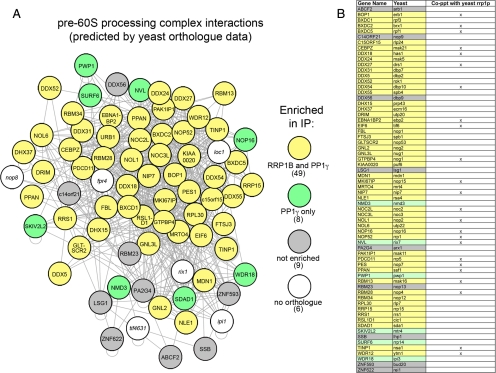
Because our results suggest RRP1B may play a role in 60S processing, we wanted to examine the overlap between our RRP1B interactome and that of pre-60S processing complexes. The latter has not yet been studied to any great extent in mammalian cells, and thus we turned to the comprehensive 60S ribosomal subunit processing complex defined by detailed interactome studies in baker's yeast system (Lebreton et al., 2008). We identified 66 mammalian orthologues to the 72 known yeast proteins depicted in this interaction diagram, of which 74% were found to be enriched with RRP1B (Figure 8A, yellow), representing a significant overlap between the yeast 60S processing complex and the mammalian RRP1B interactome. Having also noted a clear enrichment of RRP1B and other pre-60S processing proteins in the nucleolar PP1γ interactome, we compared this dataset to the yeast 60S processing complex and found enrichment of the full RRP1B interactome plus 8 additional proteins (Figure 8A, green). Proteins that were detected with PP1 but not RRP1B may be PP1-specific interaction partners but could also simply reflect differences in experimental conditions.
Figure 8.
RRP1B and PP1γ copurify an overlapping subset of pre-60S ribosomal subunit processing proteins. (A) An overlay of mammalian orthologues on their yeast counterparts in a yeast-derived map of pre-60S ribosomal subunit processing proteins demonstrates a specific enrichment of midlate processing complexes. Of the 72 proteins predicted in the yeast map, 66 have known mammalian orthologues. Of these, 49 (74%) were found in both the RRP1B and PP1γ interactomes (yellow), with an additional eight proteins only detected with the phosphatase (green). This may represent different subcomplexes or a difference in sensitivity of the experiment. Of the nine proteins that were not enriched (gray), most are peripheral to the core complex and function later in the processing/export pathway. In addition to these nonribosomal processing proteins, 36/40 (90%) of 60S subunit RPL proteins were specifically enriched with RRP1B and PP1γ. (B) 60S processing proteins identified in a recent nonquantitative screen of TAP-tagged yeast Rrp1p are indicated in this table, which includes both the yeast and mammalian gene names for each protein and uses the same color coding as in A to indicate whether a protein was found in our RRP1B and/or PP1γ IP or neither.
RRP1B is one of two suggested human orthologues for S. cerevisiae Rrp1p, a known rRNA processing protein. A direct comparison between the RRP1B and Rrp1p interactomes, the latter derived from a previous nonquantitative proteomic screen of Rrp1p (Horsey et al., 2004), revealed an overlap of 24 pre-60S processing proteins (not including RPLs) (Figure 8B). This confirms that RRP1B is found in similar complexes to its yeast counterpart. Note that the other human counterpart to Rrp1p, Nop52/NNP-1/hRRP1 (see below), is also part of the RRP1B interactome (Figure 8B).
As these interactomes represent a “snapshot” of pre-60S processing complexes at various stages of maturation, the fact that later processing and export factors are not found in either interactome suggests that the RRP1B/PP1 complex functions primarily at upstream stages of pre-60S subunit processing.
We next examined the spatial interaction of RRP1B with pre-60S ribosomal subunits, which are processed in the nucleolus and transit through the nucleoplasm to their final sites of action as mature ribosomes in the cytoplasm. In the U2OSGFP-RPL27 cell line, the 60S ribosomal subunit marker GFP-RPL27 is distributed throughout the nucleolus, nucleoplasm and cytoplasm (Figure 7C, Supplemental Figure 2B), whereas RRP1B is predominantly enriched in nucleoli, with an additional ∼20% found in the nucleoplasm (Figure 7C, Figure 3A). When GFP-RPL27 was depleted from these three subcellular fractions, it coprecipitated RRP1B from nucleolar and nucleoplasmic, but not cytoplasmic, fractions (Figure 7C). Similarly, PP1γ coprecipitated with GFP-RPL27 from these fractions. No detectable PP1γ was found with cytoplasmic 60S subunits, although a significant pool of the phosphatase is found in this subcellular compartment.
Finally, sucrose gradient fractionation confirmed the specific association of RRP1B with nuclear pre-60S (and not pre-40S) subunits (Figure 7D). PP1γ is found in both fractions, which fits with our identification of both RPSs (Figure 6D) and 40S processing proteins such as MPP10 in the nucleolar PP1 interactome. Taken together, these data suggest that PP1 is targeted to and regulates multiple steps in the ribosome biogenesis pathway (see diagram in Figure 7E).
RRP1B Is an Ancient Protein
The NOP52 domain was defined as a conserved region within a nucleolar protein of 52 kDa. Cloning of this human autoantigen revealed it to be a protein previously named novel Nuclear Protein of 52 kDa (NNP-1 or hNop52). This protein has a high sequence identify/similarity to the S. cerevisiae rRNA processing protein Rrp1p, a protein involved in the maturation of the 27S rRNA (Savino et al., 1999) suggested that this NOP52 domain-containing protein (NNP-1) was the human homologue of S. cerevisiae Rrp1p and thus renamed this gene/protein as hRRP1.
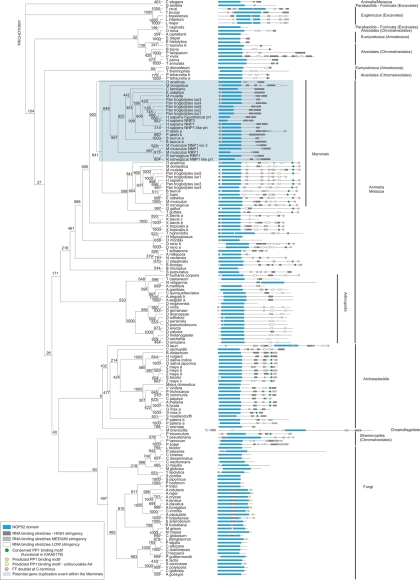
KIAA0179 was the second human NOP52 domain containing protein to be identified and was thus designated RRP1B. Here we have identified RRP1B as a PP1 binding partner that docks via its RVxF motif. To further explore the architecture and possible origins of KIAA0179/RRP1B we performed a bioinformatic analysis of the sequenced genomes of a broad range of organisms. Using truncations of RRP1B and a Hidden Markov model (HMM) for the NOP52 domain (PFAM website), we identified and aligned all NOP52 domain-containing proteins and used three independent methods (Neighbor Joining, Maximum Likelihood, Maximum Parsimony) to generate phylogenetic trees. Because primary sequences of RRP1B homologues are highly divergent, apart from the NOP52 domains, trees were built with the NOP52 domain sequences only. This yielded similar relationship patterns for all three methods.
The result of this search strategy and phylogenetic analysis is presented in Figure 9. Consistent with the NOP52 domain having been classified as a nucleolar domain of eukaryotic origin (Savino et al., 1999), we did not identify NOP52 proteins in Bacteria or Archeae. Although the NOP52 domain is highly conserved between KIAA0179/RRP1B and NNP-1/RRP1, phylogenetic analysis groups these proteins into separate branches when organisms contain both of these genes (NNP-1/RRP1 sequences are shaded blue). This analysis revealed the striking feature that the NNP-1/RRP1 protein appears only in mammals and, through gene duplication, has proliferated in several mammalian species. KIAA0179/RRP1B, on the other hand, is found in organisms throughout the eukaryotic lineage and is typically present as a single copy, with the exception of the genomes of P. tetraurelia (Chromalveolate), P. patens (spikemoss), and G. max (bean). This suggests that KIAA0179/RRP1B is an ancient protein, from which the mammalian NNP-1/RRP1 likely derived, and that the rRNA processing function accredited to the NOP52 domain is an early eukaryotic event.
Figure 9.
Phylogenetic tree of NOP52 domain-containing sequences. A rectangular cladogram was generated by comparison of conserved regions in NOP52-domain containing sequences. Multiple phylogenetic tree inferences were performed (see Materials and Methods). Tree topology shown is NJ (1000 replicates) which is largely representative for all three methods. Most notable discrepancy was M. brevicolis, which aligns with the euglenozoa (MP) or animalia (ML) in other methods. Bootstrap values are indicated at the nodes. K, presence of a conserved (RRP1B/KIAA0179) PP1 interaction motif; F, presence of a FF doublet at C-terminal end.
DISCUSSION
Ribosome subunit biogenesis is the major cellular function carried out by the nucleolus, with the very structure of this organelle intrinsically related to ongoing RNA Pol I transcription and pre-rRNA processing. Reports have also linked the nucleolus to control of a wide range of cellular pathways including cell division and DNA damage response (Pederson, 1998; Boisvert et al., 2007). It is now believed to be a major stress sensor, responding to stresses such as hypoxia and DNA damage by coordinating inhibition of ribosome biogenesis, cell cycle arrest, and, in certain cases, triggering of apoptosis (Mayer and Grummt, 2005). However, many of the regulatory events underlying these key functions remain undefined. Here we identified, via quantitative proteomics, fluorescence imaging and biochemical approaches, a nucleolar pool of PP1-RRP1B that is a component of pre-ribosomal subunit processing complexes. These results confirm our previous observations that a pool of PP1 activity accumulates within nucleoli and accounts for a large fraction of the associated Ser/Thr dephosphorylation events (Trinkle-Mulcahy et al., 2003).
With previous studies linking PP1 activity to the regulation of diverse nuclear functions ranging from gene transcription and splicing to cell growth and proliferation (Moorhead et al., 2007; MacKeigan et al., 2005), we hypothesized that this ubiquitous phosphatase would be targeted to more than one site within the nucleolus and to more than one level of the ribosome biogenesis pathway. Using an efficient nucleolar protein extraction method developed specifically for interactome analyses, we demonstrated the selective enrichment of a range of multiprotein complexes with nucleolar PP1 (Chamousset et al., 2010). Identifying RRP1B as an interaction partner for nucleolar-targeted PP1β and PP1γ was not surprising as it has long been suggested as a candidate, although the interaction had not been confirmed (Moorhead et al., 2008; Hendrickx et al., 2009). Our biochemical studies and the dominant negative effect of RRP1BKATA overexpression conclusively validate RRP1B as a PP1 interactor.
Our initial hypothesis, namely that PP1 forms more than one complex in the nucleolus, is corroborated by several observations. The first is the enrichment of additional known/putative targeting subunits and a wide range of multiprotein complexes in the nucleolar PP1 interactome studies (Chamousset et al., 2010). Another is the presence of PP1 in both pre-40S and pre-60S peaks, with PP1-RRP1B limited to pre-60S peaks. Lastly, the limited dominant-negative effect of RRP1BKATA mutant overexpression suggests that other functional nucleolar PP1 complexes are not affected by its specific displacement from pre-60S ribosomal subunits. This work thus opens novel routes to elucidate the impact of PP1 on ribosome biogenesis via a direct approach on individual complexes.
The name RRP1B derives from the protein's homology to the yeast Rrp1p protein (a.k.a. NOP52), which is involved in generation of 27S rRNA (Horsey et al., 2004). Nevertheless, the first published functional study on RRP1B identified it as a new candidate susceptibility gene for breast cancer progression and metastasis (Crawford et al., 2007). These data suggested that the protein plays a key role in regulating cell growth and proliferation, which could be a consequence of its predicted role in ribosome subunit biogenesis. More recently, RRP1B has also been linked to regulation of E2F-mediated apoptosis, and is believed to function directly in transcriptional control (Paik et al., 2010). The cellular role(s) of RRP1B thus remains open for debate, particularly in light of the existence of a second mammalian orthologue of NOP52, RRP1/NNP-1/Nop52 (Savino et al., 1999).
We provide here the first evidence for a role for RRP1B in 60S ribosome processing and also demonstrate that this protein has a functional PP1 interaction motif lacking in RRP1/NNP-1/Nop52. It is likely that RRP1B is the more complex cellular effector, as it can recruit a phosphoregulatory mechanism to a specific subset of processing complexes in the ribosome biogenesis pathway. It is also noteworthy that predicted RNA binding motifs display a different pattern between RRP1 and RRP1B, which may reflect differing rRNA affinities. These differences will likely have significant impact on their respective cellular interaction profiles and functions. Our phylogenetic analysis indicates that RRP1/NNP-1/Nop52 and KIAA0179/RRP1B are both homologues of yeast Rrp1p, yet KIAA0179/RRP1B is the true functional orthologue. This is consistent with the significant overlap between our endogenous nuclear RRP1B interactome and recently identified TAP-tagged yeast Rrp1p interaction partners (Horsey et al., 2004). This does not exclude RRP1/NNP-1/Nop52 from a role in pre-60S subunit processing, and indeed it is also found in the RRP1B and PP1 interactomes, however it does raise the question of how much, if any, functional overlap exists between these two mammalian proteins with regard to pre-60S ribosomal subunit processing.
We have shown here that the subnucleolar targeting of RRP1B throughout the cell cycle coincides with that of several GC-localized pre-60S processing proteins, consistent with a role for RRP1B in nucleolar rRNA processing. This targeting requires rRNA transcripts but not the presence of rDNA. In contrast, fibrillarin, which is a DFC protein involved in earlier processing steps, is partially lost with RNAse treatment and fully lost with DNAse treatment, reflecting its association with both rDNA and rRNA (Ochs et al., 1985). This again suggests that RRP1B is mainly involved in rRNA processing, likely interacting with these molecules after they are released from sites of transcription.
We also exploited the well-characterized segregation of nucleolar components in response to actinomycin D and DRB treatment (Scheer et al., 1984; Louvet et al., 2006) to compare RRP1B behavior to that of both rRNA transcripts and other known rRNA processing proteins. The retention of a pool of RRP1B in the remnant central body of the nucleolus was unique compared with the loss of nucleolar B23 and the “capping” of fibrillarin at the nucleolar periphery. Interestingly, the pool of RRP1B that is lost to the nucleoplasm was found to accumulate in small foci. Further work will be necessary to define these foci and determine whether they represent a link to the suggested transcriptional role of RRP1B (Paik et al., 2010). With regard to the pool of nucleolar-retained RRPB, it remains associated with both Pyronin Y-stained nucleolar RNA and GFP-RPL27 (Supplemental Figures 1C and 2C), again suggesting a structure/function relationship between RRP1B and nucleolar pre-60S processing complexes. Nucleolar-retained GFP-RRP1B in actinomycin D–treated cells is significantly less mobile than GFP-RRP1B in untreated cells, and as this pool of protein is also lost upon RNAse treatment, it may be sequestered or “trapped” in inactive RNA processing complexes when ribosome biogenesis shuts down (Supplemental Figure 1C).
In contrast, DRB treatment, which induces segregation of FC, DFC, and GC constituents into a characteristic “beads on a string” conformation while preserving RNA Pol I activity, leads to a small but significant increase in the mobility of nucleolar GFP-RRP1B. The mechanism of action of DRB is still debated, as it is a CK2 inhibitor that also indirectly inhibits RNA Pol II. Furthermore, different DRB derivatives (DMAT, TBB) give contradictory results with regard to GFP-RRP1B dynamics (Supplemental Figure 3B). Finally, CK2 targets several key nucleolar proteins, including B23 and nucleolin, and the kinase has been postulated to play a crucial role in compartmentation of nucleolar protein complexes (Louvet et al., 2006). Thus, apart from experimental differences including cell type, concentration, and treatment time, and the off-target effects of kinase inhibitors that can complicate interpretation of results (Bain et al., 2007), ribosome biogenesis has such a plethora of potential CK2-dependent effectors that direct and indirect effects are difficult to tease apart.
Our localization data strongly supported a role for RRP1B in later stages of ribosome subunit biogenesis. To validate this hypothesis, we quantitatively defined the nuclear interactome of RRP1B. We indeed found an enrichment of large ribosomal proteins (RPLs) and proteins linked to processing of the pre-60S subunit. In addition, comparison with the nucleolar interactome of PP1γ showed a significant overlap in the subset of proteins found in pre-60S processing complexes. Previously, we noted that the RRP1BKATA interactome is very similar to the wild-type RRP1B interactome. This suggests that RRP1B presence at rRNA processing complexes occurs independently of PP1, making RRP1B a bona fide PP1 targeting subunit.
Strikingly, proteins involved in very late stages of ribosome maturation and nuclear export are distinctly lacking in the RRP1B and PP1γ interactomes. Using stably expressed GFP-RPL27 as a marker for 60S subunits, we confirmed that association of RRP1B and PP1γ with these complexes occurs in both the nucleolus and nucleoplasm. Mature cytoplasmic 60S subunits, however, do not coprecipitate either RRP1B or PP1, placing the site of action of this holoenzyme complex firmly within the nucleus.
Rrp1/Nop52 is an essential gene in yeast, with knockout compromising cell growth (Horsey et al., 2004). When we reduced cellular levels of RRP1B in U2OS or HeLa cells by ∼90% using an siRNA approach (Supplemental Figure 4, B and D), little or no effect on cell growth or proliferation was evident. Although no significant changes were observed in the distribution of either nuclear pre-ribosomal subunits or mature cytoplasmic ribosomal subunits (data not shown), an increase in larger RNA species detected by Northern blot analysis may suggest an analogous role to its yeast counterpart in 28S processing (Supplemental Figure 4C). A similar lack of effect on cell growth or proliferation in response to reduction of RRP1B levels in human cells was observed recently by another group (Paik et al., 2010). This may reflect either a degree of genetic redundancy, or that levels of RRP1B are kept deliberately high to ensure it is never limiting for the essential process of ribosome subunit production and cell growth. Alternately, subtle effects may be masked by the high level of ribosome biogenesis in immortalized cell lines and/or the presence of RRP1/NNP-1/Nop52. Alternate approaches, such as analysis in primary cell lines or concurrent knockdown of RRP1B and RRP1/NNP-1, will be required to better understand the functional role of RRP1B-PP1 in regulation of pre-60S processing and, potentially, coordination of this pathway with regulation of transcription and/or proliferation.
Importantly, future work must focus on identification of RRP1B-PP1 targets within the pre-60s processing complex. Known phosphoproteins such as B23 and EIF6 are likely targets, with the latter being particularly attractive given that dephosphorylated pre-60S subunit-bound EIF6 is believed to prevent premature association of 40S and 60S subunits in the nucleus (Ceci et al., 2003). It is also important to further characterize proteins identified in the RRP1B nuclear interactome that are not directly related to pre-60S processing, as they may represent links to pathways that are controlled concurrently with ribosome biogenesis.
As discussed here, the ubiquitous nature of PP1 in cellular regulation emphasizes the importance of identifying and characterizing the specific holoenzyme complex(es) involved in each pathway. As part of our systematic dissection of the molecular mechanisms controlling targeting of PP1 activity to nucleolar substrates in ribosome biogenesis, cellular proliferation, and stress response pathways, we have identified and characterized a major nucleolar pool of PP1 targeted to pre-60S ribosomal subunit processing complexes by RRP1B. It is anticipated that this information will lead to a more direct therapeutic intervention by facilitating the targeted disruption of PP1 activity in disease states related to nucleolar dysfunction, including cancer, accelerated aging and viral infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank our colleagues in the Trinkle, Moorhead, and Lamond groups for helpful discussions and suggestions, Dr. David Tollervey (University of Edinburgh, UK) for provision of the hITS1 probe sequences for Northern blot analysis, and Dr. Martin Holcik (University of Ottawa, Canada) for use of the sucrose gradient fractionator. We also thank Drs. Douglas Lamont and Kenneth Beattie of the Fingerprints Proteomics and Mass Spectrometry Facility at the University of Dundee for technical assistance. Funding for this research was provided by an NSERC Discovery Grant (Ref: 372370) and Canadian Cancer Research/Terry Fox Foundation New Investigator Award (Ref: 20148) (to L.T.M.), NSERC and Alberta Cancer Board (to G.B.M.), and a Wellcome Trust Programme Grant (to A.I.L.) (Ref: 073980/Z/03/Z). A.I.L. is a Wellcome Trust Principal Research Fellow, and L.T.M. holds a Canadian Institutes of Health Research New Investigator Salary Support Award.
Abbreviations used:
- CK2
Casein kinase 2
- DFC
dense fibrillar component
- FC
fibrillar centre
- GC
granular component
- PP1
protein phosphatase 1
- RNA Pol I
RNA polymerase I
- RNA Pol II
RNA polymerase II
- RNA Pol III
RNA polymerase III
- RPL
ribosomal protein, large (60S) subunit
- RPS
ribosomal protein, small (40S) subunit
- RRP1B
ribosomal RNA processing 1 homolog B.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0287) on October 6, 2010.
REFERENCES
- Andreassen P. R., Lacroix F. B., Villa-Moruzzi E., Margolis R. L. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J. Cell Biol. 1998;141:1207–1215. doi: 10.1083/jcb.141.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker H. M., Brewis N. D., Street A. J., Spurr N. K., Cohen P. T. Three genes for protein phosphatase 1 map to different human chromosomes: sequence, expression and gene localisation of protein serine/threonine phosphatase 1 beta (PPP1CB) Biochim. Biophys. Acta. 1994;1220:212–218. doi: 10.1016/0167-4889(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barker H. M., Craig S. P., Spurr N. K., Cohen P. T. Sequence of human protein serine/threonine phosphatase 1 gamma and localization of the gene (PPP1CC) encoding it to chromosome bands 12q24.1-q24.2. Biochim. Biophys. Acta. 1993;1178:228–233. doi: 10.1016/0167-4889(93)90014-g. [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhauser N., Marchisio P. C., Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chamousset D., Mamane S., Boisvert F. M., Trinkle-Mulcahy L. Efficient extraction of nucleolar proteins for interactome analyses. Proteomics. 2010;10:3045–3050. doi: 10.1002/pmic.201000162. [DOI] [PubMed] [Google Scholar]
- Cohen P. T. Protein phosphatase 1–targeted in many directions. J. Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Crawford N. P., Qian X., Ziogas A., Papageorge A. G., Boersma B. J., Walker R. C., Lukes L., Rowe W. L., Zhang J., Ambs S., Lowy D. R., Anton-Culver H., Hunter K. W. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli T., Olson M. O. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Baserga S. J. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Horsey E. W., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A., Rousselle J. C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M., Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008;36:4988–4999. doi: 10.1093/nar/gkn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage B., Beullens M., Ceulemans H., Himpens B., Bollen M. Determinants of the nucleolar targeting of protein phosphatase-1. FEBS Lett. 2005;579:5626–5630. doi: 10.1016/j.febslet.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet E., Junera H. R., Berthuy I., Hernandez-Verdun D. Compartmentation of the nucleolar processing proteins in the granular component is a CK2-driven process. Mol. Biol. Cell. 2006;17:2537–2546. doi: 10.1091/mbc.E05-10-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKeigan J. P., Murphy L. O., Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- Meiselbach H., Sticht H., Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem. Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Moorhead G. B., Trinkle-Mulcahy L., Nimick M., De Wever V., Campbell D. G., Gourlay R., Lam Y. W., Lamond A. I. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem. 2008;9:28. doi: 10.1186/1471-2091-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G. B., Trinkle-Mulcahy L., Ulke-Lemee A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007;8:234–244. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs R. L., Lischwe M. A., Spohn W. H., Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Paik J. C., Wang B., Liu K., Lue J. K., Lin W. C. Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J. Biol. Chem. 2010;285:6348–6363. doi: 10.1074/jbc.M109.072074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadcap D. W., Brush M. H., Shenolikar S. Identification of cellular protein phosphatase-1 regulators. Methods Mol. Biol. 2007;365:181–196. doi: 10.1385/1-59745-267-X:181. [DOI] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J. C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino T.M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112(Pt 12):1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Scheer U., Hugle B., Hazan R., Rose K. M. Drug-induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-dichloro-beta-D-ribofuranosylbenzimidazole. J. Cell Biol. 1984;99:672–679. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Hatano Y., Chun Y. S., Sugimura T., Zhang Z., Lee E. Y., Nagao M. Identification of PP1 catalytic subunit isotypes PP1 gamma 1, PP1 delta and PP1 alpha in various rat tissues. Biochem. Biophys .Res. Commun. 1993;192:1289–1296. doi: 10.1006/bbrc.1993.1556. [DOI] [PubMed] [Google Scholar]
- Tran H. T., Ulke A., Morrice N., Johannes C. J., Moorhead G. B. Proteomic characterization of protein phosphatase complexes of the mammalian nucleus. Mol. Cell Proteomics. 2004;3:257–265. doi: 10.1074/mcp.M300115-MCP200. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andrews P. D., Wickramasinghe S., Sleeman J., Prescott A., Lam Y. W., Lyon C., Swedlow J. R., Lamond A. I. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol. Biol. Cell. 2003;14:107–117. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Chusainow J., Lam Y. W., Swift S., Lamond A. Visualization of intracellular PP1 targeting through transiently and stably expressed fluorescent protein fusions. Methods Mol. Biol. 2007;365:133–154. doi: 10.1385/1-59745-267-X:133. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Sleeman J. E., Lamond A. I. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- Wakula P., Beullens M., Ceulemans H., Stalmans W., Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.