Abstract
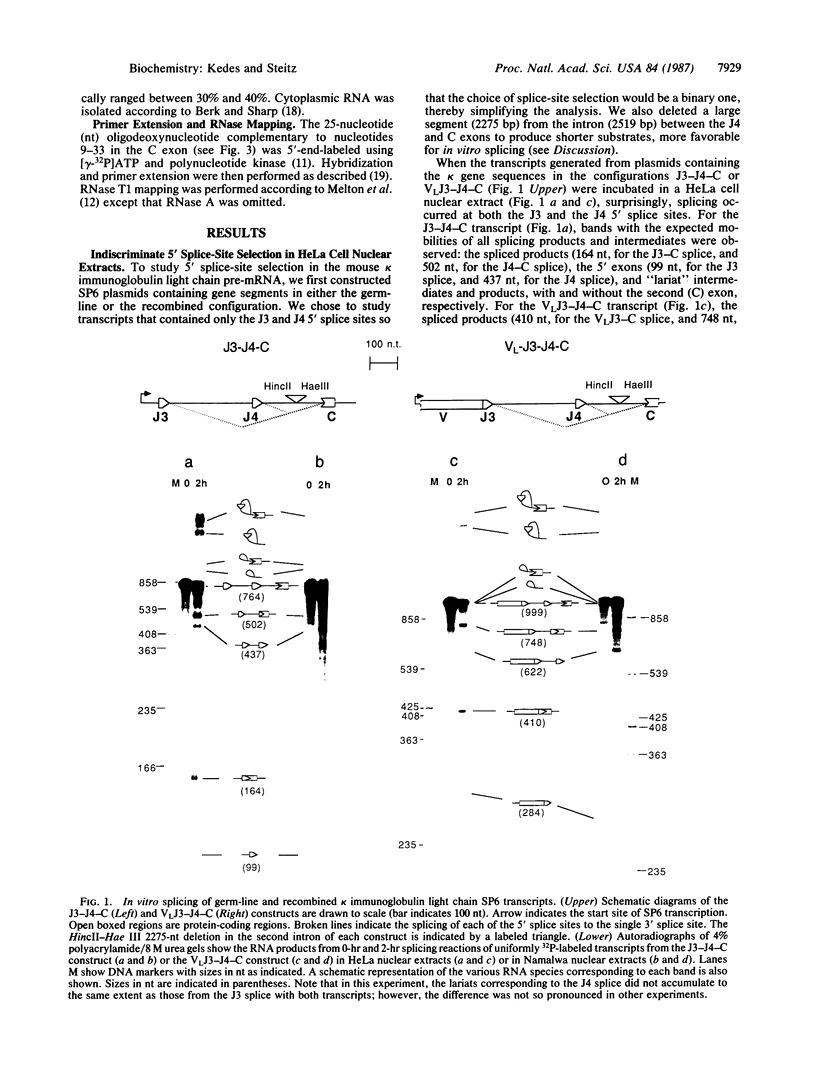
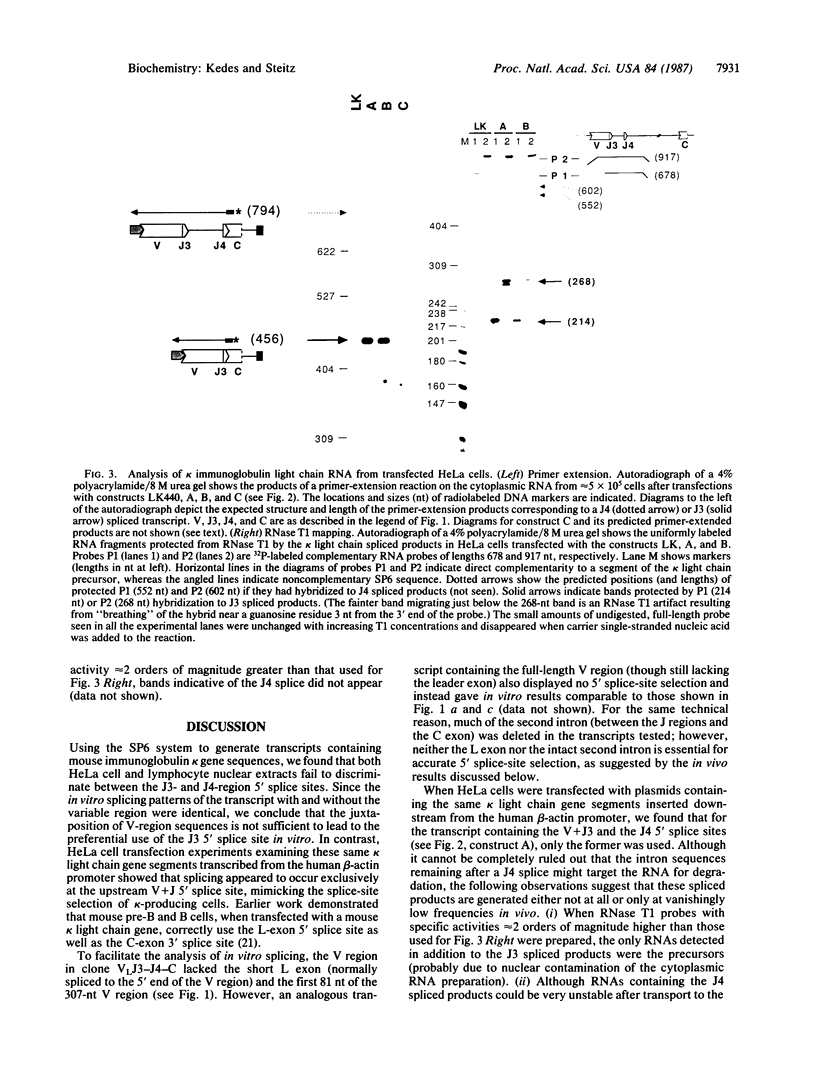
In mature mouse B lymphocytes, immunoglobulin kappa light chain transcripts contain an intervening sequence separating the recombined variable (V) plus joining (J) exon from the distant constant (C) exon. After V-J recombination, this intervening sequence can include as many as three unused but very similar J-region 5' splice sites. Each of these sites is potentially functional if the gene is appropriately recombined. It is unclear how the splicing machinery distinguishes among these 5' splice sites, always choosing the most upstream site. We used synthetic transcripts of kappa gene sequences containing J3 and J4 in both the germ-line and the recombined configurations to study the pattern of 5' splice-site selection in vitro. We find that both HeLa cell and lymphocyte nuclear extracts fail to discriminate between the J3- and J4-region 5' splice sites. In contrast, after transfection into HeLa cells, similar kappa light chain transcripts are spliced correctly at the most upstream 5' splice site--that which is used in kappa-producing cells. We conclude that accurate 5' splice-site selection in the mouse kappa light chain is neither cell-type- nor species-specific. Potential mechanisms for this controlling step in gene expression are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Crenshaw E. B., 3rd, Russo A. F., Swanson L. W., Rosenfeld M. G. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987 May 8;49(3):389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P., Leavitt J., Muscat G., Ng S. Y., Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Roeder R. G. Transcription of human histone genes in extracts from synchronized HeLa cells. Proc Natl Acad Sci U S A. 1984 May;81(9):2713–2717. doi: 10.1073/pnas.81.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T., Habu S. Origin of immune diversity: genetic variation and selection. Annu Rev Biochem. 1985;54:803–830. doi: 10.1146/annurev.bi.54.070185.004103. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Trans splicing of mRNA precursors in vitro. Cell. 1985 Aug;42(1):165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lewin B. Alternatives for splicing: recognizing the ends of introns. Cell. 1980 Nov;22(2 Pt 2):324–326. doi: 10.1016/0092-8674(80)90340-2. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gifford A., Baltimore D. Joining of V kappa to J kappa gene segments in a retroviral vector introduced into lymphoid cells. 1984 Mar 29-Apr 4Nature. 308(5958):425–428. doi: 10.1038/308425a0. [DOI] [PubMed] [Google Scholar]
- Lowery D. E., Van Ness B. G. In vitro splicing of kappa immunoglobulin precursor mRNA. Mol Cell Biol. 1987 Apr;7(4):1346–1351. doi: 10.1128/mcb.7.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D. Trans splicing of mRNA precursors. Cell. 1985 Aug;42(1):157–164. doi: 10.1016/s0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J. Presence of a polyadenylated RNA fragment encoding the membrane domain for immunoglobulin alpha chain indicates that mRNAs for both secreted and membrane-bound alpha chains can be produced from the same RNA transcript. Nucleic Acids Res. 1986 Aug 11;14(15):6129–6144. doi: 10.1093/nar/14.15.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Tsurushita N., Avdalovic N. M., Korn L. J. Regulation of differential processing of mouse immunoglobulin mu heavy-chain mRNA. Nucleic Acids Res. 1987 Jun 11;15(11):4603–4615. doi: 10.1093/nar/15.11.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Weir L., Leder P. Structure and expression of a human subgroup II immunoglobulin kappa gene. Nucleic Acids Res. 1986 May 12;14(9):3957–3970. doi: 10.1093/nar/14.9.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]






