Abstract
In this paper we define a hierarchical Bayesian model for microarray expression data collected from several studies and use it to identify genes that show differential expression between two conditions. Key features include shrinkage across both genes and studies, and flexible modeling that allows for interactions between platforms and the estimated effect, as well as concordant and discordant differential expression across studies. We evaluated the performance of our model in a comprehensive fashion, using both artificial data, and a “split-study” validation approach that provides an agnostic assessment of the model's behavior not only under the null hypothesis, but also under a realistic alternative. The simulation results from the artificial data demonstrate the advantages of the Bayesian model. The 1 – AUC values for the Bayesian model are roughly half of the corresponding values for a direct combination of t- and SAM-statistics. Furthermore, the simulations provide guidelines for when the Bayesian model is most likely to be useful. Most noticeably, in small studies the Bayesian model generally outperforms other methods when evaluated by AUC, FDR, and MDR across a range of simulation parameters, and this difference diminishes for larger sample sizes in the individual studies. The split-study validation illustrates appropriate shrinkage of the Bayesian model in the absence of platform-, sample-, and annotation-differences that otherwise complicate experimental data analyses. Finally, we fit our model to four breast cancer studies employing different technologies (cDNA and Affymetrix) to estimate differential expression in estrogen receptor positive tumors versus negative ones. Software and data for reproducing our analysis are publicly available.
1 Introduction
Microarray technologies that simultaneously measure transcriptional activity in a very large number of genes have been widely used in biology and medicine in the last decade, and the resulting data is often publicly available. To increase the reliability and efficiency of biological investigations, it can be critical to combine data from several studies. However, when considering multiple studies, variation in the measured gene expression levels is caused not only by the biological differences of interest and natural variation in gene expression within a phenotype, but also by technological and laboratory-based differences between studies (Irizarry et al., 2005; Consortium et al., 2006; Kerr, 2007). Two of the most important difficulties are the presence of both absolute and relative expression measurements, depending on the technology, and the challenges associated with cross referencing measurements made by different technologies to the genome and to each other (Zhong et al., 2007). Despite these difficulties, the results of combined analysis clearly demonstrate the potential for increased statistical power and novel discovery by combining data from several studies (Wu et al., 2002; Rhodes et al., 2002; Tomlins et al., 2005).
Most statistical work to date on combining microarray studies has focused on identifying genes that exhibit differential expression across two experimental conditions or phenotypes. We consider this problem here as well. There is now a substantial literature on Bayesian approaches to assessing differential expression across two or more experimental conditions within a single study. Both empirical and fully Bayesian models have been proposed, including parametric (Baldi and Long, 2001; Newton et al., 2001; Lönnstedt and Speed, 2002; Pan, 2002; Tseng et al., 2001; Bröet et al., 2002; Ibrahim et al., 2002; Townsend and Hartl, 2002; Gottardo et al., 2003; Ishwaran and Rao, 2003; Kendziorski et al., 2003; Ishwaran and Rao, 2005), semi-parametric (Newton et al., 2004) and non-parametric (Efron et al., 2001; Do et al., 2005) models. In each of these papers, a critical issue is shrinkage, and in particular borrowing strength across genes when estimating the gene-specific variance across samples. It is well established that shrinkage of the variance estimates provides worthwhile enhancements to single study analysis of differential expression (Liu et al., 2004).
There are several natural approaches for combining information from multiple microarray studies. One is to compute, separately for each study, statistics that summarize the relationship between each gene and the phenotype of interest. These may then be combined using methodologies such as those originally devised to integrate published results in meta-analysis (Hedges and Olkin, 1985). While initial efforts in this direction have considered combination of p-values (Rhodes et al., 2002), subsequent papers have focused on the more efficient strategy of combining effect sizes (Ghosh et al., 2003; Wang et al., 2004; Garrett-Mayer et al., 2007). At the opposite extreme of study combination are cross-study normalization methods (Wu et al., 2002; Parmigiani, 2002; Shen et al., 2004; Rhodes et al., 2004; Hayes et al., 2006; Johnson et al., 2007; Choi et al., 2007; Shabalin et al., 2008) that consider directly the sample-level measurements within each study, and merge these into a single data set, to which standard single-study analysis can be applied. A third approach, intermediate between the two above, is to integrate information about differential expression from the available studies using a joint stochastic model for all the available data (Choi et al., 2003; Conlon et al., 2006; Jung et al., 2006; Conlon, 2007; Conlon et al., 2007), in which only selected features of each study, such as parameters that capture the relationship between genes and phenotypes, are assumed to be related across studies. This perspective has the potential to offer additional efficiency over integration of summary statistics, and to allow for a more comprehensive treatment of uncertainty. At the same time it models the cross-study integration in a way that is tailored to the problem of interest, and potentially relies on fewer assumptions than direct data integration.
In this article we adopt this latter, intermediate approach, and propose a fully Bayesian hierarchical model to identify genes that exhibit differential expression between two experimental conditions, and across multiple studies. In this context, use of a fully Bayesian model has several desirable features. The model borrows strength across both genes and studies and can thereby provide better estimates of the gene-specific means, variances and effects. The model yields, through simulation, posterior probability distributions for all unobserved quantities. These distributions can be used to quantify the uncertainty of any parameter in the model, or to make joint inferences about multiple genes. Lastly, for each gene, the model yields the posterior probability that the gene is differentially expressed.
While the work of Conlon and colleagues considers several of these issues, its primary strength is in the combination of multiple studies from the same technology. We expand this and related work to address multi-platform analysis via several technical generalizations that are described in detail in the methods, and reviewed in the discussion. These include: modeling of study-specific indicators of differential expression; modeling of overall cross-platform correlations to allow shrinkage to be stronger across pairs of studies that are generally more concordant; modeling of both the mean of a gene and its phenotypic interactions with sufficient flexibility to avoid distributional modeling of the main effects; allowing for interactions between the technology and the effect; modeling of an adaptive, smooth dependence between effects and the variance terms.
Perhaps the most radical difference between our approach and its predecessors is the attention given to discordant differential expression. This occurs when a gene is more highly expressed in one phenotype than the other in some studies, while the opposite is observed in other studies. Earlier approaches would discount the gene: the high cross-study variance and cancellation of overall effects would likely position it with the uninteresting genes. However, across many meta-analyses, we have observed an excess of these discordant genes compared to what would have been predicted by chance alone, as captured by permutation of phenotype labels. When implementing shrinkage strategies, reliable assessments of concordant differential expression, which is typically of primary interest, must therefore account for the possibility of discordant differential expression across studies. We implement this by introducing a gene-specific indicator of whether a gene is different across conditions in all of the studies, but then we allow these differences to be gene and study specific.
While concordant differentially expressed genes remain the primary focus of the analysis, discordant genes can reveal important biological or technological information, and it is useful to identify them and report them. This is for at least two reasons: first, given the heterogeneous experimental designs that are encountered in microarrays, a discordant effect for a set of important genes may be the result of genetic heterogeneity of the samples across studies. For a simple example, consider the comparison of administering or not administering a certain drug in two studies which, unbeknown to the investigators, use strains of animals where the sets of biochemical pathways activated by the drug are not the same. Then certain genes' expression may be increased by the drug in one strain and decreased in the other. Another reason discordance is important is that the cross-referencing of transcripts across studies is typically gene-centric. However, as many as 40 to 60% of genes are able to produce multiple alternative transcripts (Modrek and Lee, 2002), whose expression may be positively or, as is common, negatively correlated. For example one transcript may be made primarily under normal conditions, while the other may be made mainly in response to stress. When two technologies measure a gene's expression by targeting portions of that gene that are associated with different transcripts that are negatively correlated with each other, discordant effects will be observed. In either case, important insight about technology, study designs and potentially the genetics of alternative splicing can be gained by following up on discordant genes.
The paper is organized as follows. In Section 2 we describe our Bayesian model and in Section 3 we define a Markov chain Monte Carlo algorithm for simulation from the resulting posterior distribution. Section 4 describes statistics from the Bayesian model that can be used to quantify differential expression, as well as alternative approaches for quantifying differential expression in the context of multiple studies. The datasets used in the simulation and experimental data example are described in Section 5. Sections 6 and 7 present results when applying our model to simulated and real data with comparisons to alternative methods. Concluding remarks are in Section 8. The software for fitting our Bayesian model is open source and freely available from Bioconductor (Gentleman et al., 2004).
2 Bayesian hierarchical cross-study model
In this section we introduce some basic notation and our Bayesian model. We refer to the resulting method for cross-study assessment of differential expression by the R package name, XDE.
2.1 Notation and basic assumptions
In what follows we use p and q to index studies (i.e. data sets), g to index genes, and s to index samples (arrays) within each study. Let xgsp denote the observed expression value for gene g and sample s in study p. Let P denote the number of available studies, G the number of common genes and Sp the number of samples in study p. Thus the observed expression values are
We assume that each study has been suitably normalized (and if necessary log-transformed) so that the mean expression value for each study is zero and the expression values for a given gene are approximately Gaussian under each condition. We restrict our analysis to the set of common genes in the available studies, though our model formulation can easily be extended to a situation in which there is substantial overlap, but not complete agreement, between the gene sets in different studies.
In the model described below, each sample is assumed to belong to one of two possible conditions or phenotypes. Let ψsp ∈ {0, 1} denote the phenotype of sample s in study p. (An example in which ψsp represents the estrogen receptor status of breast tumors is presented in Section 7.) In order to model differential expression, we assume that, for a subset of the available genes, the expression value xgsp has a different mean value in samples where ψsp = 0 than in samples where ψsp = 1. We have implemented two versions of the Bayesian model: one with study-specific indicators of differential expression, δgp ∈ {0, 1} for studies p ∈ {1, … , P}, and a second with a single indicator, δg ∈ {0, 1}, that assumes a gene is differentially expressed in all of the studies or in none of the studies.
2.2 Bayesian model
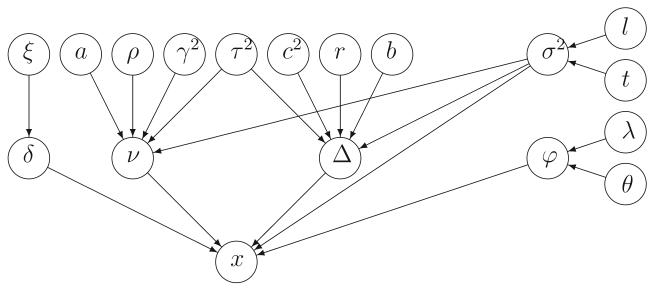
We define a hierarchical Bayesian model for the expression values xgsp. In the following discussion, the graphical model representation in Figure 1 can be used as a reference.
Figure 1.
A graphical model representation of the hierarchical Bayesian model defined for the microarray data sets.
At the lowest level we assume the expression values xgsp, conditional on some unobserved parameters, are independent and have a Gaussian distribution. For genes that are not differentially expressed, νgp denotes the mean value of xgsp, i.e., the mean value may be different for different genes and studies, but is the same for all samples in the same study. By contrast, differentially expressed genes have different means under the two phenotypic conditions. When δgp = 1 the mean of gene g in study p is equal to νgp − Δgp and νgp + Δgp for samples with ψsp = 0 and ψsp = 1, respectively. Thus Δgp represents half the average difference between expression levels across phenotypes for gene g in study p. By allowing Δ to depend on both g and p we acknowledge that the measured magnitude of an effect may depend on the technology. We impose no restriction that the Δgp should have the same sign across studies, thereby allowing for the possibility of discordant differential expression. We also allow the variance of xgsp to depend on the gene g, the study p, and the phenotypic condition ψsp. Let and denote the variances of xgsp for samples with ψsp = 0 and ψsp = 1, respectively. Our basic model may be written as follows:
| (1) |
At the next level in the model specification, prior distributions are selected for the model parameters. We begin by discussing the priors for
which represent, respectively, the means and offsets for gene g. The vectors νg and Δg are assumed to be independent across genes, and independent of each other. Furthermore, for every gene g, it is assumed that νg and Δg have a multi-Gaussian distribution. In the context of single studies, the choice of a Gaussian prior for modeling the mean expression value for a gene has been used in other settings (e.g., Baldi and Long (2001)), though many other choices are possible (Berger, 1993). As we have required that the expression values of each gene be centered around zero, we set the mean of νg and Δg equal to zero as well. Let Σg and Rg denote the covariance matrices of νg and Δg, respectively, so that
| (2) |
To specify the covariance matrices of Σg and Rg we adopt the strategy advocated in Barnard et al. (2000), namely, to assign independent prior distributions to the standard deviation and the correlation matrix of each quantity (see below for more details).
When modeling normal location and scale parameters in a hierarchical way, as we do here, two modeling choices are common. One is independence between scale and location, and the other is conjugacy. The latter is computationally more convenient, as it allows analytical expressions for the full conditional distribution. However, which of these two specifications fits better can vary from experiment to experiment, and in microarray analysis the fit is sensitive to the technology and normalization method used, see Liu et al. (2004). Recently, Caffo et al. (2004) proposed a more general family of models, one that encompasses both independence and conjugacy, by including a single parameter that indexes the distribution of location given scale. Here we extend this idea by introducing separate parameters for each individual study, and setting a common relative scale for Σg and Rg in each study. More specifically, the diagonal elements of Σg and Rg are given as follows:
| (3) |
Here , the parameters ap, bp ∈ [0, 1], and the parameters are such that . Thus γ2 and ap control the overall scale and conjugacy of νg, respectively, while c2 and bp play analogous roles for Δg, and control the relative scales of the different studies.
The correlation structure of Σg (and Rg) is assumed to be the same for all genes g. Let and denote the correlation matrices corresponding to Σg and Rg, respectively. Following Barnard et al. (2000), the prior distribution for [ρpq] is obtained by beginning with a covariance matrix having an inverse Wishart distribution with νρ degrees of freedom, and then integrating out its component variances. The prior distribution for [rpq] is of the same form, with νr degrees of freedom, and independent of the prior for [ρpq].
At the next level in the hierarchical model specification, priors are placed on the hyperparameters γ2, c2, , ap and bp. To enforce model parsimony, the prior distributions for ap and bp place positive probability mass at the values 0 and 1, corresponding to independence and conjugacy between location and scale, respectively. More specifically, independently for each study p, we let
| (4) |
and
| (5) |
Independent vague priors are assigned to the remaining hyper-parameters. For γ2 we use an (improper) uniform distribution on (0, ∞), and for c2 a uniform distribution on (0, ). Note that an improper prior can not be used for c2 as this may result in an improper posterior distribution. For we assign a joint (improper) uniform distribution under the natural restrictions , p = 1, … , P and .
In order to have a fully defined Bayesian model, it remains to specify prior distributions for the differential expression indicators δgp, and for the variances used to define . For the indicators δgp we have implemented two prior models. In prior model A, we assume that δg1, … , δgP are a priori independent given a hyperparameter ξp with
| (6) |
independently. In prior model B, we set the restriction δg1 = … = δgP = δg and assume that the G indicators are a priori independent, given a hyperparameter ξ, with
The variances are assumed to be independent for different genes g and studies p, given the other hyperparameters. However, and should be correlated for the same gene g and study p. To obtain this, we set
| (7) |
where and φgp have independent gamma prior distributions with , , E[φgp] = λp and Var[φgp] = θp. At the next level we assign independent (improper) uniform distributions on (0, ∞) for each of the hyper-parameters lp, tp, λp, θp, independently for p = 1, … , P.
The above prescriptions fully define the hierarchical Bayesian model visualized in Figure 1. The observed quantities are the expression values xgsp and the conditions ψsp. Conditioning on the observed values we get a posterior distribution for the unobserved parameters ξp, δgp, ap, ρpq, γ, , νgp, c2, rpq, bp, Δgp, , φpg, lp, tp, λp and θp. Hyper-parameters that have to be specified by the user are αa, βa, αb, βb, , , , , νρ, νr, αξp, βξp and . Default hyperparameters provided in the R package XDE work well in most instances (see Table 1).
Table 1.
Each row in the table displays parameters used to simulate an artificial dataset of three studies with S samples in each. From Equation 11, k* and c* control the location and scale of the simulated Gaussian offsets, respectively. Together, r*, k*, and c* control the degree of concordance of the simulated offsets. The probability that a gene was differentially expressed was ξ*.
| k* | S | c* | r* | ξ* | |
|---|---|---|---|---|---|
| A† | 0.5 | 4 | 0.5 | (0.1, 0.2, 0.4) | 0.10 |
| B | · | · | · | · | 0.50 |
| C | · | · | · | (0.8, 0.9, 0.92) | 0.10 |
| D | · | · | · | · | 0.50 |
| E† | · | 8 | 0.5 | (0.1, 0.2, 0.4) | 0.10 |
| F | · | · | 1 | · | 0.10 |
| G | · | · | · | · | 0.50 |
| H | · | · | · | (0.8, 0.9, 0.92) | 0.10 |
| I | · | · | · | · | 0.50 |
| J† | 0 | 16‡ | 10 | (0.1, 0.2, 0.4) | 0.10 |
| K | · | · | · | · | 0.50 |
| L | · | · | · | (0.8, 0.9, 0.92) | 0.10 |
| M | · | · | · | · | 0.50 |
| O | · | 32‡ | 20 | (0.1, 0.2, 0.4) | 0.10 |
| P | · | · | · | · | 0.50 |
| Q | · | · | · | (0.8, 0.9, 0.92) | 0.10 |
| R | · | · | · | · | 0.50 |
Ten artificial datasets were generated from one set of simulation parameters (S, k*, c*, r*, ξ*) but with different seeds for the random number generator. By varying only the seed for the random number generator, we can assess the sensitivity of performance measures such as AUC to randomly generated quantities in the simulation (e.g., the set of genes with δ* = 1). In general, simulation parameters were selected such that the AUC statistic from the simulations ranged between 0.6 and 0.9.
The Stanford dataset contained 11 stage I and II adenocarcinomas that were randomly split into 6 cases and 5 controls.
3 Posterior simulation
In order to evaluate the properties of the resulting posterior distribution, we adopt the Metropolis–Hastings algorithm (Metropolis et al., 1953; Hastings, 1970) to generate realizations from it. Nice introductions to the Metropolis–Hastings algorithm can be found in Smith and Roberts (1993) and Dellaportas and Roberts (2003). The algorithm is iterative, and each iteration consists of two parts. First, potential new values for one or a number of parameters are proposed according to a proposal distribution. Second, the proposed values are accepted with a specified probability. Depending on the mathematical form of the distribution of interest, different proposal mechanisms can be employed. In our posterior distribution we have seventeen types of parameters and to update these we combine seven types of proposal mechanisms. In the following we specify each of the proposal strategies used. In the description we use tilde to denote potential new values, e.g. δgp and δ̃gp are the current and potential new values of the differential expression indicator for gene g in study p, respectively. Note that we restrict attention to the proposal distributions, as the acceptance probabilities are then uniquely defined by the Metropolis–Hastings setup. Preliminary runs show that the parameters Δg, c2 and γ2 are often highly correlated with other parameters in the model. To cope with this strong posterior dependency and improve mixing, multiple block updates are given to the Δg, c2 and γ2 parameters. As with many Metropolis–Hastings proposal strategies, several of our proposal distributions include a “tuning” parameter that measures the amount of change proposed. In the following we consistently use the same symbol ε to denote all the tuning parameters, but, as the values used in our examples in Sections 6 and 7 suggest, one can of course use different values when updating the different parameters.
The full conditionals for νg, Δg, γ2 and ξp have standard forms and we therefore use Gibbs steps (see the references given above) to update each of these separately. The full conditionals for νg and Δg both are multiGaussians, the full conditional for γ2 is an inverse gamma distribution, and for ξp it is a beta distribution.
Separately, for each of the parameters ap and bp, we use a “truncated” random walk proposal. In particular, for ap we do the following: if ap = 0 we draw ãp from a uniform distribution on [0, ε]; if ap = 1 we draw ãp uniformly on [1 − ε, 1]; and if ap ∈ (0, 1) we draw U from a uniform distribution on [ap − ε, ap + ε] and set ãp = min(1, max(0, U)). We note that this is a reversible jump type of proposal, and to e get the correct acceptance probability, one needs to use the theory introduced in Green (1995).
Separately, for each of the parameters , φgp, lp, tp, λp and θp, we propose a multiplicative change. In particular, for we set , where U is sampled from a uniform distribution on the interval [1/(1 + ε), 1 + ε].
When updating we must ensure that the product of the proposed new values equals unity. We do this by randomly selecting two of the components, p and q say, drawing U from a uniform distribution on [1/(1 + ε), 1 + ε], and setting and
A block Gibbs update is used for c2 and all the Δg's for genes that have δgp = 0.
Separately for each g = 1, …, G, a block update is used for (δg1, …, δgP) and Δg. First, potential new values for δg1, …, δgP are set. For prior model A we do this by inverting the current value of δgp for a randomly chosen study p, i.e. δ̃gp = 1 − δgp, and keeping the other indicators unchanged. For prior model B (δg1 = … = δgP = δg) we invert all the indicators. Second, a potential new value for Δg is sampled from the associated full conditional (given the potential new values δ̃g1, …, δ̃gP ). The proposed values are then accepted or rejected jointly.
A block update is used for [ρpq] and γ2.
A similar block update is used for [rpq] and c2. For [ρpq] and γ2, potential new values for [ρpq] are obtained via the transformation
Here [Tpq] is a correlation matrix which with probability one half is generated from the prior for [ρpq], and with probability one half is set equal to unity on the diagonal with constant off diagonal elements. In the latter case, the value of the off diagonal elements is sampled from a uniform distribution on (−1/(P − 1), 1). Thereafter, the potential new value for γ2 is sampled from the associated full conditional (given the potential new values [ρ̃pq]). The proposed values are then accepted or rejected jointly.
4 Estimation of differential expression
In assessing the differential expression of genes across multiple studies, one naturally encounters a difficulty that is not present in single study analyses. This difficulty arises from the fact that a single differentially expressed gene g may be up-regulated in one or more studies, and down-regulated in others. When this occurs, we say that g is discordantly differentially expressed. If g is up-regulated in every study, or down-regulated in every study, we say that g is concordantly differentially expressed. Although concordant differential expression is the norm, discordance can arise from biological differences in the sample populations of each study, or from technological effects related to the design and implementation of specific array technologies. Discordance appears to be an unavoidable (and inconvenient) feature of multi-study analyses, one that comprehensive multi-study analyses should take into account.
4.1 Bayesian estimation
We have developed two implementations of our Bayesian model: a single indicator model that assumes differential expression occurs in all of the studies or in none of the studies, and a multiple indicator model that allows study specific indicators, δgp, of differential expression.
Single indicator implementation
In the following discussion we discuss the single indicator implementation of the Bayesian model for differential expression. Note that the indicator δg summarizes information across studies. The basis for our cross-platform analysis of differential expression is the posterior mean of δg, equivalently the posterior probability that gene g is differentially expressed. This posterior mean is not analytically available, so in practice we have to generate samples from the posterior distribution, as discussed in Section 3, and estimate the posterior mean by the empirical mean of the simulated δg's.
Let PMε(g) denote the posterior mean of δg. We view PMε(g) as a measure of the evidence for the overall differential expression of g. In particular, one may classify a gene g as differentially expressed whenever PMε(g) > a for some threshold a > 0. Concordant and discordant differential expression can also be addressed in a direct way in the context of the Bayesian model described above. A gene g for which δg = 1 is concordantly differentially expressed if each of its offsets Δgp, p = 1, …, P has the same sign, and is discordant if its offsets include both positive and negative values. Thus, indicators for concordant and discordant differential expression can be defined by
| (8) |
and
| (9) |
respectively.
Multiple indicator implementation
For each gene, we compute the number of positive (N+) and negative (N−) signed offsets. Specifically, and , where I{·} takes the value 1 when {·} is true and 0 otherwise. Then concordant and discordant indicators for differential expression are given by the relationships
where m is the minimum number of studies for which the gene is differentially expressed. The posterior mean of each indicator can again be estimated by the empirical mean of the corresponding simulated quantities. Let PM𝒞(g) and PM𝒟(g) denote the corresponding posterior mean values. Then a gene g may be classified as concordantly or discordantly differentially expressed whenever PM𝒞(g) > a or PM𝒟(g) > a for some threshold a > 0.
4.2 Alternative methods
We consider three alternatives to XDE for estimating differential expression: the implementation of the Choi et al. (2003) random effects model in the R package GeneMeta (Gentleman et 2005), and cross-study summaries of t- and SAM-statistics. While a comparison with the Conlon et al. (2006) paper would be interesting, software for fitting this model to expression data is not readily available.
The study-specific statistics from which we derive cross-study summaries of differential expression are the Welch t-statistic (tgp), SAMgp (Tusher et al., 2001), and a standardized unbiased estimate for effect size, zgp (Hedges and Olkin, 1985), discussed by Choi et al. (2003) in the context of a cross-study microarray analysis. The Welch t-statistic allows for unequal variances between the phenotypes, whereas the z statistic uses a pooled estimate that assumes equal variance between the phenotypes. In contrast to the t- and z-statistics, the SAM statistic downweights genes with small variance, favoring genes with larger effect sizes. We hereafter generically denote the study-specific statistics by Ug = (Ug1, …, UgP), and cross-study summaries of differential expression by non-negative statistics u*(g), where the subscript indicates whether the statistic measures overall differential expression (), concordant differential expression (𝒞), or discordant differential expression (𝒟). A gene g may then be classified as being appropriately differentially expressed if the corresponding statistic u*(g) exceeds a fixed constant a > 0.
For evaluating overall differential expression, we follow the discussion of Garrett-Mayer et al. (2007), and combine the elements of Ug in a linear fashion to obtain a statistic suitable for assessing differential expression:
| (10) |
Here L is the covariance loading from the first principal component of the vectors Ug, and Sp is the number of samples in Study p. Summary measures of concordance for tgp and SAM were obtained by
As an alternative, we also used the combined (across studies) estimate of effect size from the random effects model proposed by Choi et al. (2003) directly. We denote this statistic by z𝒞(g). The ‘borrowing of strength’ in the estimation of z𝒞(g) is strictly across studies (as opposed to across genes and studies), as the study-specific effect sizes for a given gene are assumed to be a draw from a Gaussian distribution in the second level of the random effects model. Assessments of discordant differential expression using the tgp, SAM and z statistics are obtained by
5 Datasets and Software
The R package XDE contains the software used to fit the Bayesian hierarchical model, as well as convenient methods to compute the alternative statistics described in this paper.
Lung cancer datasets
The following lung cancer studies are referred to by institution: Harvard (Bhattacharjee et al., 2001) (203 samples on the Affymetrix Human Genome 95A platform containing 12,453 probesets), Michigan (Beer et al., 2002) (108 samples on the Affymetrix HuGeneFL Genome Array platform containing 6663 probesets), and Stanford (Garber et al., 2001) (68 samples on the cDNA platform containing 23,100 probes). The simulation described in Section 6 uses the normalized and merged platform annotations made publicly available from the authors of a previous cross-study analysis (Parmigiani et al., 2004). Briefly, Parmigiani et al. (2004) applied a robust multichip average (Irizarry et al. (2003)) separately to the two Affymetrix platforms (Harvard and Michigan). Intensity ratios from the Cy5 and Cy3 channels for the Stanford (Garber et al., 2001) dataset (23,100 features on the cDNA platform) were log-transformed. Following normalization, probesets (Affymetrix) and image clone identifiers (cDNA) in each platform were mapped to UniGene identifiers. Many-to-one mappings (multiple probes map to one UniGene identifier) were averaged and one-to-many mappings were excluded. The studies were then merged by Uni-Gene identifiers, resulting in a common set of 3,171 features. The normalized and merged datasets were obtained from the R package lungExpression available on the Bioconductor website (http://www.bioconductor.org).
Breast cancer datasets
Four breast cancer studies containing phenotypic data on estrogen receptor (ER) status (Sorlie et al. (2001), Huang et al. (2003), Hedenfalk et al. (2001), and Farmer et al. (2005)) were normalized according to platform type. In particular, Affymetrix platforms (the Farmer and Huang datasets) were normalized by RMA, whereas cDNA platforms (Sorlie and Hedenfalk) were normalized using the methods described in Smyth and Speed (2003) and implemented in the R package LIMMA. Following normalization, platform-specific annotations were mapped to Entrez-gene identifiers and the resulting lists merged to obtain a set of 2064 genes.
6 Validation
This section, comprised of two parts, extensively evaluates two implementations of the Bayesian model. In the first part of this section, we assess the single indicator and multiple indicator implementations using two simulation scenarios: one that simulates differential expression in all of the studies or none of the studies through a single indicator , and a second that simulates differential expression in a subset of the studies through study-specific indicators of differential expression, . As the set of genes that are differentially expressed is known through simulation, we assess the performance using diagnostics such as the area under the ROC curve (AUC). In the second part of this section, we evaluate the shrinkage properties of the Bayesian model by applying XDE to multiple splits of a single study. Comparisons of XDE to alternative methods for cross-platform analysis are discussed throughout.
6.1 Simulation
6.1.1 Experimental Data
Our simulations are based on three publicly available lung cancer datasets that we refer to by institution: Harvard (Bhattacharjee et al., 2001) , Michigan (Beer et al., 2002), and Stanford (Garber et al., 2001). See Section 5 for a brief description of these datasets. We begin by describing an approach for generating artificial datasets for which the true set of differentially expressed genes is known. We append the superscript ‘⋆’ to parameters used in the simulation to distinguish the true values from the corresponding variables in the Bayesian model. The simulation uses only stage I or II adenocarcinomas in the Harvard (n=83), Stanford (n=11), and Michigan (n=61) studies. Late stage adenocarcinomas were excluded, as the heterogeneity of these tumors is typically much greater. From each available study we randomly select S samples, and then randomly assign the clinical variable φ⋆ = 0 to half of the samples in the study, and φ⋆ = 1 to the remaining half. Although there is non-trivial heterogeneity within the adenocarcinomas, these differences become small (on average) after random assignment into classes, and provide a background noise that would be difficult to simulate de novo.
simulation
Independently for each gene, we simulate from a Bernoulli distribution with parameter ξ⋆ that is common to all genes. For genes with we thereafter generate “true” offsets from a multivariate normal distribution
| (11) |
where sg1, sg2 and sg3 are the empirical standard deviations for the adenocarcinoma samples in Harvard, Michigan, and Stanford, respectively, and k⋆, c⋆ and r⋆ are parameters in the simulation procedure. Letting xgsp denote the original adenocarcinoma expression values, we generate the corresponding artificial data as
| (12) |
We consider a gene g as differentially expressed if . Differential expression is concordant if have the same sign in all studies and discordant if have opposing signs. Concordant and discordant differential expression are special cases of differential expression that we consider separately. Note that the simulation parameters r⋆, c⋆, and k⋆ control the proportion of differentially expressed genes that are concordant in the simulation. For instance, increasing r⋆ and c⋆ has the effect of increasing the percentage of concordantly differentially expressed genes. Table 1 provides a complete listing of the simulation settings evaluated. Table 2 illustrates the possible patterns of differential expression for P = 2 studies and G = 4 genes.
Table 2.
A trivial example of a dataset with four genes and two studies. For each gene, we evaluate three possible truths for differential expression defined over the set of studies: concordant differential expression in both studies (𝒞g = 1), discordant differential expression (𝒟g = 1), and differential expression that is either concordant or discordant across studies (𝓔g = 1).
| gene | δ* | sign (Δ*) | ε | 𝒞 | 𝒟 |
|---|---|---|---|---|---|
| 1 | 0 | · | 0 | 0 | 0 |
| 2 | 1 | {−,−} | 1 | 1 | 0 |
| 3 | 1 | {−, +} | 1 | 0 | 1 |
| 4 | 1 | {+,+} | 1 | 1 | 0 |
simulation
We modified the algorithm to simulate study-specific differential expression as follows. First, we simulated Δ⋆ for all of the genes. Secondly, for genes with greater than the 0.9 quantile of the distribution, we set . Notice that correlation between the elements of induces a correlation between the elements of . Finally, we simulated Δ⋆ a second time to obtain Δ⋆ independent of δ⋆. The simulated expression data was generated as in Equation 12, replacing with study-specific indicators, . Following the above algorithm, we generated four datasets using the settings of Simulations A, F, J, and O in Table 1.
6.1.2 Evaluation procedures
For each of the Simulations A-R in Table 1, we develop summary measures, referred to as scores, to quantify concordant (𝒞), discordant (𝒟), or the union of (differentially) expressed () genes. Section 4 discusses the summary statistics proposed for the Bayesian model, as well as alternative methods for summarizing differential expression. We emphasize that for a gene g, 𝒞g, 𝒟g, and g are defined on a set of studies, as opposed to differential expression in a single study.
Let score*(g) denote any of the scores defined in Sections 4.1 (PM*) and 4.2 (u*) for a gene g. If, for a fixed threshold a > 0, we classify each g as being (overall, concordantly or discordantly) differentially expressed if score*(g) > a, then we obtain a standard two-by-two table containing the number of false negatives FN*(a), false positives FP*(a), true negatives TN*(a), and true positives TP*(a) for that particular value of a. For example, in the case of differential expression () the number of true negatives is given by
| (13) |
and the remaining entries of the table for differential expression are defined in a similar fashion. The false positive and true positive rates associated with the statistics score*(g) and threshold a are given by
| (14) |
respectively. Plotting FPR*(a) against TPR*(a) as a varies produces the standard receiver operating characteristic (ROC) curve associated with the statistics score*(g). The area under the ROC curve, AUC, is a nonparametric measure of the quality of the statistic, with values close to unity (i.e., a statistic that simultaneously achieves FPR close to zero and TPR close to one) being the best.
As an alternative to ROC curves, which are based on false and true positive rates, we also considered the false discovery rate (FDR) of the statistics score*(g) as a function of the number of genes determined to be differentially expressed. Specifically, for each threshold a, we plotted the number of discoveries, , against
As expected, the FDR increases as the number of overall discoveries increases. Curves close to the horizontal axis are preferable to those having a more rapid increase of FDR with the number of discoveries. Similarly, we plotted the number of non-differentially expressed genes, , against the missed discovery rate
Again, curves close to the horizontal axis are preferable to those having a more rapid increase of MDR with the number of negative discoveries.
6.1.3 Results
Following the algorithm for simulating , we generated artificial datasets for Simulations A - R in Table 1. Initial model parameter values for single indicator implementation of XDE were chosen to specify little prior knowledge: αa = βa = αb = βb = 1, , νρ = νr = 4 and αξ = βξ = 1. The values for the tuning parameters in the Metropolis–Hastings algorithm were chosen to achieve a robust algorithm, not to optimize convergence and mixing properties for this particular data set. In all updates of type (3) we used ε = 0.01. For updates of type (4) we used ε = 0.5 in updating and φgp, and ε = 0.1 in updating lp and λp. In updates of types (5), (6) and (7) we used ε = 0.1, ε = 0.05 and ε = 0.02, respectively. To monitor convergence and mixing properties, we inspected trace plots of the various simulated variables, as in Supplementary Figure 1. We observed that most parameters converge relatively quickly, and that the model parameters coincide in many cases with the true values in the simulation. For instance, 10% of the genes were simulated to be differentially expressed in Simulation A (ξ* = 0.10) and traceplots of the ξ parameter in the Bayesian model show that this parameter has converged to a value near 0.12.
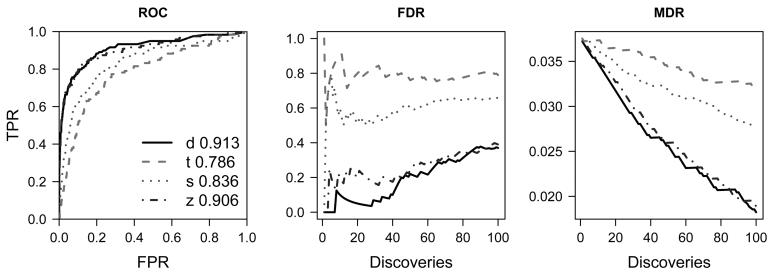
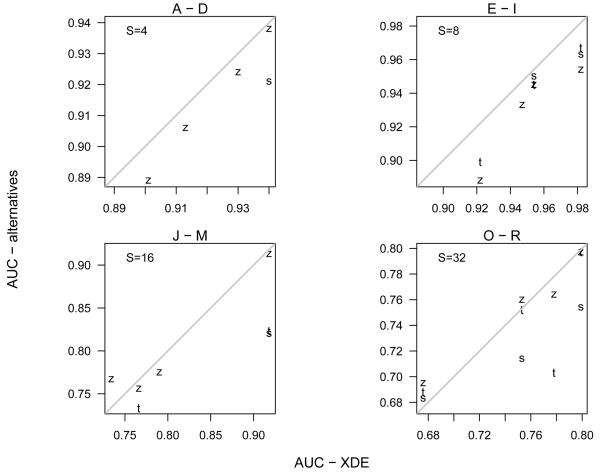
For each simulated dataset, performance of the cross-study scores were assessed by the AUC, FDR, and MDR criteria. A graphical display of the results for Simulation A is shown in Figure 2. The Bayesian model has a higher AUC (panel 1), as well as a lower FDR and MDR than the alternative scores over a range of cut-offs for evaluating 𝒞 (panels 2 and 3, respectively). Because of the extensive nature of the simulations, we visually assess the relative performance of the Bayesian method to the alternative methods via scatterplots of the AUC (e.g., Figure 3). Points beneath the identity line are simulations in which the Bayesian score had a higher AUC than an alternative method evaluated on the same dataset. Figure 3 plots the AUCs for 𝒞 in Simulations A-R. The corresponding AUC statistics for and 𝒟 are provided in Supplementary Figures 3(a) and 3(b). To assess the sensitivity of the AUC to the random number seed used for simulating and , each panel of Supplementary Figure 2 displays a scatterplot of the AUCs for multiple datasets generated from the same simulation parameters.
Figure 2.
Performance diagnostics for scores quantifying 𝒞 in Simulation A. The letter d in the legend corresponds to the Bayesian score. Although the SAM-score does markedly better than the t-score when the the individual studies are small (S = 4), considerable improvement can be obtained by a more formal borrowing of strength across studies in the Bayesian and z-scores.
Figure 3.
The AUC for concordant differential expression in Simulations A - D (top left), E - I (top right), J - M (bottom left), and O - R (bottom right) was calculated for each of the alternative methods (t, SAM, and z) and plotted against the AUC obtained from the single indicator imple-mentation of the Bayesian model. The diagonal line in each panel is the identity. The lower limit for the axes are based on the minimum of the AUC's from the Bayesian and z-scores; hence the t and SAM scores are not always plotted. See Table 1 for the simulation parameters.
To evaluate the relative performance of the single and multiple indicator implementations of the Bayesian model, we generated 8 datasets using the settings of Simulation A, F, J, and O in Table 1 and the and simulation algorithms. The single indicator implementation generally had higher sensitivity and specificity for assessing concordant differential expression than the multiple indicator implementation when differential expression was simulated for all of the studies or in none of the studies (Row 1, Figure 4). Row 2 of Figure 4 displays a scatterplot of the AUCs when differential expression was simulated in a subset of the studies (). Note that both the single and multiple indicator implementations have higher AUCs than the alternative methods, irrespective of sample size, for concordant-, discordant-, and differential-expression when differential expression was simulated in a subset of the studies.
Figure 4.
We assessed the relative performance of the single and multiple indicator implementations of the Bayesian model through simulation using AUC as a measure of performance. Row 1:. Differential expression was simulated using a single indicator of differential expression for all studies, ; row 2: study-specific indicators of differential expression, , were simulated. Each panel displays the AUC from the single indicator model versus the AUC from the multiple indicator model, “X”, and the maximum of the t-, s-, and z-scores, “o”, for simulation settings A, F, J, and O (Table 1) . When simulating differential expression in a subset of studies (row 2), the single and multiple indicator implementations of the Bayesian model have higher AUCs than the alternative methods for assessing concordant-, discordant-, and differential-expression irrespective of sample size.
In general, the Bayesian model outperforms the three alternative methods for cross-study analysis of differential gene expression across a range of simulated parameters (Figure 3). Our overall assessment does not appear to be sensitive to the random quantities simulated in these datasets (Figure 2). As the sample sizes of the individual studies increase, the relative benefit of borrowing strength across genes and studies in the hierarchical model diminishes. Instances in which the z-score has a better AUC than the corresponding Bayesian statistic (e.g., panel (2,1) in Figure 3) occurred only when differential expression was simulated in all of the studies through a single indicator, , and most often occurred when the simulated data was particularly noisy and the AUC from all methods were at the low-end of the range. In such instances, scatterplots of the study-specific effect sizes were largely uncorrelated (data not shown). Scatterplots of a study-specific statistic for effect-size, such as t, may be a useful indicator of whether the Bayesian model is likely to improve on simpler alternatives. In instances where the data is negatively correlated across studies, this may induce the ‘wrong’ borrowing of strength. When simulating study-specific differential expression, , both implementations of the Bayesian model performed better than the alternative methods over the range of simulations evaluated. In simulated datasets with no signal (data not shown), gene-specific posterior probabilities in the Bayesian model were approximately zero.
6.2 Split study validation
To assess the baseline behavior of XDE, we split the Huang study into four disjoint parts, treating each part as an independent study. We randomly assigned 5 estrogen receptor (ER) negative and 16 ER positive samples to each split. In this simplified setting, we avoid the potential difficulties of cross-platform analyses that can arise from technological and/or biological differences between studies. For instance, differences in the annotation of the probes or ethnic composition of the study populations may each contribute to discrepant results in a meta-analysis, but such concerns are reduced when splitting a single study. Split study validation has been used by others to assess meta-analytic methodologies for gene expression analysis. In particular, Gentleman et al. (2005) use split study validation to illustrate their implementation of the cross-platform statistic introduced by Choi et al. (2003).
After fitting the single indicator implementation of the Bayesian model to the four splits, traceplots for the parameters a, b, l, t, γ2, c2, τ2, ξ, ρ, and r (each of which are updated by Metropolis-Hastings proposals) were used to evaluate convergence (see Supplementary Figure 4). We define the Bayesian effect size BES for gene g and platform p, by , and use this as a study-specific Bayesian estimate of differential expression, contrasting it with the z, t, and SAM statistics. Scatterplots of the study-specific t-, z-, and BES statistics are shown in Supplementary Figure 5. If we consider the t, SAM (not shown), and z statistics as evidence of differential expression in a single study, we observe that the evidence is study-dependent with only moderate correlation of these statistics across the splits (Supplementary Figures 5(c) and 5(d)). Hence, scatterplots of the study-specific statistics provide two important pieces of information: first, even in a scenario that minimizes inter-study discordance, the variation across studies of the effect size statistics underscore the difficulty of identifying genes that show consistent evidence of differential expression; secondly, while the scatterplots do not lend themselves directly to identifying a list of genes for follow-up, the moderate correlation among the study-specific statistics does motivate an approach that uses the information from all of the studies.
A set of concordant differentially expressed genes emerges from the visualization of the BES scatterplots in Supplementary Figure 5(b). Through modeling the inter-relationships of genes and studies at higher levels of the model, the Bayesian model shrinks noisy genes to zero without requiring extensive filtering prior to the analysis. The cigar-shaped pattern in Supplementary Figure 5(b) is typical when fitting the Bayesian model, though the correlation is higher than what one may expect to observe when the studies are independent and use different platforms (see Section 7). In choosing a list of genes to follow for subsequent laboratory investigation, the PM𝒞(g), displayed in Supplementary Figure 5(a), can be used to rank the evidence of concordant differential expression.
Validation of microarray experiments typically involves assaying the RNA transcript abundance of selected genes by using low-throughput platforms, such as qRT-PCR. As the PM𝒞(g) identifies genes whose differential expression is relatively study- and/or platform-independent, validation of the gene list selected by PM𝒞(g) may be less likely to result in false discoveries, as suggested by the simulations in the previous section. Thus, meta-analysis has the potential to reduce the cost of downstream analyses. Moreover, meta-analysis enables an impartial investigator not directly associated with the primary studies to synthesize the information that the primary studies contain. The Bayesian model enables one to identify genes and pathways that are concordantly effected across studies, as well as the genes and pathways that appear discordantly regulated. Whether the goal is to produce a gene list that is likely to be validated by other platforms, or to explore the biology underlying concordance and discordance, the Bayesian model provides a useful means of achieving these goals.
7 Experimental data example
Estrogen receptor
Estrogen receptor is an important risk factor for breast cancer tumori-genesis. Several gene expression studies have collected phenotypic information on estrogen receptor (ER) status (positive or negative). In this section, we fit the Bayesian model to four publicly available datasets described in Huang et al. (2003) (Huang), Hedenfalk et al. (2001) (Hedenfalk), Farmer et al. (2005)(Farmer), and Sorlie et al. (2001) (Sorlie), using ER status as the clinical variable. Table 3 shows the distribution of ER status in the four breast cancer studies. See Section 5 for a brief description of the data and pre-processing steps. Integration of the breast cancer studies provides an opportunity to define a ranked list of differentially expressed genes that is potentially more complete, and less likely to be platform-dependent, than lists derived from a single study. Because the studies involve different gene expression platforms, we cross-reference the study-specific gene annotations by Entrez-gene identifiers and focus our discussion on the set of 2064 Entrez genes that were present in each of the four studies.
Table 3.
Distribution of the estrogen receptor in the three studies.
| platform | ER− | ER+ | |
|---|---|---|---|
| Hedenfalk | cDNA | 6 | 10 |
| Sorlie | cDNA | 30 | 81 |
| Farmer | Affymetrix hu133a | 22 | 27 |
| Huang | Affymetrix hu95av2 | 23 | 65 |
Model fit
When fitting the Bayesian hierarchical model to the breast cancer datasets, we found it unnecessary to change the hyperparameters and tuning parameters for the Metropolis–Hastings algorithm from their default values (see Table 1). To monitor the convergence and mixing properties of the Markov chain, we used visual inspection of the trace plots of the various simulated variables. The slowest convergence and mixing properties occurred for the four hyper-parameters θp, λp, tp and lp, see for example the trace plots of lp, p = 1, 2, 3 in Supplemental Figure 6. A burn-in of 5000 iterations is sufficient for convergence in most instances, but this should be evaluated on a case by case basis.
Results
We calculated posterior statistics using every 20th iteration after 2000 iterations of burn-in. The scatterplot in Figure 5 displays the distribution of the posterior means for the indicators of concordant differential expression (x-axis) and discordant differential expression (y-axis). We use a grey-scale to display the gradient of posterior means for differential expression. See Supplemental Figure 5 for a color gradient. In the discussion that follows, we discuss the gradient in terms of evidence for differential expression, ranging from uncertainty (near 0.5) in light grey to strong evidence (near 1) in black.
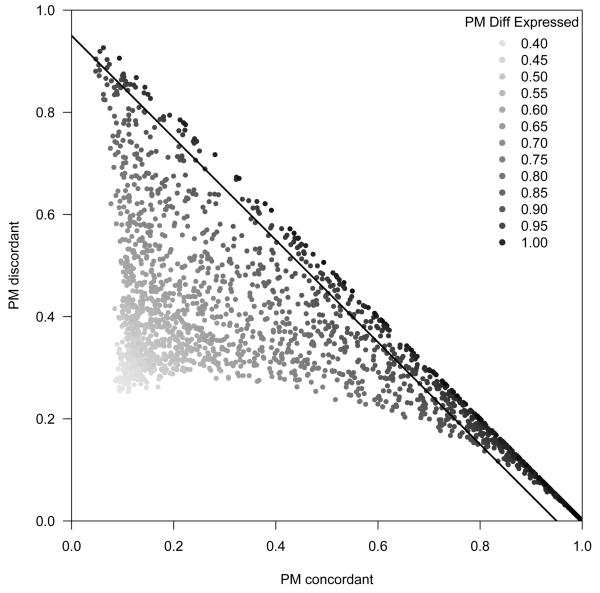
Figure 5.
A scatterplot of the posterior means (PM) for the indicators of concordant (x-axis) and discordant (y-axis) differential expression. Plotting symbols are color coded by the gradient of the PM for the differential expression indicators. In purple, are genes for which the model is uncertain regarding differential expression. Here, the model has difficulty distinguishing between low levels of differential expression and no differential expression. Genes with strong evidence of differential expression above the diagonal PMε(g) = 0.95 line are predominantly concordant across studies.
Our model finds strong evidence of differential expression in a moderate number of genes: 30.9% of the genes have PMε(g) > 0.95, and among these genes we observe more concordance than discordance, as reflected by the relative density of genes at (x, y) coordinates (1,0) versus (0, 1) in the scatterplot. The remaining genes show moderate to weak evidence (uncertainty) of differential expression. We note that our model has difficulty distinguishing between no differential expression and low levels of differential expression in the ER dataset. Recall that the prior for Δ is a multivariate normal distribution with mean zero and overall variance parametrized by c2 (Section 2). The posterior mean for c2 in the ER dataset is approximately 0.028 (Supplementary Figure 6), and this value has several related implications. First, when the variance of Δ is very small, values of δg = 1 or δg = 0 do not substantially effect the likelihood. That both δg = 0 and δg = 1 are plausible is reflected by posterior means of differential expression near 0.5. Therefore, the proportion of differentially expressed genes (ξ) is directly tied to c2, with high values of ξ corresponding to small c2 and low values of ξ corresponding to large c2.
In the ER dataset, the posterior mean of ξ is 0.77, although a much smaller proportion of genes show strong evidence of differential expression. The software implementation of the Bayesian model flags instances of small c2, alerting the user of the potential for inflated ξ. Evaluating different priors for Δ is a future direction of this research.
The main conclusions of the analysis are only affected by the ranking of the PMε(g). Estimates of concordant differential expression, the most common goal of most integration efforts, appear to be unaffected by large values of ξ. For example, the posterior expected proportion of false positives for the experimental data (as estimated using the methods in Efron and Tibshirani (2002)), was low for a range of PM𝒞(g) cutoffs. In particular, the posterior expected proportion of false positives using thresholds of 0.5 and 0.9 for PM𝒞(g) were 0.22 and 0.04, respectively.
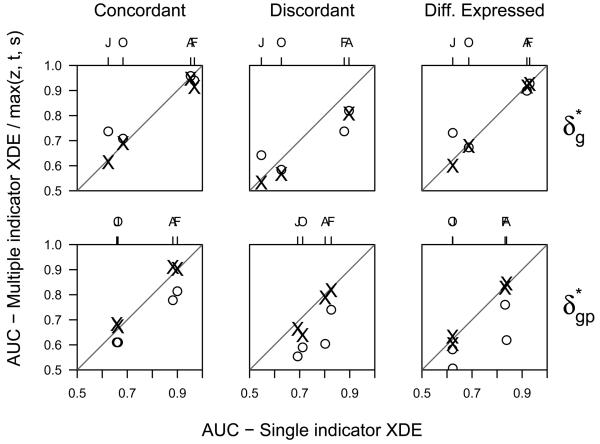
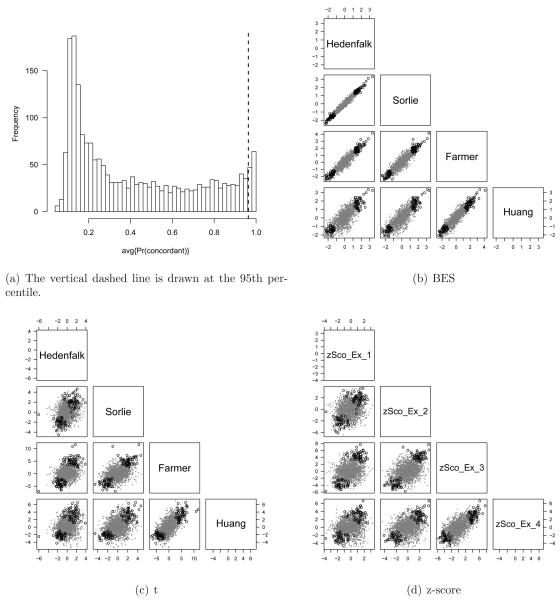
We separately explored concordant and discordant differential expression among the four ER data sets under study, combining visualizations that are effective for summarizing overall reproducibility (pairwise scatterplots of effect size) with statistics from the Bayesian model that can be used to target a specific subgroup of genes that appear to be concordantly (Figure 6) or discordantly (Figure 7) regulated in the different studies. Genes in the 95th percentile of PM𝒞(g) (to the right of the vertical dashed line in Figure 6(a)) are plotted with a different symbol (circles) and color (black) in the pairwise scatterplots of BES, t-, and z-statistics. The Bayesian model shrinks noisy estimates of the effect size towards zero (panel b, Figure 6(b)), whereas genes with stronger evidence of concordant differential expression are shrunk less and appear in the upper right and lower left quadrants of the pairwise scatterplots of BES in panel b.
Figure 6.
Ranking genes by the PM𝒞(g) is useful for exploring inter-study agreement of differential expression. Here, we threshold genes by the 95th percentile of the distribution for the posterior average of concordant differential expression, PM𝒞(g) (panel a). Pairwise scatterplots of the study-specific statistics for the four breast cancer studies are provided in panels b - d. A different plotting symbol (circles) and color (black) is used for the genes in the highest decile of PM𝒞(g). The posterior expected proportion of false positives corresponding to this threshold is approximately 0.04.
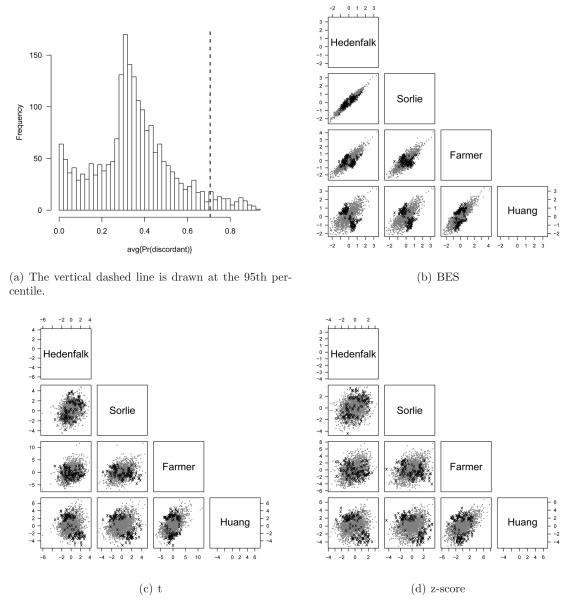
Figure 7.
The posterior average of the probability of discordant differential expression, PM𝒟(g) (panel a), can be used to explore discordance. Here we threshold pairwise scatterplots of the study-specific statistics from the four breast cancer studies (panels c and d) by the 95th percentile of the PM𝒟(g) distribution (panel a). Again, we use a different plotting symbol (x) and color (black) for genes surpassing this threshold to emphasize the discordance. In particular, note that almost all of the discordance in the scatterplots of panels b - d arise from pairwise comparisons of cDNA platforms (Sorlie and Hedenfalk) to the Affymetrix platforms (Farmer and Huang). For the two scatterplots comparing more similar platforms (Sorlie versus Hedenfalk and Farmer versus Huang), the effect size estimates of the highlighted genes are positively correlated.
Figure 7 explores discordance. Panels b - d are the same as in Figure 6, but with an emphasis on inter-study discordance identified by thresholding the upper 5% of the PM𝒟(g) distribution (genes to the right of the vertical dashed line in Figure 7(b)). Again, emphasis is placed on a subset of genes through different plotting symbols (x) and color (black). Note that almost all of the discordance in the scatterplots shown in Figure 7b - d arise from pairwise comparisons of cDNA platforms (Sorlie and Hedenfalk) to the Affymetrix platforms (Farmer and Huang). Discordance between Affymetrix and cDNA platforms may arise, for instance, as a result of probes hybridizing to different transcripts from the same gene. Note that in the scatterplots comparing like platforms, Sorlie versus Hedenfalk (both cDNA) and Farmer versus Huang (both Affymetrix), the effect size estimates of the highlighted genes are positively correlated.
8 Closing remarks
In this paper, we define a hierarchical Bayesian model for microarray expression data collected from several studies, and use it to identify genes that show differential expression between two phenotypic conditions. Two implementations of this model are available in the R package XDE available from the Bioconductor website (http://www.bioconductor.org). The first implementation uses a single indicator for differential expression that summarizes information across studies. The second implementation allows multiple indicators for differential expression, permitting differential expression of a gene in some of the studies and not in other studies.
We evaluated the performance of the single and multiple indicator models using artificial and experimental data. The simulation results from the artificial data demonstrate the advantages of a Bayesian model. Compared to a more direct combination of t- or SAM-statistics, the 1 − AUC values for the Bayesian model are roughly half of the corresponding values for the t- and SAM-statistics. Furthermore, the simulations provide guidelines for when the Bayesian model is most likely to be useful. In small studies the Bayesian model generally outperforms other methods when evaluated by AUC, FDR, and MDR across a range of simulation parameters, and these differences diminish for larger sample sizes in the individual studies. When differential expression was simulated in a subset of the studies, the Bayesian model outperformed the alternative methods irrespective of sample size. In addition, we carried out a ”split-study” validation, which provides a model-free assessment of the method's behavior in the absence of platform differences. The split-study validation illustrates appropriate shrinkage of the Bayesian model in the absence of confounding platform and annotation based differences. Split-study validation may also provide a useful context for exploring how the ranks resulting from integration efforts differ across a set of genes known to be involved in a particular pathway. For example, using the multiple indicator implementation of the Bayesian model and a split of the Farmer study into three artificial studies, we can compare the ranks of genes known to be modulated by estrogen in an analysis of estrogen receptor (ER) status (positive or negative) as the clinical covariate. Supplementary Figure 8 explores this idea, with estrogen-related genes stratified into three categories according to the direction of regulation by estrogen (up or down) and whether the gene has a known transcription factor binding sites (TFBS) in the promoter.
Using experimental data from four high-throughput gene expression studies for breast cancer and ER status as the clinical covariate, posterior averages from the Bayesian model may be used to identify subsets of genes to explore in-silico. Figure 7 identified a subset of genes in the breast studies that were discordant across platforms (cDNA versus Affymetrix) but remain positively correlated within a platform (cDNA versus cDNA and Affymetrix versus Affymetrix). Such discordance can provide information about the differential expression of alternatively transcribed genes. For example, consider the genes in the discordant set that have two or more alternative transcripts. If for any of these genes the target sequence for the cDNA platform lies on transcript A and the target sequence(s) of the Affymetrix platform lies on transcript B, discordance could indicate, for example, that transcript A is up-regulated and transcript B is down-regulated across the two biological conditions. Because such in-silico hypotheses can only be validated by laboratory based methods such as qRT-PCR, we leave this as an open thread for future investigation.
Of the models previously proposed in the literature, the model of Conlon et al. (2006) is conceptually closest to ours. The Conlon model is designed for cross-study, within-platform analyses, and is not directly applicable to the case studies in our article. However, it is useful to contrast the technical features of the two approaches. Both are hierarchical Bayesian models, and both have a differential expression indicator for each gene. Differences emerge in how each model handles the increased variation in expression values for differentially expressed genes. We assign separate distributions to the expression values of samples in each condition. Conlon et al. (2006) assume that the expression values for differentially expressed genes are independent, but with an increased variance; they do not make use of the condition information for each sample. In addition, we adopt a more refined and flexible model for the covariance structure of the expression values and an implementation that allows study-specific indicators of differential expression. A practical consequence of these differences is in the application of the models to gene expression data from different platforms. In particular, our model can be fit to multiple studies regardless of platform, whereas the model of Conlon et al. (2006) is most applicable for combining biological replicates from a single platform; their model could be viewed as a special case of our single indicator implementation in which the technological differences in scale and variation are close to zero and there is conjugacy between location and scale.
Our hierarchical model does not require that studies be measured on the same platform. This generality has advantages and disadvantages. One advantage is that we model differences in scale and variation of expression intensities across platforms directly, removing some of the need for extensive normalization and rank-based approaches to assessing differential expression. However, in any multi-study analysis, discordance can arise from biological differences in the sample populations of each study, as well as technological effects related to the design and implementation of specific array technologies. Through multi-level modeling of gene expression, we borrow strength across studies and genes, by shrinking noisy estimates to zero and capturing correlated signals from the different studies. Simple scatterplots of study-specific measures of effect size, such as the SAM statistic, are a simple diagnostic for whether there is information in the joint distribution of the signals and can be evaluated before fitting the Bayesian model. In the typical situation, one will see a cloud of effect size statistics near zero, and positive correlation evidenced by having most scatter points in the positive (+, +) and negative (−, −) quadrants. In this case, the Bayesian model will tend to shrink the cloud of noise to zero, and (correctly) provide less shrinkage of the concordant differentially expressed genes.
It is common in the analysis of high-throughput gene expression data to apply a gene-selection procedure prior to the formal analysis of differential expression. For instance, when estimating differential expression by a statistic that has in its denominator an estimate of the across-sample variation, one may wish to remove genes of low abundance that show very low across-sample variation. In our Bayesian model, each gene has a parameter representing the numerical value of its differential expression. The priors for these parameters have a point mass at zero, corresponding to no differential expression. This feature of the model reduces the need for initial gene filtering, and facilitates the direct consideration, via ranked posterior means, of the complete set of available genes. See also the discussion in Ishwaran and Rao (2003, 2005).
When fitting the Bayesian model to pure noise, the model behaves appropriately and the estimated proportion of differentially expressed genes (the union of concordant and discordant genes) is approximately zero (data not shown). Additionally, the simulated data examples illustrate that the proportion of differentially expressed genes, as estimated by the posterior mean of ξ, is typically calibrated. Nevertheless, in the experimental data example the posterior average of ξ was 0.77. Closer inspection reveals that the Bayesian model has difficulty distinguishing between genes with low levels of differential expression and no differential expression (Figure 5). This difficulty can be diagnosed by high ξ values and a small posterior mean for the variance of the offsets as discussed in Section 7. The main conclusions of our analysis are affected only by the overall ranking of the posterior means for differential expression.
Our Bayesian model can be modified and generalized in several respects. First, the possibility of missing gene expression observations can easily be included. The missing xgsp can simply be integrated out from the posterior distribution. Second, in the current model we have used the (common) genes appearing in all the studies. Partly overlapping gene sets can be handled in the model by considering expression values corresponding to genes that are not present in a study as missing. However, in order to design a more efficient computational algorithm one should integrate out both these xgsp's and the corresponding from the model, which can be carried out with minor modifications. Third, it is also straightforward to allow for missing observations in the phenotype. In this case, we need to assign an additional probabilistic model for the ψsp's and simulate the unobserved ones within the Metropolis–Hastings algorithm. This will in effect produce a prediction of the unobserved clinical variables. However, if the number of unobserved clinical variables is large, we expect it to be necessary to use block updates in the Metropolis–Hastings algorithm to avoid slow convergence and mixing.
Our results provide a strong indication that borrowing strength across both genes and studies can be effective in the analysis of multi-platform studies. As is the case for most complex multilevel models, this comes at the price of added computational effort, and an increased burden of proof that the modeling assumptions are tenable.
Supplementary Material
9 Acknowledgments
RBS was supported by the training grant 5T32ES012871 from the U. S. National Institute of Environmental Health Sciences (P. I. Thomas Louis), training grant 5T32HL007024 from the National Heart, Lung, and Blood Institute, and grant DMS034211 from the National Science Foundation (P. I. Giovanni Parmigiani). Andrew Nobel's research was supported in part by NSF Grant DMS 0406361 and EPA Grant RD-83272001.
References
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: Regularized t–test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Barnard J, McCulloch RR, Meng X-L. Modelling Covariance Matrices in Terms of Standard Deviations and Correlations, with Application To Shrinkage. Statistica Sinica. 2000;10:1281–1311. [Google Scholar]
- Beer DG, Kardia SL, Huang C-C, Giordano TJ, David E, Misek AML, Lin L, Chen G, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature Medicine. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Berger JO. Statistical Decision Theory and Bayesian Analysis. Springer; 1993. [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröet P, Richardson S, Radvanyi F. Bayesian hierarchical model for identifying changes in gene expression from microarray experiments. Journal of Computational Biology. 2002;9:671–683. doi: 10.1089/106652702760277381. [DOI] [PubMed] [Google Scholar]
- Caffo B, Dongmei L, Parmigiani G. Technical report. Vol. 62. Johns Hopkins University, Dept. of Biostatistics; 2004. 2004. Power conjugate multilevel models with applications to genomics. [Google Scholar]
- Choi H, Shen R, Chinnaiyan A, Ghosh D. A Latent Variable Approach for Meta-Analysis of Gene Expression Data from Multiple Microarray Experiments. BMC Bioinformatics. 2007;8(1):364. doi: 10.1186/1471-2105-8-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Yu U, Kim S, Yoo OJ. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19-1:I84–I90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- Conlon E. A Bayesian mixture model for metaanalysis of microarray studies. Funct Integr Genomics. 2007 doi: 10.1007/s10142-007-0058-3. [DOI] [PubMed] [Google Scholar]
- Conlon E, Song JJ, Liu J. Bayesian models for pooling microarray studies with multiple sources of replications. BMC Bioinformatics. 2006;7(1):247. doi: 10.1186/1471-2105-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EM, Song JJ, Liu A. Bayesian meta-analysis models for microarray data: a comparative study. BMC Bioinformatics. 2007;8(80) doi: 10.1186/1471-2105-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium MAQC, Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu T-M, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, hui Fan X, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li Q-Z, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaportas P, Roberts GO. An introduction to MCMC. In: J. Møller., editor. Spatial Statistics and Computational Methods. Springer; Berlin: 2003. pp. 1–41. number 173 in Lecture Notes in Statistics. [Google Scholar]
- Do K-A, Müller P, Tang F. A Bayesian mixture model for differential gene expression. Journal of the Royal Statistical Society: Series C (Applied Statistics) 2005;54(3):627–644. [Google Scholar]
- Efron B, Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genetic Epidemiology. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes Analysis of a Microarray Experiment. Journal of the American Statistical Association. 2001;96:1151–1160. [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz A-L, Brisken C, Fiche M, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24(29):4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Mayer E, Parmigiani G, Zhong X, Cope L, Gabrielson E. Cross-study validation and combined analysis of gene expression microarray data. Biostatistics. 2007 doi: 10.1093/biostatistics/kxm033. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Ruschhaupt M, Huber W. Technical Report 8. The Berkeley Electronic Press; 2005. On the synthesis of microarray experiments. http://www.bepress.com/bioconductor/paper8. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Barette TR, Rhodes D, Chinnaiyan AM. Funct Integr Genomics. 4. Vol. 3. Journal Article Meta-Analysis; 2003. Statistical issues and methods for meta-analysis of microarray data: a case study in prostate cancer; pp. 180–8.pp. 1438–793X. (Print) [DOI] [PubMed] [Google Scholar]
- Gottardo R, Pannucci J, Kuske C, Brettin T. Statistical analysis of microarray data: a Bayesian approach. Biostatistics. 2003;4:597–620. doi: 10.1093/biostatistics/4.4.597. [DOI] [PubMed] [Google Scholar]
- Green P. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Hastings W. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
- Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24(31):5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O, Wilfond B, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344(8):539–48. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Hedges LV, Olkin I. Statistical Methods for Meta-analysis. Academic Press; 1985. [Google Scholar]
- Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–6. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen MH, Gray RJ. Bayesian Models for Gene Expression with DNA Microarray Data. Journal of the American Statistical Association. 2002;97:88–99. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JGN, Geoghegan J, Germino G, Griffin C, Hilmer SC, Hoffman E, Jedlicka AE, Kawasaki E, Martnez-Murillo F, Morsberger L, Lee H, Petersen D, Quackenbush J, Scott A, Wilson M, Yang Y, Ye SQ, Yu W. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2(5):345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Rao J. Detecting differentially expressed genes in microarrays using Bayesian model selection. Journal of the American Statistical Association. 2003;98(462):438–455. [Google Scholar]
- Ishwaran H, Rao J. Spike and slab gene selection for multigroup microarray data. Journal of the American Statistical Association. 2005;100(471):764–780. [Google Scholar]
- Johnson W, Li C, Rabinovic A. Adjusting batch effects in microarray data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jung Y-Y, Oh M-S, Shin DW, Kang S-H, Oh HS. Identifying differentially expressed genes in meta-analysis via Bayesian model-based clustering. Biom J. 2006;48(3):435–450. doi: 10.1002/bimj.200410230. [DOI] [PubMed] [Google Scholar]
- Kendziorski C, Newton M, Lan H, Gould M. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Statistics in Medicine. 2003;22:3899–3914. doi: 10.1002/sim.1548. [DOI] [PubMed] [Google Scholar]
- Kerr K. Extended analysis of benchmark datasets for Agilent two-color microarrays. BMC Bioinformatics. 2007;8(1):371. doi: 10.1186/1471-2105-8-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Parmigiani G, Caffo B. Technical report. Vol. 34. Johns Hopkins University, Department of Biostatistics; 2004. 2004. Screening for Differentially Expressed Genes: Are Multilevel Models Helpful? p. 34. [Google Scholar]
- Lönnstedt I, Speed T. Replicated Microarray Data. Statistica Sinica. 2002;12(1):31–46. [Google Scholar]
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equations of state calculations by fast computing machine. J. Chem. Phys. 1953;21:1087–1091. [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30(1):13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Newton M, Noueiry A, Sarkar D, Ahlwuist P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics. 2004;5:155–176. doi: 10.1093/biostatistics/5.2.155. [DOI] [PubMed] [Google Scholar]
- Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: Improving statistical inference about gene expression changes from microarray data. Journal of Computational Biology. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- Pan W. A Comparative Review of Statistical Methods for Discovering Differentially Expressed Genes in Replicated Microarray Experiments. Bioinformatics. 2002;18:546–554. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- Parmigiani G. Measuring uncertainty in complex decision analysis models. Stat Methods Med Res. 2002;11(6):513–537. doi: 10.1191/0962280202sm307ra. [DOI] [PubMed] [Google Scholar]
- Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10(9):2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Cancer Res. 15. Vol. 62. Journal Article Meta-Analysis; 2002. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer; pp. 4427–33. 0008-5472 (Print) [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Proc Natl Acad Sci U S A. 25. Vol. 101. Journal Article Meta-Analysis; 2004. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression; pp. 9309–14. 0027-8424 (Print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA, Tjelmeland H, Fan C, Perou CM, Nobel AB. Merging two gene-expression studies via cross-platform normalization. Bioinformatics. 2008;24(9):1154–1160. doi: 10.1093/bioinformatics/btn083. [DOI] [PubMed] [Google Scholar]
- Shen R, Ghosh D, Chinnaiyan A. Prognostic meta-signature of breast cancer developed by two-stage mixture modeling of microarray data. BMC Genomics. 2004;5(1):94. doi: 10.1186/1471-2164-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AFM, Roberts GO. Bayesian Computation Via the Gibbs Sampler and Related Markov Chain Monte Carlo Methods (Disc: P53-102) Journal of the Royal Statistical Society, Series B, Methodological. 1993;55:3–23. [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Townsend J, Hartl D. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple treatements or samples. Genome Biology. 2002;3:research0071.1–71.16. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng G, Oh M, Rohlin L, Liao J, Wong W. Issues in cDNA Microarray Analysis: Quality Filtering, Channel Normalization, Models of Variations and Assessment of Gene Effects. 2001 doi: 10.1093/nar/29.12.2549. Submitted to Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Coombes KR, Highsmith WE, Keating MJ, Abruzzo LV. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: a meta-analysis of three microarray studies. Bioinformatics. 2004;20(17):3166–3178. doi: 10.1093/bioinformatics/bth381. [DOI] [PubMed] [Google Scholar]
- Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet. 2002;31(3):255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- Zhong X, Marchionni L, Cope L, Iversen ES, Garrett-Mayer ES, Gabrielson E, Parmigiani G. Technical Report 129. johns Hopkins University Department of Biostatistics; 2007. Optimized Cross-study analysis of microarray based predictors. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.