Abstract
Human DEC1 (deleted in esophageal cancer 1) gene is located on chromosome 9q, a region frequently deleted in various types of human cancers, including squamous cell carcinoma of the head and neck (SCCHN). However, only one epidemiological study has evaluated the association between DEC1 polymorphisms and cancer risk. In this hospital-based case–control study, four potentially functional single-nucleotide polymorphisms −1628 G>A (rs1591420), −606 T>C [rs4978620, in complete linkage disequilibrium with −249T>C (rs2012775) and −122 G>A(rs2012566)], c.179 C>T p.Ala60Val (rs2269700) and 3′ untranslated region-rs3750505 as well as the TP53 tumor suppressor gene codon 72 (Arg72Pro, rs1042522) polymorphism were genotyped in 1111 non-Hispanic Whites SCCHN patients and 1130 age-and sex-matched cancer-free controls. After adjustment for age, sex and smoking and drinking status, the variant −606CC (i.e. −249CC) homozygotes had a significantly reduced SCCHN risk (adjusted odds ratio = 0.71, 95% confidence interval = 0.52–0.99) compared with the −606TT homozygotes. Stratification analyses showed that a reduced risk associated with the −606CC genotype was more pronounced in subgroups of non-smokers, non-drinkers, younger subjects (defined as ≤57 years), carriers of the TP53 Arg/Arg (rs1042522) genotype, patients with oropharyngeal cancer or late-stage SCCHN. Further in silico analysis revealed that the −249 T-to-C change led to a gain of a transcription factor-binding site. Additional functional analysis showed that the −249T-to-C change significantly enhanced transcriptional activity of the DEC1 promoter and the DNA–protein-binding activity. We conclude that the DEC1 promoter −249 T>C (rs2012775) polymorphism is functional, modulating susceptibility to SCCHN among non-Hispanic Whites.
Introduction
Squamous cell carcinoma of the head and neck (SCCHN), including cancers of the oral cavity, oropharynx, hypopharynx and larynx, is one of the most commonly diagnosed cancer worldwide with about 400 000 new cases annually (1). Although the incidence and mortality rates of SCCHN have decreased slightly in the last few decades in the USA, ∼49 260 new cases and 11 480 deaths of SCCHN occurred in 2010 (2).
SCCHN is a multifactorial disease, and its etiology involves interactions of genetic variation with environmental factors. While excessive tobacco smoking and alcohol consumption are the well-known risk factors for all SCCHN and oncogenic types of human papillomavirus (HPV) for the oropharyngeal subjects of SCCHN, the fact that a substantial proportion of SCCHN patients were non-smokers or non-drinkers indicates that genetic predisposition may play an important role in the etiology of SCCHN, possibly due to polymorphisms in genes involved in cell cycle regulation, DNA repair and apoptosis (3–5). However, most of the major susceptibility genes of SCCHN still remain to be elucidated.
The human DEC1 (deleted in esophageal cancer 1, also known as CTS9) gene is located on chromosome 9q, spanning ∼2.4 kb, and it contains eight exons (National Center for Biotechnology Information reference sequences, NCBI RefSeq). Several loss of heterozygosity studies have reported frequent allelic deletions of chromosome 9q in various types of human cancers, including cancers of the esophagus (6,7), lung (8–10), head and neck (11–13) and urinary bladder (14–16). These results suggest the presence of at least one tumor suppressor gene (TSG) associated with development of these cancers may be located in this region. Furthermore, previous studies showed that expression of DEC1 was reduced or absent in esophageal cancer, and its in vitro and in vivo tumor suppressive abilities were also observed (17–19). Although the detailed biological function remains to be clarified, DEC1, as a candidate TSG for esophageal cancer, may play roles in suppression of SCCHN development because these two cancers share the same risk factors such as smoking.
As of May 2010, a total of 2542 variants in human DEC1 gene have been reported according to the single-nucleotide polymorphism database (dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/). However, only one epidemiological study has been conducted to investigate the association between genetic polymorphisms of DEC1 and cancer risk (20). To examine the association between those common (with minor allele frequencies of at least 5%) and possibly functional DEC1 single-nucleotide polymorphisms (SNPs) and SCCHN susceptibility, we first used the bioinformatics tool of SNP Function Prediction (FuncPred, http://snpinfo.niehs.nih.gov/snpfunc.htm) to identify their potential functional relevance. As a result, four potential regulatory SNPs, namely 5′ untranslated region (UTR): −1628 G>C (rs1591420) and −606 T>C [rs4978620, tagging both of −249 T>C (rs2012775) and −122 G>A (rs2012566) with complete linkage disequilibrium]; exon 7: 179 C>T (rs2269700) and 3′ UTR: c.*170G>A (rs3750505), were chosen to be genotyped in 1111 non-Hispanic Whites SCCHN patients and 1130 age-and sex-matched cancer-free controls.
We also genotyped the TP53 tumor suppressor gene codon 72 polymorphism (Arg72Pro, rs1042522), which has been reportedly associated with susceptibility to tobacco-related cancers (21–23), to evaluate its joint effect with the DEC1 SNPs. High-order interactions between genetic and environmental factors for SCCHN risk were further evaluated using the classification and regression tree (CART) methodology. Finally, we performed laboratory experiments to analyze functional relevance/significance of any of the regulatory SNPs found to be associated with SCCHN risk.
Materials and methods
Study subjects and sample collection
The recruitment of study subjects for the present study was described previously (24). Briefly, during an 8 years period between October 1999 and October 2007, patients with newly diagnosed, histopathologically confirmed SCCHN were consecutively recruited at The University of Texas . D. Anderson Cancer Center. Patients who had received prior surgery (other than diagnostic biopsies), chemotherapy or radiation therapy before recruitment, any blood transfusion during the preceding 6 months, any malignancies other than SCCHN, second SCCHN primary tumors, primary tumors of nasopharynx or sinonasal tract or primary tumors outside the upper aerodigestive tract were excluded. An additional 1130 age- (±within 5 years) and sex-matched, self-reported cancer-free controls, who agreed to participate in the study, were also enrolled during the same period from those hospital visitors who were not seeking health care but accompanying other cancer patients to our outpatient clinics. The controls were frequency matched to cases by age (±within 5 years), sex and ethnicity and were not genetically related to the cases or one another included in this study. Because the expected differences in genotype frequencies between ethnic groups, the few minority patients enrolled were excluded from the analysis. Thus, our final analysis included 1111 SCCHN patients and 1130 controls of non-Hispanic Whites.
During an in-person survey, all potential subjects were interviewed to identify their willingness to participate in research studies and to collect their demographic and risk factors information, such as age, sex, ethnicity and the history of tobacco and alcohol consumption using a standardized, structured questionnaire (25). Among those subjects who we had been contacted for interview and recruitment, the response rates for SCCHN patients and cancer-free controls were ∼92 and 85%, respectively. Each subject provided a one-time 30 ml venous blood samples (after the diagnosis and before the initiation of treatment for the cases), and samples were kept frozen till DNA extraction for genotyping. An informed consent was obtained from all recruited individuals, and the research protocol was approved by the M. D. Anderson Institutional Review Board.
Selection of potential functional polymorphisms
We first used the computational tool of SNP Function Prediction (FuncPred, http://snpinfo.niehs.nih.gov/snpfunc.htm) to select any SNP with any of the following predicted functionalities: (i) affecting transcription factor-binding sites (TFBSs) activity in the putative promoter region (here, we defined as 2 kb upstream from the first exon), (ii) the introduction of premature termination codons, (iii) single-amino-acid substitutions or changing the frame of the protein-coding region, resulting in alteration of protein structures or properties or (iv) affecting the microRNA-binding sites activity. After we limited the SNPs to those with minor allele frequency >5% in Hapmap CEU population, six potential functional SNPs (of a total 15 SNPs) were identified, that is −1628 G>C (rs1591420), −606 T>C (rs4978620), −249 T>C (rs2012775) and −122 G>A (rs2012566), c.179 C>T p.Ala60Val (rs2269700) and c.*170G>A (rs3750505). In a small subset of our study population (n = 100), we found that the three SNPs −606 T>C (rs4978620), −249 T>C (rs2012775) and −122 G>A (rs2012566) were in complete linkage disequilibrium (r2 = 1); thus, only −606 T>C (rs4978620) was selected for further genotyping.
Genotyping of DEC1 polymorphisms
The genomic DNA was extracted from peripheral blood leukocytes using the DNA Blood Mini Kit (Qiagen, Valencia, CA), according to the protocol of the manufacturer. The quantification of DNA was determined using a Nanodrop analyzer (ND-1000) spectrophotometer (Nano Drop Technologies, Wilmington, DE).
Among the four selected SNPs, two SNPs [c.179 C>T p.Ala60Val (rs2269700) and c.*170G>A (rs3750505)] were genotyped using the TaqMan assays (Applied Biosystems, Foster City, CA) and the other two DEC1 promoter SNPs, −1628 G>C (rs1591420), −606 T>C (rs4978620) was genotyped using the SNPlex genotyping system (Applied Biosystems) in the DNA Core Lab at M. D. Anderson Cancer Center. The data of genotypes from the SNPlex assays were analyzed by the GeneMapper Software (version 4.0; Applied Biosystems). The method of polymerase chain reaction (PCR)–restriction fragment length polymorphism was also used in a subsequent effort to obtain the missing genotyping data for samples failed in the SNPlex assays (11% of all samples). Briefly, PCR was used to amplify the target fragments containing the twopromoter SNPs using primers sequence as follows: (i) −1628G>C (forward) 5′-TGTAAATATAAGATGCTAACAAAATC-3′ and (reverse) 5′-CATTCAGAGAAGCATTTCTAAT-3′ and (ii) −606T>C (forward) 5′-TGTTAAATCTGGAATGAACTTCAGA-3′ and (reverse) 5′-TCAATCACAACATGCTTTCCTAC-3′. The PCRs were performed using 35 cycles with an annealing temperature of 57°C (for −1628 G>C) and 60°C (for −606 T>C). After digested with TaqI restriction enzyme (New England Biolabs, Beverly, MA) at 65°C overnight, the 158 bp PCR products containing the −1628G>C site were cleaved into 131 and 27 bp fragments in the presence of C allele, whereas G alleles remained uncut. Similarly, the 165 bp PCR products containing −606 T>C site were cleaved into 139 and 26 bp fragments in the presence of T alleles, whereas C allele remained uncut after digested with EcoRV (New England Biolabs) at 37°C overnight. For these two SNPs, we randomly selected ∼10% of the samples and repeated with the PCR–restriction fragment length polymorphism method for confirmation, and the error rate was <1%. Those samples (one case and six controls) with either failed genotyping results in all selected SNPs or inconsistent results in repeated genotyping assays were further excluded from the final analysis. All outputs of genotype results were incorporated into Microsoft Excel 2003 spreadsheets for further statistical analysis.
Construction of promoter–reporter plasmids
Genomic DNA of both common and variant homozygous for the SNPs of −606 T>C (i.e. −249 T>C and −122 G>A) were amplified by PCR using specific forward primer 5′-AAGGTACCCAGAATGATTTGCTGCAAGG-3′ contained a KpnI site and reverse primer 5′-AAGCTAGCGCCAGGGTGAGGTAGAAACA-3′ contained a NheI sites (the underlined sequence represented each restriction enzyme site). To generate the luciferase reporter plasmid DEC1-Leu, the 1202 bp fragment (from −1178 to +24 bp relative to the transcription start site) of the DEC1 putative promoter region containing −606 T or C was cloned into a KpnI–NheI-digested pGL3-Basic firefly luciferase vector (Promega, Madison, WI). After cloning, the plasmids were sequenced to confirm the orientation and integrity in both of allelic reporter constructs.
Cell culture, transient transfection and luciferase reporter assay
The human colon cancer cell line HCT116 (a gift from Dr Bert Vogelstein of John Hopkins University School of Medicine) and head and neck carcinoma cell line MDA-1386Ln (a gift from Dr Jeffrey N.Myers of M. D. Anderson) were routinely maintained in 1× Dulbecco’s modified Eagle’s medium, and head and neck carcinoma cell lines UM-SCC-17B and UM-SCC-22A (a gift from Dr Reuben Lotan of M. D. Anderson) were regularly cultured in Dulbecco’s modified Eagle’s medium/F-12 medium, supplemented with 10% fetal bovine serum (Sigma–Aldrich, St Louis, MO) at 37°C in a humidified, 5% CO2 incubator. A total of 2 × 105 cells for HCT116, UM-SCC-17B and UM-SCC-22A and 1 × 105 cells for MDA-1386Ln were seeded onto each well of the 24-well plates and transiently transfected with 1.0 μg of plasmid DNA with both of common and variant of −606 T>C/−249T>C/−122 G>A reporter constructs using FuGENE HD Transfection Reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s protocol. As a transfection efficiency internal control, all plasmids were co-transfected with 20 ng of p-TK renilla luciferase (pRL-TK) (Promega). After 48 h of incubation, the cells were lysed in 100 μl of lysis buffer, and 20 μl of the supernatant was then measured for luciferase activities with a dual-luciferase reporter assay system (Promega) and normalized by the activity of Renilla luciferase. The data was expressed as mean ± standard errors of at least three replicates obtained from three independent experiments.
Identification of putative TFBS
The flanking sequences (∼40 bp) of the promoter SNPs −606 T>C/−249 T>C/−122 G>A were all analyzed for the presence of predicted TFBSs by using computational tools of Transcription Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess) and TF search (http://www.cbrc.jp/research/db/TFSEARCH). The putative functional impact of this promoter SNP(s) on TFBSs was assessed by identifying any formation of a new TFBS or loss of a TFBS due to the base pair change. By this in silico analysis, we identified that the −249 T-to-C change would completely gain a TFBS.
Nuclear extract preparation and electrophoretic mobility shift assay
Nuclear proteins were extracted from human head and neck carcinoma cell lines UM-SCC-17B and UM-SCC-22A as described previously (26). Probes comprising the sequence from −268 to −230 with complementary strands corresponding to −249 T allele (common) and −249 C allele (variant) were 5′-TAAATTTGGTAACATTTGCAACTGGAAAAT-3′ and 5′-TAAATTTGGTAACACTTGCAACTGGAAAAT-3′ (the -249 position is highlighted in italics). Single-stranded synthetic oligonucleotides were biotin-labeled using the 3′-end biotin labeling kit (Thermo Scientific, Rockford, IL) and re-annealed to double strand and identical but unlabeled oligonucleotides with the same sequences were used as competitors.
The EMSA assay was performed using the LightShift Chemiluminescent electrophoretic mobility shift assay kit (Thermo Scientific) according to the experimental procedures provided by the manufacturer. Briefly, 10 μg of nuclear protein extracts were incubated with 3′ biotin-labeled at room temperature for 30 min. The specificity of the Nkx-2.5 and DNA-binding activity was determined by adding a 50-fold excess of unlabeled oligonucleotides for competition reactions. DNA–protein complexes were subsequently fractionated on a non-denaturing 6% polyacrylamide gel and transferred onto a 0.45 μm nylon membrane (Thermo Scientific). After blocking, the membrane was incubated with LightShift stabilized streptavidin–horseradish peroxidase conjugate and developed using the luminol/enhancer and stable peroxide solutions included in the kit (Thermo Scientific). The Nkx-2.5 and DNA complexes were visualized by exposure of the membrane to X-ray film. Supershift experiments were performed by adding anti-Nkx-2.5 antibodies (Santa Cruz biotechnology, Santa Cruz, CA) or non-specific rabbit IgG (Santa Cruz Biotechnology). NKE-2, the highly conserved Nkx-2.5 response element in the proximal region of the cardiac atrial natriuretic factor promoter (27,28), was used as a positive control for Nkx-2.5 binding (Nkx-2.5 consensus binding sequence: 5′-CCTTTGAAGTGGGGGCCTCTTGAGGCAA-3′).
Statistical analysis
Chi-square or Fisher’s exact test (when an expected cell count was <5) were computed to test the differences in the distribution of category variables, including demographic characteristics, smoking and drinking status, DEC1 genotype/allele frequencies between cases and controls. The deviation from Hardy–Weinberg equilibrium among controls was tested by chi-square goodness-of-fit test, and the linkage disequilibrium coefficient r2 for each of the SNP pairs was also examined. We reconstructed haplotypes for the DEC1 promoter SNPs using the PHASE program described previously (29,30), and Pearson’s chi-square test was performed to test for the difference in haplotype distributions between cases and controls. To assess the association between DEC1 genotypes/haplotypes and disease status, the crude and adjusted odds ratios (ORs) with the 95% confidence intervals (95% CIs) were estimated using unconditional logistic regression analyses. Additional stratified analyses of associations of DEC1 genotypes with SCCHN risk by subgroups of age, sex, smoking and drinking status and tumor sites were also performed, followed by analyses of gene–environment interactions, which were evaluated by the P value for the interaction term in multivariate logistic regression models with adjustment for age, sex, smoking and drinking status. Cochran-Mantel-Haenszel chi-square test was carried out to examine the homogeneity and trend of ORs across strata of each variable, after controlling for possible confounding effects of age and sex. The statistical significance of fold induction between tworeporter constructs in different cancer cell lines was calculated and tested using Student’s t-test.
To identify groups with a higher probability of developing SCCHN and evaluate higher-order interactions between genetic and environmental factors, we performed the nonparametric CART analysis using the CART software (version 5.0; Salford systems, San Diego, CA) described elsewhere (31,32). Briefly, the CART method builds a decision tree based on binary recursive partitioning to identify subgroups at higher risk (33). The recursive-partitioning algorithm starts with the root node that contains all the subjects’ data and uses a statistical hypothesis-testing method to determine the first optimal split with the smallest multiplicity-adjusted P value (<0.05) for the root node. The branch of the decision tree was pruned, if there was no statistically significant split for each subsequent node or there were less than the pre-specified minimum size in the terminal nodes. The reference was the one with the lowest percentage of SCCHN cases. The higher-order interactions of genetic and environmental factors identified by the CART were further analyzed using the multivariate logistic regression to calculate the adjusted ORs and 95% CIs in each terminal node of the tree. All statistical tests were two sided with a statistical significance level of a P value <0.05. Data were analyzed using Statistical Analysis System/Genetics software program (SAS/STAT version 9.1.3; SAS Institute Inc., Cary, NC).
Results
Characteristics of the study population
We examined associations between the studied DEC1 SNPs and risk of SCCHN in 1111 cases (mean age ± standard deviation: 57.1 ± 11.1 years; range18–90 years) and 1130 controls (56.8 ± 11.0 years; range 20–87 years) of non-Hispanic Whites. The distribution of demographic characteristics of all subjects is shown in Table I. The cases and controls were adequately matched by age and sex (P = 0.419 and 0.671, respectively). However, there were more current-smokers (38.0 versus 14.6%, P < 0.0001) and drinkers (50.9 versus 40.3%, P < 0.0001) among cases than controls. Therefore, these variables were further adjusted for in subsequent multivariate analyses. Among patients with primary tumors included in the analysis, 560 (50.4%) had oropharyngeal cancers (the site having the strongest association with HPV infection), 323 (29.1%) had oral cancers and 216 (19.9%) had laryngeal cancers (including 43 hypopharyngeal cancers) (Table I).
Table I.
Frequency distribution of demographic characteristics of SCCHN cases and cancer-free controls
| Variables | Cases, No. (%) | Controls, No. (%) | P valuea |
| Total subjects | 1111 (100.0) | 1130 (100.0) | |
| Ethnicity | |||
| Non-Hispanic Whites | 1111 (100.0) | 1130 (100.0) | |
| Age group (years) | |||
| Range, median | 18–90, 57 | 20–87, 57 | |
| Mean ± SD | 57.1 ± 11.1 | 56.8 ± 11.0 | 0.419 |
| <50 | 303 (27.3) | 318 (28.1) | 0.648 |
| 50–57 | 290 (26.1) | 270 (23.9) | |
| 57–65 | 274 (24.7) | 294 (26.0) | |
| >65 | 244 (22.0) | 248 (22.0) | |
| Sex | 0.671 | ||
| Female | 274 (24.7) | 270 (23.9) | |
| Males | 837 (75.3) | 860 (76.1) | |
| Smoking status | <0.0001 | ||
| Never | 308 (27.7) | 553 (48.9) | |
| Former | 381 (34.3) | 412 (36.5) | |
| Current | 422 (38.0) | 165 (14.6) | |
| Alcohol status | <0.0001 | ||
| Never | 304 (27.4) | 494 (43.7) | |
| Former | 242 (21.8) | 181 (16.0) | |
| Current | 565 (50.9) | 455 (40.3) | |
| Tumor sitesb | |||
| Oral cavity | 323 (29.1) | — | |
| Oropharyngeal | 560 (50.4) | — | |
| Hypopharyngeal | 43 (3.9) | — | |
| Larynx | 173 (15.6) | — | |
| Clinical stagec | |||
| Stage I | 119 (10.7) | — | |
| Stage II | 157 (14.1) | — | |
| Stage III | 190 (17.1) | — | |
| Stage IV | 644 (58.0) | — | |
Two-sided χ2 test for categorical variables or t-test for a continuous variable.
Excluded subjects with nose/paranasal sinus and multiple sites.
The numbers for clinical stage may not add up to the total numbers due to the missing information from some subjects.
Association between DEC1 SNPs and risk of SCCHN
The allele and genotype distributions of the studied DEC1 SNPs are presented in Table II. All observed genotype distributions among controls were in agreement with Hardy–Weinberg equilibrium (all P > 0.05). When the DEC1 −606TT genotype was used as the reference, the −606CC homozygous genotype was found to be associated with a significantly reduced risk of SCCHN (adjusted OR = 0.71; 95% CI = 0.52–0.99) after adjustment for age, sex, smoking and drinking status. For variant genotypes of other SNPs, however, no significantly altered SCCHN risk was observed compared with their common genotypes (GG for rs1591420, TT for rs2269700, GG for rs3750505 and GG for TP53 codon 72 (Table II). In the haplotype analysis, we did not observe any statistically significant association between six common haplotypes (frequencies ≥ 5%) of these four independent SNPs (rs1591420, rs4978620, rs2269700 and rs3750505) and risk of SCCHN (global test P = 0.946, data not shown). In the stratified analysis, the association between DEC1 −606T>C SNP and SCCHN risk was further evaluated by subgroups of age, sex, smoking and drinking statuses, tumor sites and tumor stages. As shown in Table III, a reduced risk associated with the −606CC genotype was particularly more pronounced in strata of non-smokers (adjusted OR = 0.48, 95% CI = 0.26–0.89), non-drinkers (adjusted OR = 0.55, 95% CI = 0.32–0.95), younger subjects (defined as subjects ≤ 57 years, adjusted OR = 0.63, 95% CI = 0.41–0.97), carriers of TP53 codon 72 (rs1042522) Arg/Arg genotypes (adjusted OR = 0.61, 95% CI = 0.39–0.94), patients with oropharyngeal cancer (adjusted OR = 0.64, 95% CI = 0.43–0.95) and patients with late-stage SCCHN (Stage IV, adjusted OR = 0.68, 95% CI = 0.47–0.98). These results suggested potential interactions between age, smoking and drinking status, TSG TP53 and the DEC1 −606T>C SNP in the etiology of SCCHN. However, there was no statistical evidence in further multivariate logistic regression models to support an interaction between DEC1 variant genotypes and these risk factors on SCCHN risk (all P > 0.05, Table III).
Table II.
Genotypes distribution of the DEC1 polymorphisms among SCCHN cases and cancer-free controls and their associations with SCCHN risk
| Genotypes | Cases, No. (%) | Controls, No. (%) | P value | Crude OR (95% CI) | Adjusted OR (95% CI)a |
| All subjects | 1111 (100.0) | 1130 (100.0) | |||
| DEC1 −1628 G>C (rs1591420) | 0.526b | ||||
| GG | 1008 (91.8) | 1021 (91.6) | 1.00 | 1.00 | |
| CG | 89 (8.1) | 90 (08.1) | 1.00 (0.74–1.36) | 1.03 (0.75–1.42) | |
| CC | 1 (0.1) | 4 (0.4) | 0.25 (0.03–2.27) | 0.33 (0.03–3.16) | |
| CG + CC | 90 (8.2) | 94 (8.4) | 0.842c | 0.97 (0.72–1.31) | 1.00 (0.73–1.37) |
| C allele frequency | 0.041 | 0.044 | 0.680d | ||
| DEC1 −606 T>C (rs4978620)f | 0.267e | ||||
| TT | 529 (47.7) | 537 (47.5) | 1.00 | 1.00 | |
| CT | 492 (44.3) | 481 (42.6) | 1.04 (0.87–1.24) | 0.95 (0.79–1.14) | |
| CC | 89 (08.0) | 112 (09.9) | 0.81 (0.60–1.09) | 0.71 (0.52–0.99) | |
| TT + CT | 1021 (92.0) | 1018 (90.1) | 0.117c | 0.79 (0.59–1.06) | 0.73 (0.54–0.99) |
| C allele frequency | 0.302 | 0.312 | 0.462d | ||
| DEC1 T>C (Ala→Val, rs2269700) | 0.127e | ||||
| TT | 403 (37.7) | 429 (38.8) | 1.00 | 1.00 | |
| CT | 522 (48.8) | 499 (45.2) | 1.11 (0.93–1.34) | 1.11 (0.92–1.35) | |
| CC | 144 (13.5) | 177 (16.0) | 0.87 (0.67–1.12) | 0.87 (0.66–1.14) | |
| TT + CT | 925 (86.5) | 928 (84.0) | 0.094c | 0.82 (0.64–1.04) | 0.82 (0.64–1.05) |
| C allele frequency | 0.379 | 0.386 | 0.629d | ||
| DEC1 G>A (rs3750505) | 0.731e | ||||
| GG | 859 (78.2) | 882 (78.9) | 1.00 | 1.00 | |
| AG | 221 (20.1) | 222 (19.9) | 1.02 (0.83–1.26) | 1.10 (0.88–1.36) | |
| AA | 18 (1.7) | 11 (1.2) | 1.32 (0.65–2.67) | 1.46 (0.70–3.03) | |
| AG + AA | 239 (21.8) | 236 (21.1) | 0.706c | 1.04 (0.85–1.27) | 1.12 (0.90–1.38) |
| A allele frequency | 0.117 | 0.112 | 0.585d | ||
| TP53 Arg72Pro (rs1042522 G>C) | 0.307e | ||||
| GG | 576 (54.7) | 617 (54.6) | 1.00 | 1.00 | |
| CG | 398 (37.8) | 445 (39.4) | 0.96 (0.80–1.14) | 0.96 (0.79–1.15) | |
| CC | 80 (07.6) | 68 (06.0) | 1.26 (0.90–1.78) | 1.28 (0.89–1.83) | |
| C allele frequency | 0.265 | 0.257 | 0.566c |
Statistically significant results (P < 0.05) are highlighted in bold.
ORs were obtained from logistic regression models with adjustment for age, sex, smoking and alcohol status.
Fisher’s exact test for the distribution of three genotypes.
Two-sided χ2 test for distribution of combined genotypes.
Two-sided χ2 test for allele difference between cases and controls.
Two-sided χ2 test for the distribution of three genotypes.
SNP rs4978620 was in complete linkage disequilibrium with rs4978620 and rs2012566.
Table III.
Stratification analysis of the association between DEC1 -249 T > C SNP and SCCHN risk
| Variables |
DEC1 (rs4978620 T>C) |
|||||||
| (Cases/Controls) |
Pa | OR (95% CI) |
PIntc | |||||
| TT+CT |
CC |
TT+CT (ref) versus CC |
||||||
| No. | % | No. | % | Crude | Adjustedb | |||
| All subjects | 1021/1018 | 92.0/90.1 | 89/112 | 8.0/9.9 | 0.117 | 0.79 (0.59–1.06) | 0.73 (0.54–0.99) | |
| Age (median) | 0.796 | |||||||
| ≤57 | 548/527 | 92.6/89.6 | 44/61 | 7.4/10.4 | 0.076 | 0.69 (0.46–1.04) | 0.63 (0.41–0.97) | |
| >57 | 437/491 | 91.3/90.6 | 45/51 | 8.7/9.4 | 0.682 | 0.92 (0.60–1.40) | 0.89 (0.56–1.40) | |
| Sex | 0.991 | |||||||
| Female | 255/249 | 93.1/92.2 | 19/21 | 6.9/7.8 | 0.706 | 0.88 (0.46–1.68) | 0.62 (0.31–1.26) | |
| Male | 766/769 | 91.6/89.4 | 70/91 | 8.4/10.6 | 0.121 | 0.77 (0.56–1.07) | 0.76 (0.54–1.06) | |
| Smoking status | ||||||||
| Never | 293/503 | 95.4/91.0 | 14/50 | 4.6/9.0 | 0.016 | 0.48 (0.26–0.89) | 0.48 (0.26–0.89) | |
| Former | 348/367 | 91.3/89.1 | 33/45 | 8.7/10.9 | 0.286 | 0.77 (0.48–1.24) | 0.80 (0.49–1.29) | 0.433 |
| Current | 380/148 | 90.0/89.7 | 42/17 | 10.0/10.3 | 0.899 | 0.96 (0.53–1.74) | 0.96 (0.52–1.78) | 0.176 |
| Drinking status | ||||||||
| Never | 284/440 | 93.4/89.1 | 20/54 | 6.6/10.9 | 0.040 | 0.57 (0.34–0.98) | 0.55 (0.32–0.95) | |
| Former | 222/161 | 91.7/88.9 | 20/20 | 8.3/11.1 | 0.333 | 0.73 (0.38–1.39) | 0.64 (0.33–1.26) | 0.677 |
| Current | 515/417 | 91.3/91.6 | 49/38 | 8.7/8.4 | 0.849 | 1.04 (0.67–1.63) | 0.95 (0.59–1.53) | 0.495 |
| TP53 (codon 72) | 0.070 | |||||||
| Arg/Arg | 537/557 | 93.2/90.3 | 39/60 | 6.8/9.6 | 0.065 | 0.64 (0.44–1.03) | 0.61 (0.39–0.94) | |
| Arg/Pro+ Pro/Pro | 431/461 | 90.4/89.9 | 46/52 | 9.6/10.1 | 0.795 | 0.95 (0.62–1.44) | 0.89 (0.57–1.40) | |
| Tumor site | — | |||||||
| Oral cavity | 295/1018 | 91.3/90.1 | 28/112 | 8.7/9.9 | 0.504 | 0.86 (0.56–1.33) | 0.87 (0.55–1.40) | |
| Oropharynx | 521/1018 | 93.2/90.1 | 38/112 | 6.8/9.9 | 0.034 | 0.66 (0.45–0.97) | 0.64 (0.43–0.95) | |
| Hypopharynx + larynx | 196/1018 | 90.7/90.1 | 20/112 | 9.3/9.9 | 0.768 | 0.93 (0.56–1.53) | 0.85 (0.49–1.49) | |
| TNM stage | — | |||||||
| I | 108/1018 | 90.8/90.1 | 11/112 | 9.2/9.9 | 0.816 | 0.93 (0.48–1.77) | 0.91 (0.47–1.77) | |
| II | 143/1018 | 91.1/90.1 | 14/112 | 8.9/9.9 | 0.694 | 0.89 (0.50–1.59) | 0.91 (0.50–1.67) | |
| III | 173/1018 | 91.0/90.1 | 17/112 | 9.0/9.9 | 0.679 | 0.89 (0.52–1.53) | 0.85 (0.49–1.49) | |
| IV | 596/1018 | 92.7/90.1 | 47/112 | 7.3/9.9 | 0.065 | 0.72 (0.50–1.02) | 0.68 (0.47–0.98) | |
| Nodal stage | ||||||||
| N0 | 372/1018 | 91.4/90.1 | 35/112 | 8.6/9.9 | 0.440 | 0.86 (0.58–1.23) | 0.82 (0.54–1.26) | |
| N1–3 | 648/1018 | 92.3/90.1 | 54/112 | 7.7/9.9 | 0.108 | 0.76 (0.54–1.06) | 0.71 (0.50–1.01) | |
| Tumor stage | ||||||||
| T1–2 | 623/1018 | 91.3/90.1 | 59/112 | 8.7/9.9 | 0.374 | 0.86 (0.62–1.20) | 0.83 (0.59–1.17) | |
| T3–4 | 397/1018 | 93.0/90.1 | 30/112 | 7.0/9.9 | 0.078 | 0.69 (0.45–1.05) | 0.64 (0.41–1.01) | |
Statistically significant results (P < 0.05) are highlighted in bold.
P value of the comparison with a two-sided χ2 test.
ORs were obtained from a logistic regression model with adjustment for age, sex, smoking and alcohol status accordingly.
Pint: P value of a chi-square test for the cross product of two variables with adjustment for age, sex, smoking and drinking status in the logistic regression model.
CART analysis of the interactions of genetic and environmental factors with SCCHN risk
The final tree structure generated by CART analysis was presented in Figure 1, which included environmental variables (smoking and drinking status) and genotypes of two SNPs (DEC1 −606T>C and TP53 Arg72Pro G>C) that were found either to be significantly associated with SCCHN risk or has been suggested to be associated with susceptibility to tobacco-related cancers. The first split on the decision tree was smoking status, confirming that smoking status was the most influential predictor for SCCHN risk among those factors considered in this study population.
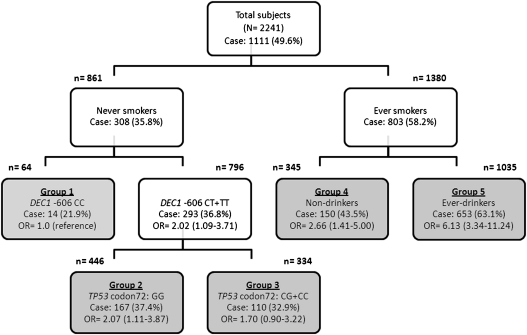
Fig. 1.
Interactions among genetic and environmental factors by CART analysis. The DEC1 −606T>C SNP was classified as having common genotypes (CT + TT) and protective variant (CC); smoking and drinking status was defined as never or ever (former and current) users; Five terminal nodes are highlighted in gray. The numbers in each node may not add up to the total numbers due to the missing information from some subjects. The reference group was the least percentage of cases, and ORs and 95% CIs for each factor combination were obtained from logistic regression models adjusted by age and sex.
We observed additional two distinct patterns of SCCHN risk in the tree structure, resulting five terminal nodes (groups 1 to 5) flanking the tree. The subsequent models were made for each of the never-smokers or ever smokers separately, suggesting that the most important risk factor was the genetic factor of the DEC1 −606T>C SNP in never-smokers, but was drinking status in ever smokers (Figure 1). Never-smokers with the variant DEC1 −606CC genotype exhibited the lowest percentage of SCCHN cases (group 1, 14 cases in the 64 subjects). The final CART model was made for two groups of subjects with common genotypes (−606CT+TT): those who carried TP53 Arg72Pro GG (group 2; OR = 2.07, 95% CI = 1.11–3.87) or CG+CC genotypes (group 3; OR = 1.70, 95%CI = 0.90–3.22). Using node 1 as the reference group, the subgroup of ever smokers and ever drinkers exhibited the highest risk of SCCHN development (group 5, OR = 6.13, 95%CI = 3.34–11.24), with a 63.1% (653/1035) case ratio.
The promoter SNP −249 T>C affected the DEC1 promoter activity
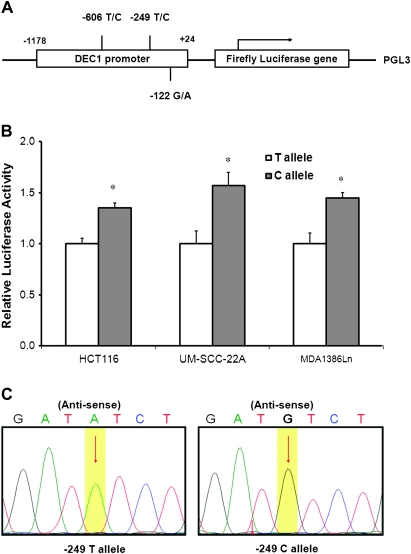
To identify the allele-specific effect of the −249 T>C SNP on the DEC1 promoter activity, we constructed two different luciferase reporter plasmids (common and variant alleles), containing 1202 bp fragment (from −1178 to +24 bp relative to the transcription start site) of the DEC1 promoter region (Figure 2A). The transfection experiments showed that the plasmid containing the protective variant C allele exhibited significantly approximately 35–60% increased luciferase expression than that with the common T allele in two human head and neck cell lines MDA-1386Ln and UM-SCC-22A and a colon cancer cell line HCT116 (all P < 0.05; Figure 2B), indicating that the allelic T-to-C change at the −249 site in the DEC1 promoter resulted in increased promoter activity in a non-tissue specific manner.
Fig. 2.
The DEC1 promoter reporter gene constructs and DEC1-luciferase assays. (A) Schematic presentation of a linealized reporter gene construct containing a 1202 bp fragment of the DEC1 promoter (−1178 to +24 relative to the transcriptional starting site) with both variants at the −606/−249/−122 sites was inserted into the pGL3-basic luciferase expression vector. (B) Luciferase assay for the two DEC1 promoter constructs containing either −249T and −249C alleles, respectively. Both the constructs were sequenced to confirm the orientation and integrity (C). Significant differences between two alleles were indicated by asterisk (*P < 0.01).
The −249 T-to-C allelic change enhanced transcription factor affinity to the promoter
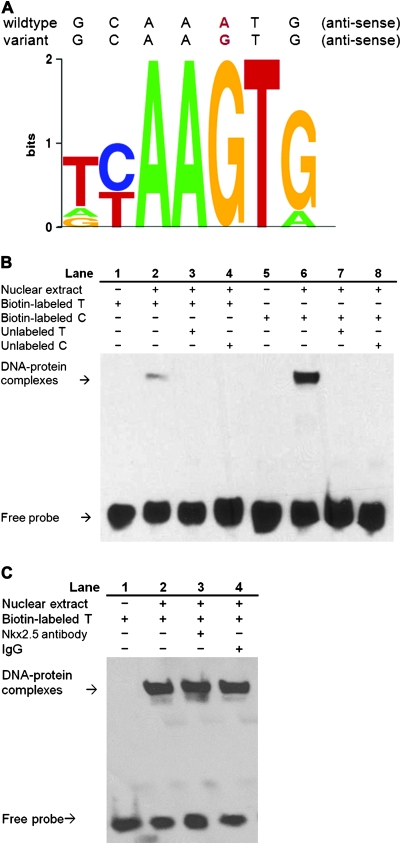
To determine whether the DEC1 −249 T>C SNP alters the binding ability of any transcriptional factor, we used two computational tools of TF Search and TESS to predict the potential TFBS(s) in the flanking region of this SNP. Base on the in silico analysis, we found that the −249 T-to-C allelic change would gain a putative Nkx-2.5-binding site (−251 to −245), which may have a functional impact on DEC1 regulation. The schematic diagram of predicted Nkx-2.5-binding motif from computational tool TESS is presented in Figure 3A.
Fig. 3.
Nuclear factor binding to the −249 T>C polymorphic region of the DEC1 promoter. (A) The putative Nkx-2.5 TFBS at −249 T and C of the DEC1 promoter was predicted by TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess). (B) EMSA was performed using UM-SCC-22A nuclear extracts with biotin-labeled probes corresponding to both −249 T and C of DEC1 gene (T allele: lanes 1–4 or C allele: lanes 5–8), along with or without competition from unlabeled probes. (C) Biotin-labeled probe containing the T allele at −249 site and UM-SCC-22A nuclear extracts in the presence of Nkx-2.5 antibody (lane 3) or normal IgG (lane 4).
Therefore, we performed EMSA to verify whether the −249T>C SNP may affect the TF-binding affinity to the DEC1 promoter and subsequently cause altered transcription. As shown in Figure 3B, the nuclear proteins derived from UM-SCC-22A cells were able to bind to both probes containing either the −249T or −249C alleles, and the biotin-labeled probes with the variant −249C allele had a significantly stronger binding capacity to the nuclear protein extracts than those with −249T allele (Figure 3B, lanes 2 versus lane 6). Competition assays showed that the formation of the DNA–protein complexes was completely eliminated by the 50-fold excess of unlabeled probes containing either −249T or −249C alleles (lanes 3, 4, 7 and 8), suggesting that these binding activities are specific between our designed DNA sequences and nuclear proteins. Similar results were also observed in assays with nuclear proteins extracted from the UM-SCC-17B cell line (data not shown).
To further characterize whether the Nkx-2.5 differentially binds to the surrounding sequences near DEC1 −249T or −249C alleles, gel supershift experiments were further performed by adding anti-Nkx-2.5 antibodies or non-specific rabbit IgG as control antibodies. However, we were unable to detect Nkx-2.5-specific binding to the promoter sequence around SNP −249 T>C because the DNA–protein complex bands formed in EMSA were neither partially abolished nor supershifted by anti-Nkx-2.5 antibodies (Figure 3C, lanes 2 and 3).
Discussion
In this hospital-based case–control study, we presented statistical evidence that the DEC1 promoter variant −606CC (i.e. −249CC) genotype was significantly associated with a reduced risk of developing SCCHN in non-Hispanic Whites. Stratification analyses showed that the protective effect was more evident in subgroups of subjects ≤57 years, non-drinkers, non-smokers and patients with oropharyngeal cancer or with late-stage SCCHN and in the presence of the common genotype of TP53 Arg/Arg (rs1042522). Additional functional experiments further demonstrated that the DEC1 SNP −249 T-to-C allelic change contributed to the increased promoter activity and higher variant allele-specific DNA-protein-binding affinities of certain transcriptional factor(s), supporting the hypothesis that genetic polymorphism influencing DEC1 transcription may play a role in human SCCHN carcinogenesis. In the subsequent experiments, however, we ruled out the possibility that the elevated promoter activity caused by the variant −249C could be attributed to the differential binding of Nkx-2.5, the putative transcription factor predicted by the computational tools.
The human DEC1 was first isolated by Nishiwaki et al. (18) from the TSG locus on chromosome 9q, a region that loss of heterozygosity was frequently observed in cancers originated from several developmentally related tissues, including the esophagus (6,7), lung (8–10), urinary bladder (14–16) and head and neck (11–13). Studies reported that the DEC1 expression was greatly reduced or even absent in esophageal cancer cells (17–19), and its tumor suppression ability was shown in the growth inhibition effects in the in vitro colony formation ability (17,18) and in vivo in nude mice model (19). Using the complementary DNA microarray hybridization approach alone with subsequent validation of reverse transcription–PCR analysis, Leung et al. (17) had further identified a set of genes associated with the DEC1 expression, such as human tissue factor pathway inhibitor-2 (TFPI-2), growth differentiation factor 15 (GDF15), dual specificity phosphatase 6 (DUSP6), Quiescin Q6 (QSCN6) and insulin-like growth factor-binding protein 2 (IGFBP2), and these genes were all shown previously to have roles in tumor progression in different cancers. These findings indicated that DEC1 acts as a TSG in cancer, possibly in SCCHN.
To date, neither transcriptional regulation nor epigenetic modifications of DEC1 expression have been fully understood. One study examined the methylation status of DEC1 from tumor samples of esophageal and lung cancers and did not find any significant difference in the methylation status of the 5′ CpG islands, although a significant reduction in the DEC1 expression was observed (18). Furthermore, multiple lines of evidence have implicated that a genetic polymorphism in the promoter or 3′-UTR may influence transcriptional and post-transcriptional gene expression in cancers (34,35). In a recent association study of genetic variation of DEC1 and esophageal cancer risk, the DEC1 3′-UTR G>A (rs3750505) AG heterozygotes were found to have a decrease risk (20). However, we did not observe an association between DEC1 3′-UTR G>A (rs3750505) SNP and SCCHN risk in the present study. Likewise, it has been proposed that genetic variation in the promoter-regulatory region may affect gene expression or transcription activity. Indeed, in the present study, we found that the variant C allele of DEC1 −249 T>C SNP could affect the DEC1 transcription by enhancing DNA–protein-binding activity, thereby modulating SCCHN susceptibility.
It has been reported that the TP53 codon 72 may play a role in SCCHN susceptibility. The TP53 codon 72 G>C polymorphism causes an Arg→Pro (arginine→proline) substitution, and a higher risk of SCCHN in carriers of the variant Pro/Pro homozygotes than those with Arg/Arg homozygotes had been observed in multiple studies (36–44). In the present study, only a few Pro/Pro homozygotes were observed, and the associated risk was not statistically significant. In the stratified analysis, despite there was no statistical support of an interaction between the DEC1 −249 T>C polymorphism and various risk factors for SCCHN risk (P for interaction terms >0.05 for all, Table III), it is interesting to note that the DEC1 −249 T>C polymorphism was associated with a greater protective effect when coupled with common genotype of TP53 codon 72 (rs1042522) Arg/Arg. Therefore, future mechanistic studies are needed to decipher biological interactions between functional SNPs of the DEC1 −249 T>C and TP53 codon 72 (Arg72Pro).
Because SCCHN is a multifactorial disease with complex interactions among genetic variation and environmental factors, we therefore performed an exploratory CART analysis to elucidate possible high-order interactions between genetic and environmental factors in SCCHN development. In the CART analysis, smoking variable was identified to be the most important risk factor. Subgroups of subjects with different risk patterns were then identified, indicating potential gene–environment interactions. However, we only observed the protective effect of the DEC1 −606CC (−249CC) genotype among never-smokers but not in ever smokers, for which the most plausible interpretation may be that tobacco and alcohol consumptions probably mask the small to moderate effects of genetic variation in complex human diseases. However, the results should be interpreted with caution due to the post hoc data-mining nature of the CART approach and the limited sample size and statistical power in the stratified analyses. Therefore, future studies with a larger sample size would be needed to confirm our finding.
In the current study, we demonstrated that the variant −249C allele affected the DEC1 transcription. Specifically, luciferase assay results showed an allele-dependent transcriptional regulation of DEC1, and EMSA further suggested that a higher promoter activity of the sequences containing the variant C allele may be regulated by the direct DNA–protein binding. Our in silico analysis predicted that a putative Nkx-2.5 TFBS binds to the surrounding sequences (−251 to −245 to the start codon) of the variant −249C allele only. Nkx-2.5 (NK2 transcription factor related, locus 5), originally identified as a potential vertebrate homologue of the Drosophila gene tinman (tin), is a homeobox-containing transcription factor that functions during heart formation and development (45,46). Furthermore, recent studies have shown that Nkx-2.5 is involved in transcription regulation of various human cancer cell lines, including hepatocellular carcinoma (47,48), breast cancer (49), prostate cancer (50) and ovarian yolk sac tumor (51). However, our EMSA results did not support Nkx-2.5 as the potential transcription factor that may contribute to the elevated promoter activity via the DNA–protein binding. It is probably that a combination of several DNA-binding transcription factors may be required for regulating gene expression in some cases. For example, previous studies have shown that Nkx-2.5 can cooperate with other transcription factors to activate transcription of various human promoters, such as serum response factor (52,53), GATA-4 (GATA-binding protein 4) (54–56) and Tbx5 (T-box transcription factor TBX5) (57). Taken together, it is still unclear what transcription factor is involved in the DEC1 regulation through the allelic change at the −249 polymorphic site or additional transcription factor(s) may be essential to synergistically cooperate with Nkx-2.5 to drive efficient transcription of DEC1. Finally, the reduced risk-associated DEC1 variants observed in subgroups of never-smokers and never drinkers, patients with TP53 Arg/Arg genotype, oropharyngeal cancer or late-stage SCCHN cancer suggests that genetic variation of DEC1 could modulate HPV-induced carcinogenesis through gene–environment interaction. This speculation needs to be further elucidated in future studies.
In summary, the DEC1 promoter −249 T>C SNP (rs2012775) was identified to be functional, modulating susceptibility to SCCHN among non-Hispanic Whites. To the best of our knowledge, this is the first functional explanation for an association study to elucidate the roles of DEC1 promoter polymorphisms on SCCHN susceptibility and the impact on its biological activity. Nevertheless, some limitations in the current study must also be considered. The selection of control subjects may not be fully representative of the general population because of the inherent limitations of a hospital-based case–control study. Thus, large-scale well-designed population-based studies are needed to further validate the observed associations and to best define the at-risk population for SCCHN. Although we have demonstrated the association between the DEC1 promoter SNP −249 T>C and SCCHN risk, we were not able to analyze the impact of this DEC1 promoter polymorphism on the messenger RNA expression level due to the lack of tumor tissue samples available to us. Moreover, only a small subset of patients with oropharyngeal cancer included in this study had available data on HPV infection, and this limitation should be overcome in our future studies, when our study population grows larger over time.
Funding
National Institute of Health (R01 CA131274, R01 ES011740 to Q.W.), (P50 CA097007 to S.L.), (P30 CA016672) (The University of Texas M. D. Anderson Cancer Center).
Acknowledgments
We thank Margaret Lung, Kathryn Tipton, Jessica Fiske and Ana Neumann for their assistance in recruiting the subjects and gathering the questionnaire information, Yawei Qiao, Jianzhong He, Kejing Xu and Min Zhao for laboratory assistance and Dakai Zhu for his technical support. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CART
classification and regression tree
- CI
confidence interval
- DEC1
deleted in esophageal cancer 1
- EMSA
electrophoretic mobility shift assay
- HPV
human papillomavirus
- OR
odds ratio
- PCR
polymerase chain reaction
- SCCHN
squamous cell carcinoma of the head and neck
- TFBS
transcription factor-binding site
- TSG
tumor suppressor gene
- TESS
transcription element search system
- UTR
untranslated region
References
- 1.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, et al. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Deshpande AM, et al. Molecular mechanisms of head and neck cancer. Expert Rev. Anticancer Ther. 2008;8:799–809. doi: 10.1586/14737140.8.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho T, et al. Epidemiology of carcinogen metabolism genes and risk of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:682–699. doi: 10.1002/hed.20570. [DOI] [PubMed] [Google Scholar]
- 5.Neumann AS, et al. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol. Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 6.Hu N, et al. Genome-wide loss of heterozygosity and copy number alteration in esophageal squamous cell carcinoma using the Affymetrix GeneChip Mapping 10 K array. BMC Genomics. 2006;7:299. doi: 10.1186/1471-2164-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miura K, et al. Deletion mapping in squamous cell carcinomas of the esophagus defines a region containing a tumor suppressor gene within a 4-centimorgan interval of the distal long arm of chromosome 9. Cancer Res. 1995;55:1828–1830. [PubMed] [Google Scholar]
- 8.Froudarakis ME, et al. Microsatellite instability and loss of heterozygosity at chromosomes 9 and 17 in non-small cell lung cancer. Chest. 1998;113:1091–1094. doi: 10.1378/chest.113.4.1091. [DOI] [PubMed] [Google Scholar]
- 9.Merlo A, et al. Frequent loss of chromosome 9 in human primary non-small cell lung cancer. Cancer Res. 1994;54:640–642. [PubMed] [Google Scholar]
- 10.Takamochi K, et al. Loss of heterozygosity on chromosomes 9q and 16p in atypical adenomatous hyperplasia concomitant with adenocarcinoma of the lung. Am. J. Pathol. 2001;159:1941–1948. doi: 10.1016/S0002-9440(10)63041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ah-See KW, et al. An allelotype of squamous carcinoma of the head and neck using microsatellite markers. Cancer Res. 1994;54:1617–1621. [PubMed] [Google Scholar]
- 12.el-Naggar AK, et al. Sequential loss of heterozygosity at microsatellite motifs in preinvasive and invasive head and neck squamous carcinoma. Cancer Res. 1995;55:2656–2659. [PubMed] [Google Scholar]
- 13.Field JK, et al. Allelotype of squamous cell carcinoma of the head and neck: fractional allele loss correlates with survival. Br. J. Cancer. 1995;72:1180–1188. doi: 10.1038/bjc.1995.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habuchi T, et al. A novel candidate tumour suppressor locus at 9q32-33 in bladder cancer: localization of the candidate region within a single 840 kb YAC. Hum. Mol. Genet. 1997;6:913–919. doi: 10.1093/hmg/6.6.913. [DOI] [PubMed] [Google Scholar]
- 15.Hirao S, et al. Loss of heterozygosity on chromosome 9q and p.53 alterations in human bladder cancer. Cancer. 2005;104:1918–1923. doi: 10.1002/cncr.21423. [DOI] [PubMed] [Google Scholar]
- 16.Kimura F, et al. Destabilization of chromosome 9 in transitional cell carcinoma of the urinary bladder. Br. J Cancer. 2001;85:1887–1893. doi: 10.1054/bjoc.2001.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung AC, et al. Frequent decreased expression of candidate tumor suppressor gene, DEC1, and its anchorage-independent growth properties and impact on global gene expression in esophageal carcinoma. Int. J. Cancer. 2008;122:587–594. doi: 10.1002/ijc.23144. [DOI] [PubMed] [Google Scholar]
- 18.Nishiwaki T, et al. Isolation and mutational analysis of a novel human cDNA, DEC1 (deleted in esophageal cancer 1), derived from the tumor suppressor locus in 9q32. Genes Chromosomes Cancer. 2000;27:169–176. [PubMed] [Google Scholar]
- 19.Yang L, et al. Tumor suppressive role of a 2.4 Mb 9q33-q34 critical region and DEC1 in esophageal squamous cell carcinoma. Oncogene. 2005;24:697–705. doi: 10.1038/sj.onc.1208179. [DOI] [PubMed] [Google Scholar]
- 20.Canova C, et al. Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res. 2009;69:2956–2965. doi: 10.1158/0008-5472.CAN-08-2604. [DOI] [PubMed] [Google Scholar]
- 21.Hancox RJ, et al. Accelerated decline in lung function in cigarette smokers is associated with TP53/MDM2 polymorphisms. Hum. Genet. 2009;126:559–565. doi: 10.1007/s00439-009-0704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. A meta-analysis of TP53 codon 72 polymorphism and lung cancer risk: evidence from 15,857 subjects. Lung Cancer. 2009;66:15–21. doi: 10.1016/j.lungcan.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Zemleduch T, et al. Contribution of polymorphism in codon 72 of TP53 gene to laryngeal cancer in Polish patients. Oral Oncol. 2009;45:683–686. doi: 10.1016/j.oraloncology.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Niu J, et al. Genetic polymorphisms in the PTPN13 gene and risk of squamous cell carcinoma of head and neck. Carcinogenesis. 2009;30:2053–2058. doi: 10.1093/carcin/bgp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LE, et al. Reduced DNA repair capacity for removing tobacco carcinogen-induced DNA adducts contributes to risk of head and neck cancer but not tumor characteristics. Clin. Cancer Res. 2010;16:764–774. doi: 10.1158/1078-0432.CCR-09-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews NC, et al. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, et al. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell. Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiojima I, et al. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J. Biol. Chem. 1999;274:8231–8239. doi: 10.1074/jbc.274.12.8231. [DOI] [PubMed] [Google Scholar]
- 29.Stephens M, et al. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens M, et al. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava K, et al. Polymorphisms in ERCC2, MSH2, and OGG1 DNA repair genes and gallbladder cancer risk in a population of Northern India. Cancer. 2010;116:3160–3169. doi: 10.1002/cncr.25063. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg D, et al. CART: Tree-Structured Non-Parametric Data Analysis. San Diego, CA: Salford Systems; 1997. [Google Scholar]
- 33.Zhang H, et al. Use of classification trees for association studies. Genet. Epidemiol. 2000;19:323–332. doi: 10.1002/1098-2272(200012)19:4<323::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Chen K, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 35.Yu Z, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortezzi SS, et al. Analysis of human papillomavirus prevalence and TP53 polymorphism in head and neck squamous cell carcinomas. Cancer Genet. Cytogenet. 2004;150:44–49. doi: 10.1016/j.cancergencyto.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Drummond SN, et al. TP53 codon 72 polymorphism in oral squamous cell carcinoma. Anticancer Res. 2002;22:3379–3381. [PubMed] [Google Scholar]
- 38.Katiyar S, et al. Polymorphism of the p53 codon 72 Arg/Pro and the risk of HPV type 16/18-associated cervical and oral cancer in India. Mol. Cell. Biochem. 2003;252:117–124. doi: 10.1023/a:1025546610920. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, et al. 172G>T variant in the 5' untranslated region of DNA repair gene RAD51 reduces risk of squamous cell carcinoma of the head and neck and interacts with a P53 codon 72 variant. Carcinogenesis. 2007;28:988–994. doi: 10.1093/carcin/bgl225. [DOI] [PubMed] [Google Scholar]
- 40.Shen H, et al. P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Lett. 2002;183:123–130. doi: 10.1016/s0304-3835(02)00117-9. [DOI] [PubMed] [Google Scholar]
- 41.Sousa H, et al. Linkage of TP53 codon 72 pro/pro genotype as predictive factor for nasopharyngeal carcinoma development. Eur. J. Cancer Prev. 2006;15:362–366. doi: 10.1097/00008469-200608000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Summersgill KF, et al. p53 polymorphism, human papillomavirus infection in the oral cavity, and oral cancer. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2000;90:334–339. doi: 10.1067/moe.2000.107359. [DOI] [PubMed] [Google Scholar]
- 43.Tiwawech D, et al. The p53 codon 72 polymorphism in Thai nasopharyngeal carcinoma. Cancer Lett. 2003;198:69–75. doi: 10.1016/s0304-3835(03)00283-0. [DOI] [PubMed] [Google Scholar]
- 44.Twu CW, et al. Association of p53 codon 72 polymorphism with risk of hypopharyngeal squamous cell carcinoma in Taiwan. J. Formos. Med. Assoc. 2006;105:99–104. doi: 10.1016/S0929-6646(09)60330-2. [DOI] [PubMed] [Google Scholar]
- 45.Komuro I, et al. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl Acad. Sci. USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lints TJ, et al. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 47.Apergis GA, et al. A novel nk-2-related transcription factor associated with human fetal liver and hepatocellular carcinoma. J. Biol. Chem. 1998;273:2917–2925. doi: 10.1074/jbc.273.5.2917. [DOI] [PubMed] [Google Scholar]
- 48.Kajiyama Y, et al. Regulation of alpha-fetoprotein expression by Nkx2.8. Mol. Cell. Biol. 2002;22:6122–6130. doi: 10.1128/MCB.22.17.6122-6130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dentice M, et al. Transcription factor Nkx-2.5 induces sodium/iodide symporter gene expression and participates in retinoic acid- and lactation-induced transcription in mammary cells. Mol. Cell. Biol. 2004;24:7863–7877. doi: 10.1128/MCB.24.18.7863-7877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwabi-Addo B, et al. Age-related DNA methylation changes in normal human prostate tissues. Clin. Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 51.Shibata K, et al. Establishment and characterization of an ovarian yolk sac tumor cell line reveals possible involvement of Nkx2.5 in tumor development. Oncology. 2008;74:104–111. doi: 10.1159/000139138. [DOI] [PubMed] [Google Scholar]
- 52.Chen CY, et al. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev. Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 53.Chen CY, et al. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dentice M, et al. The different cardiac expression of the type 2 iodothyronine deiodinase gene between human and rat is related to the differential response of the Dio2 genes to Nkx-2.5 and GATA-4 transcription factors. Mol. Endocrinol. 2003;17:1508–1521. doi: 10.1210/me.2002-0348. [DOI] [PubMed] [Google Scholar]
- 55.Sepulveda JL, et al. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol. Cell. Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small EM, et al. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: kx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev. Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- 57.Bimber B, et al. Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev. Biol. 2007;302:230–242. doi: 10.1016/j.ydbio.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]