Abstract
Here, we describe a new mechanism of host defense that involves the nuclear factors associated with dsRNA (NFAR1 [90 kDa] and NFAR2 [110 kDa]), which constitute part of the shuttling ribonuclear protein (RNP) complex. Activation of the dsRNA-activated protein kinase PKR by viral RNA enabled phosphorylation of NFAR1 and NFAR2 on Thr 188 and Thr 315, an event found to be evolutionarily conserved in Xenopus. Phosphorylated NFAR1 and NFAR2 became dissociated from nuclear factor 45 (NF45), which was requisite for NFAR reshuttling, causing the NFARs to be retained on ribosomes, associate with viral transcripts, and impede viral replication. Cre-loxP animals with depletion of the NFARs in the thymus were exquisitely sensitive to the cytoplasmic replicating virus VSV (vesicular stomatitis virus). Thus, the NFARs constitute a novel, conserved mechanism of host defense used by the cell to detect and impede aberrant translation events.
Keywords: NFAR, mRNP export, translation, PKR, host defense
The dsRNA-binding protein (DRBP) family consists of evolutionarily conserved proteins that interact in a sequence-nonspecific manner with dsRNA species and regulate a diverse array of cellular processes in the cell (Saunders and Barber 2003). DRBP members include RNase III, DICER DGCR8, and TRBP (HIV-1 TAR RNA-binding protein), which are known to regulate microRNA processing and function, and the ADAR (adenosine deaminase acting on RNA) family, which converts adenosine to inosines in dsRNA substrates and can influence protein coding (Bernstein et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Gregory et al. 2004; Han et al. 2004; Chendrimada et al. 2005; Melcher et al. 1996). A further member of the dsRNA-binding domain (DRBD) family, which plays an important role in host defense, is the interferon-inducible dsRNA-dependent kinase PKR, which exists as a latent 68-kDa ribosome-associated molecule in human cells (Meurs et al. 1990). Following interaction with dsRNA species such as viral RNA, PKR autophosphorylates on multiple serine and threonine residues, and in turn phosphorylates substrate targets, the best characterized being the α subunit of eukaryotic protein synthesis initiation factor 2 (eIF2α) (Levin and London 1978). Phosphorylated eIF2α sequesters eIF2B, a rate-limiting component of translation, leading to the repression of protein synthesis initiation in the cell (Panniers and Henshaw 1983; Choi et al. 1992). Aside from exerting eIF2α-mediated antiviral activity, PKR has been reported to be involved in the regulation of NF-κB activity and JNK as well as p38 function (Chu et al. 1999; Goh et al. 2000). Animals lacking PKR are predisposed to lethal intranasal (i.n.) infection by pathogens such as vesicular stomatitis virus (VSV) (Balachandran et al. 2000).
To further delineate PKR function, we attempted to isolate proteins that associated with this kinase. Two such products, referred to as NFAR1 and NFAR2 (for nuclear factors associated with dsRNA; also known as nuclear factor 90 [NF90], DRBP76, or TCP80), were found to be related, spliced variants encoding major products of 90 and 110 kDa, respectively, that were transcribed from a single human gene on chromosome 19 (Corthesy and Kao 1994; Patel et al. 1999; Xu and Grabowski 1999; Saunders et al. 2001a). NFAR1 and NFAR2 each contain two DRBDs in their C terminus regions and exhibit 98% homology with one another at the amino acid level. NFAR2 comprises an additional 192 amino acid at the C-terminal region that is encoded by an extra three exons present in its corresponding transcript (Saunders et al. 2001a,b).
NFAR1 and NFAR2 have been reported to be predominantly nuclear proteins that associate with mRNA ribonuclear protein complex (mRNP) species to facilitate mRNA export and translation (Saunders et al. 2001b; Shim et al. 2002; Pfeifer et al. 2008). Recent data indicate that NFAR1 and NFAR2 facilitate this process largely through the TAP/NXF1 pathway, although additional studies suggest that the NFARs may also regulate the export of mRNAs through the Crm1 pathway (Shim et al. 2002; Pfeifer et al. 2008). Furthermore, the NFARs have been reported to associate with the karyopherin β family member exportin 5 and to play a role in regulating microRNA transportation and function (Brownawell and Macara 2002; Gwizdek et al. 2004; Sakamoto et al. 2009). Plausibly, the NFARs may also facilitate the stabilization of select mRNAs by binding to UTRs (untranslated regions) that harbor AU-rich elements (Shim et al. 2002; Shi et al. 2005). Studies have demonstrated that loss of NFAR1 and NFAR2 leads to an increase in protein synthesis rates through enhancing mRNP nuclear export and/or translation, indicating that the NFARs may be negative regulators of this process (Pfeifer et al. 2008). RNAi knockdown of NFAR1 and NFAR2 renders mouse embryonic fibroblasts (MEFs) susceptible to VSV and influenza virus infection (Pfeifer et al. 2008), suggesting a putative role for the NFARs in cellular host defense by mechanisms that remain to be clarified. Here, we demonstrate that PKR uses the NFAR proteins to suppress the translation of foreign mRNAs in an eIF2α-independent manner, a process that accentuates a novel mechanism of cellular defense against viral replication.
Results
Identification of Thr 188 (T188) and Thr 315 (T315) in NFAR1 and NFAR2 as phosphorylation sites for PKR
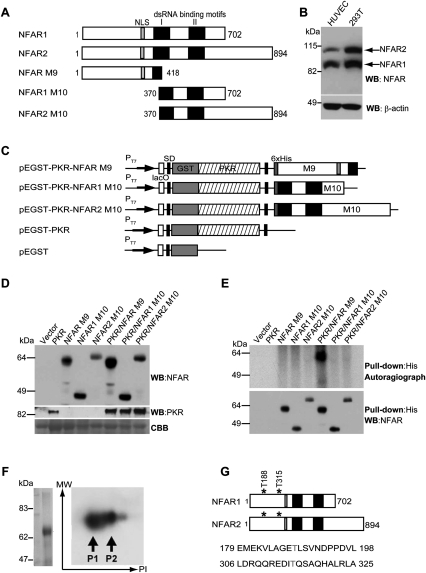
We showed previously that the two major alternatively spliced NFAR proteins of 90 and 110 kDa (Fig. 1A,B) are substrates for the dsRNA-dependent protein kinase PKR in vitro (Saunders et al. 2001b). However, the identity of the phosphorylation site(s) remains unknown, as does the physiological relevance of these post-translational modifications in eukaryotic cells. To identify the amino acids of NFAR that are phosphorylated by PKR, we exploited the ability of PKR to autophosphorylate and become active when heterologously expressed in Escherichia coli (Barber et al. 1991). A dual expression plasmid was generated that expressed both GST-tagged PKR and the first 418 amino acids of the NFAR proteins (histidine-tagged and referred to as NFAR M9), or amino acids 370–702 of NFAR1 and 370–894 of NFAR2 (histidine-tagged and referred to as NFAR1 M10 or NFAR2 M10, respectively) (Fig. 1A,C). Control plasmids comprising PKR (pEGST-PKR) or NFAR M9 alone without PKR were similarly constructed using the same expression vector. Expression of PKR or M9 or M10 alone, or coexpression of both within the same bacteria, was confirmed by immunoblot using antibody to PKR or the NFARs (Fig. 1D). In vivo 32P labeling of bacteria carrying the dual expression plasmids confirmed that PKR autophosphorylated effectively in E. coli, presumably by associating with activating bacterial RNA species or its own transcript (Barber et al. 1991; data not shown). This analysis indicated that only NFAR M9 exhibited phosphorylation, and only when coexpressed with PKR, not when expressed alone (Fig. 1E). In contrast, NFAR1 or NFAR2 M10 did not appear to be phosphorylated by PKR. These data indicated that PKR can effectively phosphorylate NFAR in bacteria, and that the sites of phosphorylation are likely retained in the N-terminal region (amino acids 1–418) of the NFAR proteins.
Figure 1.
PKR phosphorylates NFAR1 and NFAR2 on T188 and T315. (A) Schematic of the NFAR proteins (NFAR1 and NFAR2) and variants (NFAR M9 and M10) expressed in E. coli. (B) Immunoblot of the NFAR proteins in HUVECs or 293T cells using NFAR antiserum. (C) Schematic of histidine-tagged NFAR variants cloned into a pEGST-based prokaryotic expression vector coexpressing wild-type PKR. (D) Immunoblot using NFAR antiserum (top panel) or anti-PKR antibody (middle panel) demonstrating that the NFAR variants were successfully coexpressed with PKR in E. coli. (Bottom panel) Coomassie Brilliant Blue staining (CBB) of electrophoresed proteins was used to confirm equal quantitative analysis of bacterial lysates. (E) E. coli containing dual expression pEGST vectors expressing NFAR variants with PKR was cultured in the presence of 32P-H3PO4. (Top panel) Bacterial lysates were precipitated using anti-histidine antibody, electrophoresed on SDS/polyacrylaide gels, and subjected to autoradiography. Immunoblot analysis of precipitated proteins using NFAR antiserum ensured that all NFAR variants were expressed. (F) Phosphorylated NFAR M9 was purified using anti-histidine beads and phosphorylated proteins were enriched using a phosphoprotein-enriching column. (Left panel) Purified phosphorylated proteins were subjected to SDS/polyacrylamide electrophoresis and stained with Coomassie Blue. Purified proteins were then separated using two-dimensional gel electrophoresis and phosphorylated proteins were detected and isolated following autoradiography. (G) Mass spectrometry analysis indicates that PKR phosphorylates NFAR1 and NFAR2 on amino acids T188 and T315.
This coexpression strategy enabled the purification of sufficient quantities of phosphorylated NFAR M9 to be obtained to determine the site(s) of phosphorylation by mass spectrometer analysis. Thus, following coexpression of PKR and His-tagged M9, bacteria were lysed and M9 was purified by cobalt chromatography (Fig. 1F). Using a phosphor enrichment kit, M9 material was further purified and subjected to two-dimensional isoelectric focusing analysis, which indicated the presence of two major phosphorylated proteins (Fig. 1F). Cobalt-purified M9 was subjected to mass spectrometric analysis, which resulted in the identification of two phosphorylation sites: T188 and T315 (Fig. 1G; Supplemental Fig. S1). Thus, PKR phosphorylates the NFAR proteins at T188 and T315 in prokaryotes.
Phosphorylation of T188 and T315 occurs in eukaryotic cells in response to RNA viruses
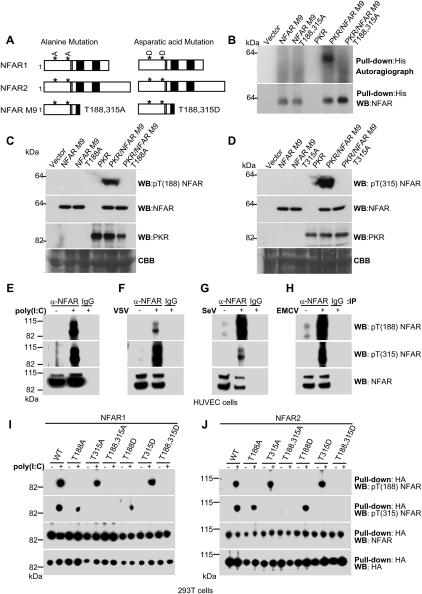
To confirm that T188 and T315 were the sites of PKR-mediated phosphorylation, these amino acids were substituted for alanine in NFAR M9 (T188,315A) and coexpressed with PKR in bacteria. This analysis indicated that NFAR M9 T188,315A, unlike NFAR M9, was not able to be phosphorylated in the presence of PKR (Fig. 2A,B). These data confirm that the NFARs are indeed phosphorylated by PKR on amino acids T188 and T315. To further characterize T188 and T315 phosphorylation, we generated individual phosphorylation site-specific antibodies against T188 or T315 [termed anti-pT(188) NFAR and anti-pT(315) NFAR antibody, respectively]. NFAR M9 variants comprising T188 or T315 substitutions for alanine (NFAR M9 T188A and NFAR M9 T315A) were subsequently coexpressed with PKR in bacteria and analyzed by immunoblot using the anti-pT(188) NFAR or anti-pT(315) NFAR antibody or NFAR antiserum. Our study indicated that the anti-pT(188) NFAR and anti-pT(315) NFAR antibody only recognized NFAR M9 when coexpressed with PKR and not when expressed alone, and not when T188 or T315 was substituted for alanine (Fig. 2C,D). Our data confirm that both T188 and T315 are targeted by PKR for phosphorylation in E. coli.
Figure 2.
Phosphorylation of NFAR1 and NFAR2 on T188 and T315 occurs in eukaryotic cells. (A) Schematic of NFAR proteins with T188 and T315 mutated to alanine or aspartic acid. (B) Coexpression of histidine-tagged NFAR M9 or M9 variants with PKR in E. coli. Cultures were labeled with 32P-H3PO4 and bacterial lysates were precipitated using anti-histidine beads. (Top panel) Purified proteins were separated using SDS/polyacrylamide gels and were subjected to autoradiography. Precipitated extracts were analyzed by immunoblot using NFAR antiserum to ensure quantitative loading. (C) Phosphorylation site-specific antibody against T188 [termed anti-pT(188) NFAR antibody] was generated and used to analyze immunoblots of bacterial lysates coexpressing PKR and NFAR M9 or NFAR M9 T188A. Blots were reprobed with NFAR antiserum or anti-PKR antibody to ensure expression of all proteins. (D) Phosphorylation site-specific antibody against T315 [termed anti-pT(315) NFAR antibody] was generated and used to analyze immunoblots of bacterial lysates coexpressing PKR and NFAR M9 or NFAR M9 T315A. Blots were reprobed with NFAR antiserum or anti-PKR antibody to ensure expression of all proteins. (E) HUVECs were transfected with synthetic dsRNA [poly(I:C)] or infected with VSV (F), SeV (G), or EMCV (H). Cell lysates were immunoprecipitated using NFAR antiserum and immunoblotted. Blots were incubated with anti-pT(188) NFAR or anti-pT(315) NFAR antibody. Reprobing blots with NFAR antiserum confirmed equal loading. (I) Wild-type HA-tagged NFAR1 or variants were transfected into 293T cells and treated with poly(I:C). Cell lysates were precipitated using anti-HA beads and immunoblotted using anti-pT(188) NFAR antibody. (J) Wild-type HA-tagged NFAR2 or variants were transfected into 293T cells and treated with poly(I:C). Cell lysates were precipitated using anti-HA beads and immunoblotted using anti-pT(315) NFAR antibody.
Given this, we next evaluated whether the NFARs were phosphorylated on T188 or T315 in eukaryotic cells. Under nonstimulated conditions, the NFAR proteins in normal human umbilical vascular endothelial cells (HUVECs) appeared weakly phosphorylated on T188 or T315, as determined by immunoprecipitating endogenous NFAR using NFAR antiserum and immunoblotting using anti-pT(188) NFAR or anti-pT(315) NFAR antibody (Fig. 2E–H). However, HUVECs and HeLa cells transfected with synthetic dsRNA (poly[I:C]), which activates endogenous PKR, exhibited a profound increase in NFAR T188 and T315 phosphorylation (Fig. 2E; Supplemental Fig. S2A). Significantly, a similar increase in NFAR T188 and T315 phosphorylation was observed in HUVECs and HeLa cells infected with VSV, Sendai virus (SeV), and encephalomyocarditis virus (EMCV) (Fig. 2F–H; Supplemental Fig. S2B–D). These data confirm that the NFARs are phosphorylated on both T188 and T315 in eukaryotic cells in response to synthetic dsRNA and select RNA virus infection. To complement this study, we also transfected HA-tagged NFAR variants with T188 and/or T315 substituted for alanine (to prevent phosphorylation) or aspartic acid (to mimic phosphorylation) into 293T cells treated with or without the PKR activator poly(I:C). Our investigation confirmed that anti-pT(188) NFAR and anti-pT(315) NFAR antibodies only recognized their specific phosphorylated targets corresponding to T188 or T315 in eukaryotic cells (Fig. 2I,J). These data confirm that both NFAR1 and NFAR2 are also phosphorylated on T188 and T315 in eukaryotic cells, predominantly in response to dsRNA transfection and select cytoplasmic replicating viruses.
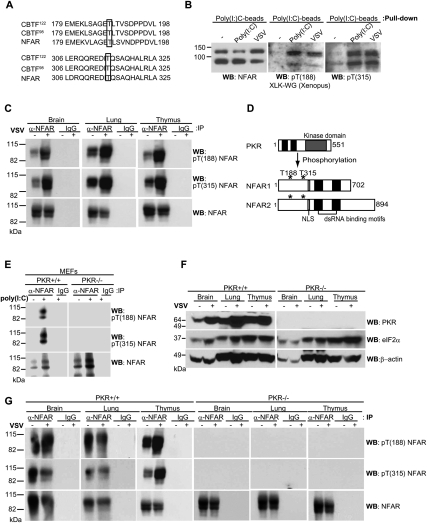
Since T188 and T315 appear to be physiologically relevant targets for phosphorylation, we carried out databank searches and noted that these residues were identically conserved in Xenopus laevis NFAR-related proteins, referred to as CCAAT box transcription factors CBTF98 and CTBF122 (Fig. 3A; Brzostowski et al. 2000). The NFARs are ∼80% conserved with CBTF98 and CTBF122 at the amino acid level. We observed that CBTF98 and CTBF 122 could be successfully precipitated from X. laevis XLK-WG cells using poly(I:C) beads, as determined by immunoblotting using cross-reacting NFAR antiserum (Fig. 3B). Significantly, infection of XLK-WG cells with VSV-GFP or transfection with dsRNA [poly(I:C)], lead to phosphorylation of CBTF98 and CTBF122 on T188 and T315 (Fig. 3B). PKR homologs in zebrafish and X. laevis have been reported previously, and have been shown capable of phosphorylating yeast eIF2α on Ser 51(Rothenburg et al. 2008). Our data here demonstrate that CBTF98 and CTBF122 are phosphorylated on T188 and T315, perhaps by a X. laevis PKR-homolog, in response to virus infection, plausibly indicating a conserved mechanism of cellular antiviral host response.
Figure 3.
NFAR is phosphorylated by PKR on T188 and T315 in vivo. (A) T188 and T315 are conserved in X. laevis CBTF98 and CBTF122. (B) XLK-WG cells were infected with VSV-GFP (m.o.i: 10) and transfected with poly(I:C) for 6 h, and lysates were incubated with poly(I:C) agarose beads and immunoblotted using anti-NFAR antiserum or anti-pT(188) NFAR or anti-pT(315) NFAR antibody. (C) Six-week-old to 8-wk-old mice (C57BL/6) were inoculated i.n. with (+) or without (−) VSV Indiana strain (1 × 107 pfu per mouse). After 24 h, the brain, lung, and thymus were retrieved from the animals, and cell lysates were immunoprecipitated using NFAR antiserum before being analyzed by immunoblot using anti-pT(188) NFAR anti-pT(315) NFAR antibody. (D) Schematic of PKR and NFAR1 and NFAR2. (E) MEFs from wild-type or PKR-deficient animals were transfected with (+) or without (−) poly(I:C), and cell lysates were immunoprecipitated using NFAR antiserum before being analyzed by immunoblot using anti-pT(188) NFAR or anti-pT(315) NFAR antibody. (F) Wild-type (+/+) or PKR-deficient (−/−) mice were inoculated i.v. with (+) or without (−) VSV Indiana strain (1 × 108 pfu per mouse). After 24 h, the brain, lung, and thymus were retrieved from the animals, and cell lysates were examined by immunoblot using antibody to PKR or eIF2α. (G) Wild-type (+/+) or PKR-deficient (−/−) mice were inoculated i.v. with (+) or without (−) VSV Indiana strain (1 × 108 pfu per mouse). After 24 h, the brain, lung, and thymus were retrieved from the animals, and cell lysates were immunoprecipitated using NFAR antiserum before being analyzed using immunoblot using anti-pT(188) NFAR or anti-pT(315) NFAR antibody.
T188 and T315 on NFAR1 and NFAR2 are phosphorylated specifically by PKR
We next evaluated whether the NFAR proteins were phosphorylated in vivo on T188 and/or T315. Mice were inoculated with VSV Indiana strain (1 × 107 plaque-forming units [pfu] per mouse) i.n., and various organs were collected 24 h post-infection. Endogenous NFARs were precipitated using NFAR antiserum and immunoblotting was carried out using anti-pT(188) NFAR or anti-pT(315) NFAR antibody. Our study indicated an increase in T188 and T315 phosphorylation in the brain, lung, and thymus following infection, indicating that these sites can also be phosphorylated in vivo (Fig. 3C). Given that we observed previously that NFAR1 and NFAR2 are phosphorylated by PKR in vitro, and that select RNA virus infection can promote an increase in T188 and T315 phosphorylation in eukaryotic cells, we evaluated the importance of PKR in these post-translational modifications. Thus,MEFs containing (+/+) or lacking (−/−) PKR were transfected with poly(I:C) and the NFAR proteins immunoprecipitated using NFAR antiserum or IgG control. After SDS/gel electrophoresis, immunoblots were incubated with anti-pT(188) NFAR or anti-pT(315) NFAR antibody. Our results indicated that, while NFAR1 and NFAR2 appeared to undergo phosphorylation in PKR+/+ MEFs on T188 and T315 in the presence of dsRNA, no phosphorylation appeared evident on T188 or T315 in PKR−/− MEFs (Fig. 3D,E). Blots were reprobed with NFAR antiserum to ensure that the NFAR proteins were precipitated efficiently in the PKR−/− MEFs (Fig. 3E). We next infected PKR+/+ or PKR−/− animals intravenously (i.v.) with VSV Indiana strain (1 × 108 pfu per mouse). Immunoblotting protein extracts from a variety of murine organs retrieved from PKR+/+ or PKR−/− mice confirmed loss of PKR expression (Fig. 3F). After 24 h post-infection, the brain, lung, and thymus were removed from infected PKR+/+ or PKR−/− animals and NFAR proteins were immunoprecipitated from organ lysates using NFAR antiserum. Following SDS/gel electrophoresis, immunoblotting was carried out using anti-pT(188) NFAR or anti-pT(315) NFAR antibody. This study confirmed that a fraction of the NFAR proteins was phosphorylated on T188 and T315 even in uninfected PKR+/+ animals, and that this event increased in infected animals (Fig. 3G). Significantly, no phosphorylation of T188 and T315 was observed in PKR−/− mice. Thus, PKR is responsible for phosphorylating NFAR1 and NFAR2 on T188 and T315 in vitro and in vivo. This event was found to occur independently of eIF2α phosphorylation (Supplemental Fig. S2E,F).
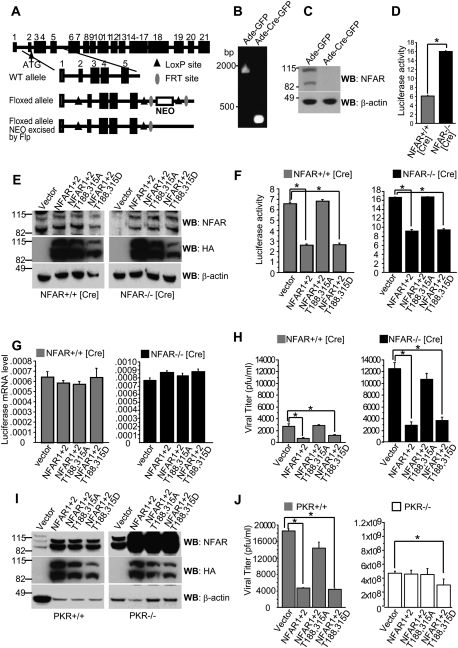
Loss of NFAR1 and NFAR2 renders cells susceptible to RNA virus replication
Previous attempts to develop an NFAR homozygous-null mouse were unsuccessful due to the embryo dying during development (Pfeifer et al. 2008). We therefore developed, using homologous recombination strategies and Cre/loxP technology (Lakso et al. 1992), a mouse model in which exons 2–5 of the NFAR gene, including the ATG codon, were floxed (loxP) (Fig. 4A). MEFs were developed from an NFAR floxed animal and subjected to infection using adenovirus expressing the Cre recombinase and GFP genes (Ade-Cre-GFP). Following exposure of NFAR floxed MEFs to Ade-Cre-GFP, we confirmed loss of NFAR gene by genomic PCR (Fig. 4B) and protein expression by immunoblot analysis (Fig. 4C). NFAR-lacking MEFs (NFAR−/− [Cre]) or control treated with Ade-GFP (NFAR+/+ [Cre]) were transfected with a eukaryotic SV40-luciferase plasmid. The results indicated that loss of the NFARs enabled an increase in luciferase expression compared with controls, as we demonstrated previously using RNAi approaches to knock down NFAR expression (Fig. 4D; Pfeifer et al. 2008). PKR-null cells exhibited a similar phenotype (Supplemental Fig. S2G). We next cotransfected NFAR-lacking MEFs (NFAR−/− [Cre]) or control (NFAR+/+ [Cre]) with SV40-luciferase and HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D. After 24 h, cell lysates were analyzed by immunoblot to confirm expression of reconstituted heterologous NFAR proteins using NFAR antiserum and anti-HA antibody (Fig. 4E). Luciferase expression was also measured. The results indicated that overexpression of NFAR1 and NFAR2 reduced luciferase protein expression by approximately fourfold to fivefold in control (NFAR+/+ [Cre]) or NFAR-lacking MEFs (NFAR−/− [Cre]) (Fig. 4F). This effect was similarly observed following reconstitution of NFAR1 and NFAR2 T188,315D, but not NFAR1 and NFAR2 harboring mutations at T188,315A (Fig. 4F). Since luciferase RNA levels were comparable in the experiments, our data indicate that the effect of the NFARs on luciferase gene expression likely occurred at the post-transcriptional level, as we described previously (Fig. 4G; Pfeifer et al. 2008). Thus, wild-type NFAR1 and NFAR2 are able to exert an inhibitory effect on the translation of transfected luciferase genes that is dependent on the phosphorylation of T188 and T315.
Figure 4.
Phosphorylated NFAR1 and NFAR2 inhibit VSV replication. (A) Diagram of the targeting LoxP vector and deletion strategy. NEO denotes the neomycin selection cassette, flanked by FRT recombination sites. (B) PCR analysis of genomic DNA extracted from MEFs with loxP sites flanking exons 2–5 of the NFAR gene (NFAR [Cre]) were infected with an adenovirus vector expressing GFP (Ade-GFP) or Cre recombinase (Ade-Cre-GFP). (C) Cells lysates treated as in B were subjected to immunoblot analysis using NFAR antiserum 7 d after infection. (D) NFAR floxed MEFs treated with Ade-GFP (NFAR+/+ [Cre]) or Ade-Cre-GFP (NFAR−/− [Cre]) were transfected with a Luciferase expression vector. After 24 h, cells were transfected with poly(I:C). After 16 h, cell lysates were measured for Luciferase expression. Data are shown as the mean ± standard error of the mean (SEM). (*) P < 0.05. (E) NFAR+/+ [Cre] or NFAR−/− [Cre] MEFs were transfected with HA-tagged wild-type NFAR1 and NFAR2; NFAR1 and NFAR2 T188,T315A; or NFAR1 and NFAR2 T188,315D. After 48 h, immunoblot analysis was carried out using NFAR antiserum or anti-HA antibody. (F) NFAR+/+ [Cre] or NFAR−/− [Cre] MEFs were cotransfected with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,T315A; or NFAR1 and NFAR2 T188,315D together with a Luciferase reporter plasmid. After 24 h, cells were transfected with poly(I:C). After 16 h, cell lysates were measured for Luciferase expression. Data are shown as the mean ± SEM. (*) P < 0.05. (G) Cells treated as in F were examined for Luciferase mRNA levels by quantitative RT–PCR. Data are shown as the mean ± SEM. (H) After 24 h of transfection, cells were infected with VSV-GFP (m.o.i.: 1) and were examined for VSV replication by plaque assay 12 h post-infection. Data are shown as the mean ± SEM. (*) P < 0.05. (I) Wild-type (+/+) or PKR-deficient (−/−) MEFs were transfected with HA-tagged wild-type NFAR1 and NFAR2; NFAR1 and NFAR2 T188,T315A; or NFAR1 and NFAR2 T188,315D. After 48 h, immunoblot analysis was carried out using NFAR antiserum or anti-HA antibody. (J) Twenty-four hours after transfection, cells were infected with VSV-GFP (m.o.i.: 1) and virus titer was measured by plaque assay at 12 h post-infection. Data are shown as the mean ± SEM. (*) P < 0.05.
To extend this study, we next transfected control (NFAR+/+ [Cre]) and NFAR-lacking MEFs (NFAR−/− [Cre]) with NFAR1 and NFAR2 or variants and infected them with VSV (multiplicity of infection [m.o.i.]: 1). The results indicated that expression of NFAR1 and NFAR2 modestly inhibited VSV replication in the control (NFAR+/+ [Cre]) MEFs (Fig. 4H). A similar effect was observed following transfection of NFAR 1 and NFAR2 T188,315D. However, no suppression of VSV replication was observed following transfection of NFAR1 and NFAR2 T188,315A. Similar though more dramatic findings were observed using the NFAR−/− [Cre] MEFs, which were considerably more sensitive to VSV replication. Significantly, reintroduction of NFAR1 and NFAR2 or NFAR1 and NFAR2 T188,315D but not T188,315A rescued antiviral activity (Fig. 4H).
These data indicate that phosphorylation of NFAR1 and NFAR2 on T188 and T315 by PKR is required for inhibiting translation. To evaluate this further, we transfected control (PKR+/+) or PKR-lacking (PKR−/−) MEFs with NFAR constructs, as described above. After 36 h, cell lysates were analyzed by immunoblot to confirm expression of reconstituted heterologous NFAR proteins using NFAR antiserum and anti-HA antibody (Fig. 4I). The results indicated that PKR−/− were more sensitive than PKR+/+ MEFs to virus infection, as described previously (Balachandran et al. 2000). In addition, NFAR1 and NFAR2 could not reduce antiviral replication in PKR−/− MEFs (Fig. 4J). Reintroduction of NFAR1 and NFAR2 T188,315D did not significantly reduce viral expression in the PKR−/− MEFs, suggesting that the presence of PKR was required for full NFAR antiviral activity. Phosphorylation of eIF2α by PKR was unaffected in the absence of the NFARs (Supplemental Fig. S2E). Thus, our data suggest that PKR targets NFAR1 and NFAR2 on T188 and T315 for phosphorylation, a consequence that assists cellular host defense.
To further evaluate the importance of the NFARs in vivo, we developed a murine model from embryonic stem cells containing floxed exons (2–5) of the NFAR gene (Fig. 4A). Following the generation of homozygous NFAR loxP animals (Nf/f), we developed animals that lacked the NFAR protein in the thymus (Nf/f/−-Cre+) by crossing with transgenic mice expressing the Cre recombinase gene under control of the Lck promoter (Lck-Cre) (Gu et al. 1994). Genomic PCR demonstrated the floxed and wild allele (Fig. 5A). Thymocytes from control (Nf/f and LCK-Cre) and NFAR-lacking (Nf/f-Cre+) animals were cultured and analyzed by immunoblot using NFAR antiserum to confirm loss of NFAR1 and NFAR2 (Fig. 5B). Thymocytes from control (Nf/f and LCK-Cre) or NFAR-lacking (Nf/f-Cre+) were then infected with VSV at m.o.i. of 1 and 10 and, after 48 h, virus replication was measured. This study found that thymocytes lacking the NFARs (Nf/f-Cre+) were significantly sensitive to virus infection (fivefold) compared with control, confirming our earlier results using MEFs lacking the NFAR protein (Fig. 5C). Using VSV-expressing luciferase, we then infected control (LCK-Cre) and NFAR-lacking (Nf/f-Cre+) animals by i.v. This approach enabled us to monitor VSV-Luc replication in vivo using luminescent analyses, and indicated a high level of Luciferase activity in the thymus of infected animals at 24 and 48 h post-infection (Fig. 5D,E). Significantly, animals lacking the NFARs in the thymus (Nf/f-Cre+) were found to be more sensitive to lethal VSV infection and died within 2 d of exposure (Fig. 5F). These data indicate that the NFARs are critical components of innate defense against VSV infection.
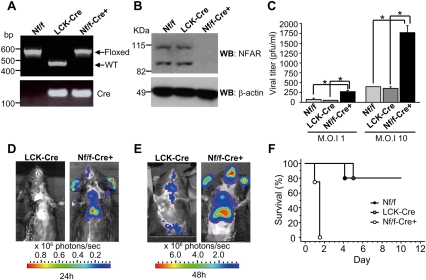
Figure 5.
Cre/loxP mice lacking thymic expression of NFAR1 and NFAR2 are susceptible to VSV infection. (A) Mice with the first four exons of the NFAR gene flanked by loxP sites (Nf/f) were crossed with transgenic mice expressing the Cre recombinase gene under control of the Lck promoter (Lck-Cre) to generate mice lacking NFAR expression in the thymus (Nf/f-Cre+). PCR analysis of genomic tail DNA-derived wild type, Lck-Cre, and Nf/f-Cre+. (B) Immunoblot of thymocytes retrieved from 6- to 8-wk Nf/f-Cre+ or control mice using NFAR antiserum. (C) Thymocytes from Nf/f-Cre+ or control mice were infected with VSV at an m.o.i. of 1 or 10 for 48 h and virus replication was measured by plaque assay. Data are shown as the mean ± SEM. (*) P < 0.05. (D) Nf/f-Cre+ or control mice were infected i.v. with 5 × 107 pfu per mouse recombinant VSV expressing a luciferase gene (VSV-Luc). Images were taken using an IVIS imaging system (Xenogen) at 24 h post-infection. (E) Animals are as in D at 48 h post-infection. (F) Survival curve of animals (n = 5; 6–8 wk of age) infected i.v. with VSV Indiana strain at 2 × 108 pfu per mouse.
Phosphorylated NFAR1 and NFAR2 are retained in the cytoplasm
Evidence suggests that the NFARs are nuclear shuttling proteins that associate with newly transcribed mRNA species to facilitate mRNA export and translation (Saunders et al. 2001b; Shim et al. 2002; Pfeifer et al. 2008). To evaluate the effect of phosphorylation on NFAR localization, 293T and HeLa cells were transfected with or without poly(I:C) and stained with NFAR antiserum. The results indicated that the NFARs appeared to undergo dramatic trafficking to the cytoplasm following exposure to dsRNA (Fig. 6A; Supplemental Fig. S3A). This observation was confirmed by analyzing nuclear and cytoplasmic fractions of poly(I:C)-treated cells (Fig. 6B; Supplemental Fig. S3B). Next, HeLa cells were infected with VSV-GFP and stained with NFAR antiserum. The results indicated that NFARs similarly localized to the cytoplasm of these cells following infection (Fig. 6C), as well as 293T or MEF cells (data not shown). Following this, we transfected normal MEFs and 293T and HeLa cells with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D. Transfected cells were also treated with or without poly(I:C). Immunofluorescent analysis indicated that, similar to the situation in 293T cells, NFAR1 and NFAR2 were localized predominantly in the nucleus of MEFs and 293T and HeLa cells in the absence of poly(I:C). However, after transfection of poly(I:C), HA-NFAR1 and HA-NFAR2 localized to the cytoplasm of the cell (Fig. 6D). In contrast, NFAR1 and NFAR2 T188,315A were unable to localize to the cytoplasm, even in the presence of poly(I:C). NFAR1 and NFAR2 T188,315D, however, were found to localize largely in the cytoplasm, even in non-poly(I:C)-treated cells (Fig. 6D; Supplemental Fig. S3C–E). We thus conclude that phosphorylation of the NFARs likely results in their sequestration in the cytoplasm and their inability to reshuttle to nuclear compartments. PKR was found to be important for NFAR cytoplasmic retention, since HA-tagged NFAR1 and NFAR2 (or the T188,315D variants) were found to be localized in the nucleus of PKR−/− MEFs in the presence or absence of poly(I:C) treatment (Fig. 6E). This event might explain why reconstitution of NFAR1 and NFAR2 T188,315D into PKR−/− MEFs had only a modest effect on the inhibition of viral replication (Fig. 4J). Thus, PKR likely associates with NFAR1 and NFAR2 phosphorylated on T188 and T315.
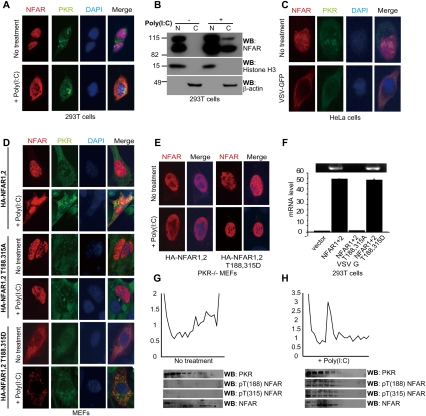
Figure 6.
NFAR1 and NFAR2 phosphorylated on T188 and T315 are retained in the cytoplasm. (A) Immunofluorescence analysis using NFAR antiserum or anti-PKR antibody of 293T cells transfected with (+) or without (−) poly(I:C). (B) Fractionation analysis of 293T cells transfected with (+) or without (−) poly(I:C). Fractions were analyzed by immunoblot using NFAR antiserum or anti-histone H3 antibody. (N) Nuclear fractions; (C) cytoplasmic fractions. (C) Immunofluorescence analysis using NFAR antiserum or anti-PKR antibody of HeLa cells infected with VSV-GFP (m.o.i. 1). (D) MEFs were transfected with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D. After 48 h, cells were transfected with (+) or without (−) poly(I:C) for 3 h and cells were examined by immunofluorescence using anti-HA antibody or anti-PKR antibody. (E) PKR-deficient MEFs (−/−) were treated as in C. (F) 293T cells were transfected with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D. After 48 h, cells were infected with VSV (m.o.i.: 20) for 6 h. Cells were cross-linked by 1% formaldehyde, cell lysates were precipitated using anti-HA beads, and viral mRNA level was measured by quantitative RT–PCR. Data are shown as the mean ± SEM. (G,H) HeLa cells were transfected without (−) (G) or with (+) (H) poly(I:C), and cells were lysed and subjected to polysome analysis after 3 h. Fractions were electrophoresed and immunoblotted using anti-PKR antibody, anti-pT(188) NFAR, anti-pT(315) NFAR antibody, or NFAR antiserum.
As part of these studies, we observed that in vitro transcribed viral mRNA has the ability to bind NFAR1 and NFAR2 T188,315A or NFAR1 and NFAR2 T188,315D (Supplemental Fig. S3F). Thus, T188 and T315 are not important for the NFARs' association with dsRNA species, presumably since these residues lie outside of the DRBDs. Fractionation analysis of VSV-infected cells indicated that cytoplasmic NFAR1 and NFAR2 (T188,315D) bound predominantly to viral RNA (VSV replicates in the cytoplasm) (Fig. 6F; Supplemental Fig. S4A–G). This would suggest that phosphorylation of NFAR1 and NFAR2 by activated PKR causes the former molecules to become retained in the cytoplasm, where they are more readily available to associate with viral RNA on ribosomes and interfere with viral replication. To examine this further, we transfected HeLa cells with or without poly(I:C) and isolated polysome fractions that were subjected to immunoblotting using NFAR antiserum or anti-pT(188) NFAR or anti-pT(315) NFAR antibody (Fig. 6G,H). The results indicated low levels of NFAR1 and NFAR2 on engaged polysomes, as we demonstrated previously (Pfeifer et al. 2008). However, phosphorylated NFAR1 and NFAR2 were strongly detected in polysome fractions at the initiation stages of translation in poly(I:C) transfected cells. PKR protein was detected in both poly(I:C)-treated and untreated fractions. Subsequently, NFAR−/− [Cre] MEFs were transfected with NFAR1 and NFAR2 or NFAR1 and NFAR2 T188,315A, and polysome analysis was carried out following VSV infection. The results indicated an increased association of the NFARs with viral RNA on lighter polysomes. However, a shift in the polysome distribution profile was observed in the T188,315A transfected cells (to heavier polysomes), possibly indicating a derepression of viral mRNA translation that would conceivably help to explain the increase in viral replication (Supplemental Fig. S4H–J). Thus, PKR-mediated phosphorylation of NFAR1 and NFAR2 prevents the immediate shuttling of the latter molecules into the nucleus after delivery of mRNA to ribosomes. This enables phosphorylated NFAR1 and NFAR2 to associate directly with viral RNA, and results in the inhibition of viral translation and replication.
NF45 is dissociated from phosphorylated NFAR complexes
The NFARs have been reported to be associated with a cofactor referred to as NF45, although the role of NF45 in NFAR function remains to be fully clarified (Kao et al. 1994). Using tandem affinity purification (TAP) and NFAR2 as bait, followed by mass spectrometric analysis, we similarly identified NF45 as an NFAR-associating protein (Supplemental Fig. S5). To start to evaluate the role of NF45 in NFAR function, RNAi was used to deplete NF45 protein levels. Immunofluorescence and immunoblot analysis confirmed a significant loss of NF45 expression, which was found to be a predominantly nuclear protein, in treated 293T and HeLa cells (Fig. 7A–D). Of significance is that this study indicated that the NFAR proteins were localized primarily to the cytoplasm and not the nucleus in NF45-lacking cells (Fig. 7A,D). These observations suggest that NF45 may play a role in facilitating the shuttling of the NFARs back into the nucleus following the export of mRNA to ribosomal compartments. Given this, we reasoned that loss of NF45 may impede viral replication due to an abundance of NFARs being available in the cytoplasm that could bind viral RNA species and inhibit translation. We thus infected MEFs and HeLa cells lacking NF45 with VSV and measured viral replication. This study confirmed that loss of NF45 inhibited the replication of VSV in control-treated MEFs as well as HeLa cells by at least one log (Fig. 7E,F; Supplemental Fig. S6A,B). This effect was found to be dependent on the presence of the NFARs (Supplemental Fig. S6C,D). In the absence of NF45, polysome profiles shifted from heavier to lighter polysomes in VSV-infected MEFs, possibly due to an abundance of NFAR being relocalized to the cytoplasm to impede viral translation (Fig. 7G,H). Our experiments further indicated that NF45's ability to associate with NFAR1 and NFAR2 was severely reduced when the NFARs were phosphorylated on T188 and T315, suggesting that NFAR1 and NFAR2 phosphorylation results in the dissociation of NF45 from NFAR complexes. These observations may explain how the NFARs are retained in the cytoplasm following phosphorylation (Fig. 7I). To evaluate these findings further, we transfected 293T and HeLa cells treated with NF45-specific RNAi with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D and carried out immunofluorescence studies. Cells were also treated with or without poly(I:C) (Fig. 7J; Supplemental Fig. S6E). Results confirmed that, in the absence of NF45, wild-type NFAR1 and NFAR2 relocated to the cytoplasm. However, it was interesting to note that the previously nuclear retained NFAR1 and NFAR2 T188,315A was also found to be in the cytoplasm in the absence of NF45. These data additionally underscore the importance of NF45 in facilitating the reshuttling of the NFARs back into the nucleus. In summary, we conclude that phosphorylation of the NFARs by PKR dissociates NF45 from NFAR complexes and enables NFAR1 and NFAR2 to be retained in the cytoplasm, where they are able to bind to viral RNAs and impede viral replication (Fig. 8). This process represents a new mechanism used by PKR to prevent the unwarranted translation of foreign mRNAs in the cell.
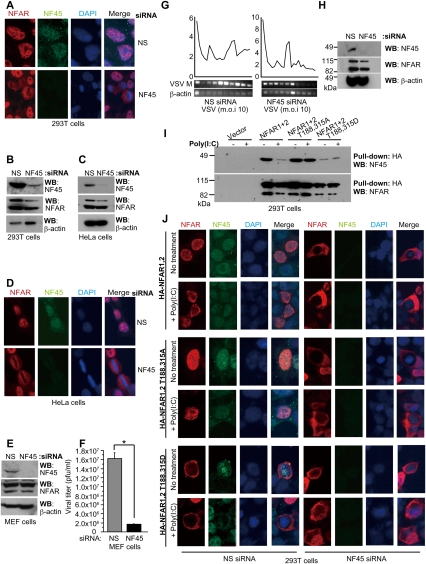
Figure 7.
NFAR cofactor NF45 retains unphosphorylated NFAR1 and NFAR2 in the nucleus. (A) 293T cells were treated with siRNA to NF45 or control. After 2 d, cells were examined by immunofluorescence using NFAR antiserum or anti-NF45 antibody. (B) Immunoblot of siRNA-treated 293T cells as in A. (C) HeLa cells were treated with siRNA to NF45 or control. After 2 d, cells were examined by immunoblot using NFAR antiserum or anti-NF45 antibody. (D) HeLa cells as in C were examined by immunofluorescence using NFAR antiserum or anti-NF45 antibody. (E) MEFs were treated with siRNA to NF45 or control. After 2 d, cells were examined by immunoblot using NFAR antiserum or anti-NF45 antibody. (F) MEFs were treated with siRNA to NF45 or control. After 2 d, cells were infected with VSV-GFP (m.o.i.: 1) for 24 h. Virus replication was measured using plaque assay. Data are shown as the mean ± SEM. (*) P < 0.05. (G) MEFs were treated with control or NF45 siRNA. After 2 d, cells were infected with VSV-GFP (m.o.i.: 10) for 12 h and cells were lysed and subjected to polysome analysis. RNA was isolated from each fraction and analyzed for VSV-M or β-actin mRNA by RT–PCR. (H) Immunoblot of control or NF45 siRNA-treated MEFs as in G. (I) 293T cells were transfected with HA-tagged NFAR1 and NFAR2; NFAR1 and NFAR2 T188,315A; or NFAR1 and NFAR2 T188,315D. After 48 h, cells were transfected with (+) or without (−) poly(I:C), and cell lysates were immunoprecipitated using anti-HA beads. Pull downs were examined by immunoblot using NFAR antiserum or anti-NF45 antibody. (J) 293T cells were treated with siRNA to NF45. After 24 h, cells were transfected with HA-tagged wild-type NFAR1 and NFAR2; NFAR1 and NFAR2 T188,T315A; or NFAR1 and NFAR2 T188,315D. After 24 h, cells were transfected with (+) or without (−) poly(I:C), and cells were examined after 3 h by immunofluorescence using anti-HA antibody or anti-NF45 antibody.
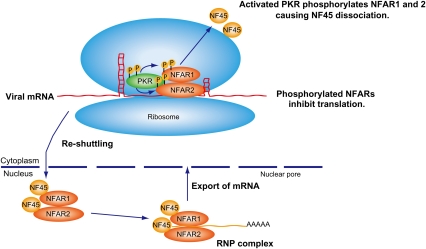
Figure 8.
Model for NFAR's role in host defense. NFAR1 and NFAR2, associated with NF45, form part of the host RNP complex, responsible for exporting cellular mRNA out of the nucleus for translation. Following delivery of mRNA to ribosomes, the NFARs are reshuttled to the nucleus in an NF45-dependent manner. Following viral infection, PKR is activated (autophosphorylates) by association with viral-specific RNAs. Activated PKR subsequently phosphorylates NFAR1 and NFAR2 on T188 and T315, an event that dissociates NF45, allowing the NFARs to be retained on polysomes, where they are able to directly bind viral RNA and prevent translation.
Discussion
NFAR1 and NFAR2 are predominantly nuclear shuttling proteins that can associate with mRNP complexes to escort cellular mRNAs out of the nucleus for translation (Saunders et al. 2001b; Shim et al. 2002; Pfeifer et al. 2008). Following this event, the NFARs return to the nucleus. However, should foreign dsRNA species, such as viral mRNA, attempt to usurp the host translational machinery to manufacture viral protein, such RNA may inadvertently interact with the ribosome-associated PKR. This incident is known to cause PKR to autophosphorylate (Galabru and Hovanessian 1987) and in turn phosphorylate eIF2α, which inhibits translation (Levin and London 1978; Panniers and Henshaw 1983; Choi et al. 1992). However, we report here that activated PKR can also phosphorylate the NFAR proteins on both Thr 188 and Thr 315. This event may be facilitated by viral RNAs bridging PKR and NFAR proteins via their DRBDs, although the DRBDs of the NFARs are not required for PKR-mediated phosphorylation. Phosphorylated NFAR1 and NFAR2 appear to be retained in the cytoplasm as ribosome-associated molecules complexed with PKR, where they are able to associate directly with viral RNA and prevent translation. Loss of NFAR function facilitates the replication of VSV, a cytoplasmic virus, and is extremely detrimental to the host, as we clearly demonstrate. Thus, we describe a new translational mechanism for host defense that is eIF2α-independent and is essential for protection of the cell against certain virus replication (Fig. 8).
It remains unclear as to whether the two NFAR proteins exhibit significantly different properties from one another, but our data confirm that they likely exist as a complex with a cofactor referred to as NF45 to help escort mRNAs and perhaps other types of cellular RNA out of the nucleus. NFAR1 and NFAR2 are essentially identical for the first 700 amino acids, but NFAR2 contains an extra 192 amino acids at its C-terminal, which probably enables it to interact with additional cofactors (Parker et al. 2001; Saunders et al. 2001b). Both NFAR1 and NFAR2 are substrates for PKR, and both appear to be retained in the cytoplasm when they are in a phosphorylated state. Mutation of T188 and T315 to alanine residues ablated the ability of the NFAR proteins to be retained in the cytoplasm. Our data here indicate that NF45 association with the NFARs is required for the reshuttling of the NFARs back into the nucleus. Phosphorylation of NFAR1 and NFAR2 causes the dissociation of NF45 from the NFARs, a process that accordingly facilitates NFAR cytoplasmic retention, which we observed was also PKR-dependent. Indeed, in the absence of NF45, even NFAR1 and NFAR2 T188,315A failed to be retained in the nucleus. Thus, NF45 is essential for the efficient reshuttling of both NFAR1 and NFAR2 into the nucleus following delivery of mRNP complexes to ribosomes for translation. Phosphorylation of NFAR1 and NFAR2 by PKR enables the NFARs to associate with viral RNA and prevent viral translation. RNAi knockdown of NF45 inhibited viral replication, presumably since more NFAR was retained in the cytoplasm to impede viral replication processes.
We demonstrated previously that loss of the NFARs can lead to enhanced viral replication in the cell (Pfeifer et al. 2008). Here, we confirm that loss of the NFAR expression in thymocytes in vivo enables rapid viral replication in these cells and facilitates viral spread throughout the host, leading to death within 2 d. Thus, the cause of death is likely to be a T-cell-independent event, since the adaptive immune response normally functions after this time period. Preliminary data indicate only a slight decrease in CD4- and CD8-positive T-cell subsets in Nf/f-Cre+ mice compared with control mice, at least under noninfected conditions (data not shown). A mouse model with the NFAR gene floxed and crossed with a transgenic animal expressing Cre recombinase under control of the Lck promoter (Nf/f-Cre+ mice) was generated, since our previous attempts to develop a viable murine model with homozygous deletion of the NFAR gene was unsuccessful due to early embryonic lethality (Pfeifer et al. 2008). Further analysis of these Nf/f-Cre+ animals when exposed to pathogens will enable the study of the response of the T-cell repertoire. Previous reports have suggested that the NFARs may specifically regulate IL-2 mRNA stability (Shim et al. 2002; Shi et al. 2007). However, our data indicate a much more comprehensive role for NFAR1 and NFAR2 in the regulation of cellular gene expression, and indicate that the NFAR proteins, which are widely expressed in many cell types, can control the export of numerous cellular RNA species through the TAP/NXF1 as well as the Crm1 pathway (Shim et al. 2002; Pfeifer et al. 2008). In addition, the NFARs have been reported to associate with the karyopherin-β nuclear receptor exportin 5, required for efficient microRNAi export and to negatively regulate select microRNA function (Brownawell and Macara 2002; Gwizdek et al. 2004; Yi et al. 2003; Sakamoto et al. 2009). Other DRBD-containing proteins such as DICER, TRBP, and PACT have been shown to be important molecules in RNAi and microRNA function (Bernstein et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Chendrimada et al. 2005; Lee et al. 2006). Collectively, the DRBD family plays critical roles in controlling post-transcriptional, translational, and epigenetic events, and has significant importance in host defense (Saunders and Barber 2003).
DRBDs such as the NFARs are known to bind dsRNA species as small as 11–16 base pairs in a sequence-independent manner (Manche et al. 1992; Ryter and Schultz 1998). A number of viral RNA species have now been reported to associate with the NFAR proteins, including those encoded by poliovirus, hepatitis B virus (HBV), bovine diarrhea virus (BDV), hepatitis C virus (HCV), adenovirus, and human immunodeficiency virus (HIV1) (Shin et al. 2002; Isken et al. 2003; Merrill and Gromeier 2006; Urcuqui-Inchima et al. 2006; Isken et al. 2007; Agbottah et al. 2007). These events have been described to interfere with virus replication, and support our findings that the NFARs are critically important for effective host defense countermeasures. Genetically engineered mice lacking functional PKR are known to be sensitive to lethal VSV infection (Balachandran et al. 2000). Our data here additionally indicate that the antiviral activity manifested by PKR may not depend solely on the inhibition of translational regulation through phosphorylation of eIF2α. Thus, PKR appears to have evolved to use two independent cellular mechanisms to control viral translation.
The DRBP family appears evolutionarily conserved and NFAR homologs are conserved, at least in X. laevis, although it remains unclear whether they have any role in host defense. In Xenopus, NFAR-like proteins CBTF98 and CBTF122 or 4F.1 and 4F.2 have been reported to associate with translationally quiescent mRNP complexes and regulate embryo development (Orford et al. 1998; Brzostowski et al. 2000; Scarlett et al. 2004). Of interest is that T188 and T315 are evolutionarily conserved in CBTF98 and CBTF122 and undergo phosphorylation in response to virus infection. PKR homologs have been reported in amphibians (Rothenburg et al. 2008). Thus, phosphorylation of CBTF98 and CBTF122 on T315 and T188 may have been evolutionarily conserved. T315 but not T188 was also found to be conserved in two other mammalian DRBPs, referred to as SPNR (spermatid perinuclear RNA-binding protein) and ZFR1 (a zinc finger DRBP) (Gene ID: 51663). SPNR is expressed predominantly in the testes and may play a role in spermatogenesis (Schumacher et al. 1995; Pires-daSilva et al. 2001), while no function has yet been assigned to ZFR1. It remains to be determined whether PKR regulates T315 in SPNR or ZFR1. Finally, it is likely that the NFARs are phosphorylated by other kinases, although on other sites, although little is presently known regarding this potential form of NFAR post-translational control (Parrott et al. 2005). In summary, our data indicate that PKR uses the NFAR proteins to control translation of foreign mRNAs in an eIF2α-independent manner, a process that accentuates a novel mechanism of cellular defense against viral replication.
Materials and methods
Plasmids
HA-tagged NFAR proteins expressing plasmids pcDNA3.1(−) NFAR1 and NFAR2, which were also constructed in our laboratory, were mutated at nucleotide positions 562 (A–G), 562–564 (ACG–GAC), 943 (A–G), and 943 (ACA–GAC) of the NFAR coding sequence using the Site-Directed Mutagenesis kit (Stratagene).
Generation of antibody to phosphorylated T188 and T315
Affinity-purified antibody was made to phosphomimetic T188 (EMEKVLAGE[pT]LSVNDC) and T315 (DRQQREDI[Pt]QSAQHAC) by New England Peptide, Inc.
Bacterial expression
pEGST, pESGT-PKR, pEGST-PKR-NFAR M9, pEGST-PKR-NFARM9 T188, 315A, pEGST-PKR-NFAR1 M10, and pEGST-PKR-NFAR2 M10 were used to transform E. coli BL21(DE3) (Novagen) (Matsui et al. 2001). Bacterial proteins were labeled as described previously (Barber et al. 1991).
Identification of phospho sites
His-tagged NFAR M9 was purified from PKR-coexpressing bacteria using Cobalt columns (Qiagen), and was analyzed using microcapillary HPLC nano-electro spray tandem mass spectrometry (Harvard Microchemistry Facility, Harvard University).
Cells and transfection of DNA plasmids
MEF, HeLa, 293T, and XLK-WG cells were maintained according to American Type Culture Collection protocols. Transfections were carried out using Plus reagent or Lipofectamine 2000 (Invitrogen) transfection reagents in Opti-MeM (Invitrogen) according to manufacturer's protocol. MEFs were transfected using AMAXA Nucleofector Apparatus (program A-023) and MEF Nucleofected Kit 1 according to the manufacturer's recommendations (AMAXA Biosystems).
Immunoprecipitation
Cells or tissues were lysed with RIPA buffer or M-PER supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail 1 (Sigma-Aldrich). Immunoprecipitation was performed with NFAR polyclonal rabbit antiserum or rabbit IgG as a control or anti-HA beads (Covance).
Production of a conditional NFAR knockout
NFAR Cre-loxP mice were generated by inGenious Targeting Laboratory, Inc., and the Transgenic Facility at the University of Miller School of Medicine. A 9-kb fragment containing a neomycin selection cassette flanked by FRT sites was electroporated into embryonic stem cells. MEF isolation was obtained as described previously (Venkataraman et al. 2007). For inactivation of NFARs, NFARFlox/Flox MEFs were plated into a 10-cm dish (1 × 106 cells per dish) and grown for 1 d. Cells were infected with 2.5 × 108 pfu of Ad-GFP or Ad-Cre-GFP, an adenovirus that expresses Cre from the cytomegalovirus promoter (Vector Biolabs), for 3 d, after which time the virus was removed and fresh medium was added. To generate T-cell-specific NFAR-deficient mice, NFARFlox/Flox (Nf/f) mice were bred to transgenic mice expressing Cre recombinase under the direction of the Lck promoter (LCk-Cre) (Jackson Laboratory) to generate NFARFlox/Flox/Lck-Cre+ (Nf/f-Cre+) mice. Screening of tail DNA for inheritance of the floxed NFAR gene was performed by PCR.
In vivo imaging of mice
Imaging of was carried out by the Oncogenomic Core Facility for their help with the IVIS Spectrum small animal imager.
Luciferase reporter assay and mRNA level
Firefly luciferase (250 ng) and renilla luciferase (50 ng) constructs were transfected into MEFs and assays were carried out as described previously (Pfeifer et al. 2008).
Virus infections
Cells were grown in six-well plate to 70% or in 10-cm dish to 80% and were infected with VSV-GFP, SeV, or EMCV using the indicated m.o.i. For determination of viral titer, BHK cells were employed for VSV-GFP. Plaques were fixed and visualized with 0.1% crystal violet and 30% methanol. Viral titers are expressed in pfu per milliliter.
Confocal microscopy
HeLa, 293T, and MEF cells were fixed in 2% paraformaldehyde and were permeabilized with 0.2% Triton X-100. For immunofluoresence, NFAR antiserum (1:100), anti-PKR antibody (1:100) (Santa Cruz Biotechnology), and anti-HA antibody (1:250) (Abcam) were used, and DAPI was used as a nuclear marker.
Cell fractionation
Cells fractionation was carried out as described (Parrott et al. 2005).
RNA pull-down
RNA pull-down assay was carried out as described (Pfeifer et al. 2008).
RNA–protein interaction in vivo
RNA–protein interaction assay was carried out as described (Niranjanakumari et al. 2002).
RNAi
The anti-human NF45 (L-017599) and mouse NF45 (L-060911) ON-TARGETplus SMARTpool were purchased from Dharmacon. Cells grown in six-well or 24-well plates to 30%–40% confluence were transfected using RNAiMAX (Invitrogen).
Polysome analysis
Cells were cultured as above and polysome analysis was carried out as described previously (Mamane et al. 2007).
Acknowledgments
We thank Dr. T. Date for bacterial duel expression vectors, Dr. R.J. Kaufman for eIF2α (S51A) MEFs, and Dr. T.M. Alain, Dr. N. Sonenberg, and Dr. J. Bell for research relating to the use of PKR-deficient mice.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1965010.
Supplemental material is available at http://www.genesdev.org.
References
- Agbottah ET, Traviss C, McArdle J, Karki S, St Laurent GC III, Kumar A 2007. Nuclear factor 90 (NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology 4: 41 doi: 10.1186/1742-4690-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13: 129–141 [DOI] [PubMed] [Google Scholar]
- Barber GN, Tomita J, Hovanessian AG, Meurs E, Katze MG 1991. Functional expression and characterization of the interferon-induced double-stranded RNA activated P68 protein kinase from Escherichia coli. Biochemistry 30: 10356–10361 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG 2002. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol 156: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostowski J, Robinson C, Orford R, Elgar S, Scarlett G, Peterkin T, Malartre M, Kneale G, Wormington M, Guille M 2000. RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. EMBO J 19: 3683–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Scherer BJ, Schnier J, Davies MV, Kaufman RJ, Hershey JW 1992. Stimulation of protein synthesis in COS cells transfected with variants of the α-subunit of initiation factor eIF-2. J Biol Chem 267: 286–293 [PubMed] [Google Scholar]
- Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M 1999. JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity 11: 721–731 [DOI] [PubMed] [Google Scholar]
- Corthesy B, Kao PN 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem 269: 20682–20690 [PubMed] [Google Scholar]
- Galabru J, Hovanessian A 1987. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem 262: 15538–15544 [PubMed] [Google Scholar]
- Goh KC, deVeer MJ, Williams BR 2000. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J 19: 4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K 1994. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science 265: 103–106 [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Evers S, Macara IG, Dargemont C 2004. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem 279: 884–891 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J 22: 5655–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, Behrens SE 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13: 1675–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem 269: 20691–20699 [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H 1992. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci 89: 6232–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D, London IM 1978. Regulation of protein synthesis: Activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci 75: 1121–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N 2007. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE 2: e242 doi: 10.1371/journal.pone.0000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 12: 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Tanihara K, Date T 2001. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem Biophys Res Commun 284: 798–807 [DOI] [PubMed] [Google Scholar]
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M 1996. A mammalian RNA editing enzyme. Nature 379: 460–464 [DOI] [PubMed] [Google Scholar]
- Merrill MK, Gromeier M 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol 80: 6936–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62: 379–390 [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Orford RL, Robinson C, Haydon JM, Patient RK, Guille MJ 1998. The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double-stranded-RNA-binding subunit. Mol Cell Biol 18: 5557–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panniers R, Henshaw EC 1983. A GDP/GTP exchange factor essential for eukaryotic initiation factor 2 cycling in Ehrlich ascites tumor cells and its regulation by eukaryotic initiation factor 2 phosphorylation. J Biol Chem 258: 7928–7934 [PubMed] [Google Scholar]
- Parker LM, Fierro-Monti I, Mathews MB 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J Biol Chem 276: 32522–32530 [DOI] [PubMed] [Google Scholar]
- Parrott AM, Walsh MR, Reichman TW, Mathews MB 2005. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J Mol Biol 348: 281–293 [DOI] [PubMed] [Google Scholar]
- Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, Williams BR, Sen GC 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem 274: 20432–20437 [DOI] [PubMed] [Google Scholar]
- Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN 2008. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci 105: 4173–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A, Nayernia K, Engel W, Torres M, Stoykova A, Chowdhury K, Gruss P 2001. Mice deficient for spermatid perinuclear RNA-binding protein show neurologic, spermatogenic, and sperm morphological abnormalities. Dev Biol 233: 319–328 [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Deigendesch N, Dey M, Dever TE, Tazi L 2008. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: Varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol 6: 12 doi: 10.1186/1741-7007-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC 1998. Molecular basis of double-stranded RNA–protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 17: 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y 2009. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol 29: 3754–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Barber GN 2003. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J 17: 961–983 [DOI] [PubMed] [Google Scholar]
- Saunders LR, Jurecic V, Barber GN 2001a. The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics 71: 256–259 [DOI] [PubMed] [Google Scholar]
- Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, Barber GN 2001b. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem 276: 32300–32312 [DOI] [PubMed] [Google Scholar]
- Scarlett GP, Elgar SJ, Cary PD, Noble AM, Orford RL, Kneale GG, Guille MJ 2004. Intact RNA-binding domains are necessary for structure-specific DNA binding and transcription control by CBTF122 during Xenopus development. J Biol Chem 279: 52447–52455 [DOI] [PubMed] [Google Scholar]
- Schumacher JM, Lee K, Edelhoff S, Braun RE 1995. Spnr, a murine RNA-binding protein that is localized to cytoplasmic microtubules. J Cell Biol 129: 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem 280: 18981–18989 [DOI] [PubMed] [Google Scholar]
- Shi L, Godfrey WR, Lin J, Zhao G, Kao PN 2007. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med 204: 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Lim H, Yates JR III, Karin M 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell 10: 1331–1344 [DOI] [PubMed] [Google Scholar]
- Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM 2002. Host cell proteins binding to the encapsidation signal ɛ in hepatitis B virus RNA. Arch Virol 147: 471–491 [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, Castano ME, Hernandez-Verdun D, St-Laurent G III, Kumar A 2006. Nuclear factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology 3: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178: 6444–6455 [DOI] [PubMed] [Google Scholar]
- Xu YH, Grabowski GA 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid β-glucosidase and other mRNAs. Mol Genet Metab 68: 441–454 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]