Abstract
Background
While quantification of viral loads has been successfully employed in clinical medicine and has provided valuable insights and useful markers for several viral diseases, the potential of measuring bacterial DNA load to predict outcome or monitor therapeutic responses remains largely unexplored. We tested this possibility by investigating bacterial loads in Acinetobacter baumannii bacteremia, a rapidly increasing nosocomial infection characterized by high mortality, drug resistance, multiple and complicated risk factors, all of which urged the need of good markers to evaluate therapeutics.
Methods and Findings
We established a quantitative real-time PCR assay based on an A. baumannii-specific gene, Oxa-51, and conducted a prospective study to examine A. baumannii loads in 318 sequential blood samples from 51 adults patients (17 survivors, 34 nonsurvivors) with culture-proven A. baumannii bacteremia in the intensive care units. Oxa-51 DNA loads were significantly higher in the nonsurvivors than survivors on day 1, 2 and 3 (P = 0.03, 0.001 and 0.006, respectively). Compared with survivors, nonsurvivors had higher maximum Oxa-51 DNA load and a trend of increase from day 0 to day 3 (P<0.001), which together with Pitt bacteremia score were independent predictors for mortality by multivariate analysis (P = 0.014 and 0.016, for maximum Oxa-51 DNA and change of Oxa-51 DNA, respectively). Kaplan-Meier analysis revealed significantly different survival curves in patients with different maximum Oxa-51 DNA and change of Oxa-51 DNA from day 0 to day 3.
Conclusions
High Oxa-51 DNA load and its initial increase could predict mortality. Moreover, monitoring Oxa-51 DNA load in blood may provide direct parameters for evaluating new regimens against A. baumannii in future clinical studies.
Introduction
The successful employment of quantitative PCR to measure viral load in clinical medicine has not only advanced our understanding of pathogenesis of many viral diseases but also provided useful markers for predicting outcome and/or monitoring therapeutic responses [1]. The possibility of measuring bacterial DNA load to predict outcome or evaluate therapeutics in bacterial diseases, especially those with high mortality, antimicrobial resistance, multiple and complicated risk factors, has been raised but never been fully explored [1], [2].
Acinetobacter baumannii (A. baumannii), a nonfermenting Gram-negative aerobic coccobacillus which can survive for prolonged periods in various environmental conditions, is an increasingly important nosocomial pathogen [3]. Among the A. calcoaceticus- A. baumannii complex, which included genospecies 1, 2, 3 and 13TU, A. baumannii (genospecies 2) is primarily associated with human diseases [4]. Nosocomial infection caused by A. baumannii has increased substantially, especially among patients in the ICU [3]–[6] or immunocompromized hosts [3], [4], [6]. It is a life-threatening opportunistic infection among critical ill patient [7], [8]. Patients with A. baumannii infection have been reported to be associated with higher mortality and longer hospital stay compared with other infections [3], [7], [8]. In addition to pneumonia, bacteremia is the most common focus of A. baumannii infection [4]. Due to rapid increase in the drug resistance of A. baumannii, the choices of antimicrobial agents were limited [3], [4], [9]. Previous studies have identified multiple and complicated risk factors for the high mortality of A. baumannii bacteremia [5], [6], [10]–[15], but none has investigated the roles of bacterial load in the disease outcome.
The relationship between the magnitude of bacteremia and clinical outcome was originally demonstrated by semiquantitative blood culture [16], [17]. The drastic decrease in the sensitivity of blood culture after antimicrobial treatment and the time required for blood culture limited the application of this approach [2], [18]. Recently, several studies using quantitative PCR assays have shown high initial bacterial DNA loads in blood correlated with disease severity and mortality in infection caused by Streptococcus pneumoniae, Neisseria meningitides, and methicillin-resistant Staphylococcus aureus (MRSA) [19]–[22]. However, the change in bacterial DNA load during the course of infection and its relationship to disease outcome or therapy remain largely unclear. We reported previously that high levels of mecA DNA in blood after 3 and 7 days of antibiotic therapy were associated with mortality in MRSA bacteremia [21], suggesting sequential bacterial DNA load in blood can be used to evaluate therapeutic responses. In this study, we established a quantitative real-time PCR assay by using an A. baumannii-specific gene, Oxa-51, to measure both the initial and sequential A. baumannii loads in blood from patients with A. baumannii bacteremia and to investigate the relationships to disease outcome [23].
Methods
Patients and blood samples
This prospective observational study was conducted in accordance with the principles expressed in the Declaration of Helsinki at the National Taiwan University Hospital, a medical center with a 2,200-bed capacity in Taiwan. With the approval of the Institutional Review Board (No. 200809045R), adult patients admitted to the medical or surgical ICU between 1 April 2008 and 28 February 2009 were enrolled once a positive blood culture for A. baumannii was reported. Blood cultures were taken when clinical symptoms/signs of infection such as fever, shock, etc. were observed. The Board waived the need for informed consent, since this study required no additional blood drawing from the study patients in ICU and all the data were analyzed anonymously (Text S1 and S2). All blood cultures were performed by the Bactec 9240 system (Becton Dickson, Sparks, MD) in the microbiological laboratory. A. baumannii complex was identified by classic methods and verified by the Phoenix system (BD Diagnostics, Sparks, MD) [24], followed by a disk diffusion susceptibility test with gentamicin, amikacin, ciprofloxacin, levofloxacin, cefepime, ceftazidime, aztreonam, ticarcillin/clavulanate, meropenem, and ampicillin/sulbactam [25]. The susceptibility of tigecycline was determined by the agar dilution method, in which MIC ≤ 2 µg/mL was considered the break point [26], and that of colistin by E-test [25], [27]. Genospecies A. baumannii was defined by A. baumannii complex positive for the Oxa-51 by real-time PCR assay [23]. The course of A. baumannii bacteremia in each patient was carefully monitored by clinical parameters and routine laboratory tests. Whole blood samples in EDTA-containing tubes taken before and on the day of reporting A. baumannii bacteremia were retrospectively obtained from the central laboratory. After that, blood samples were collected daily for the first week and two to three times a week thereafter.
Clinical data and definitions
The demographic, clinical, laboratory and culture data for each patient were collected (Tables 1 and 2). The underlying illness was assessed by the Charlson score [28], and clinical severity by the acute physiology and chronic health evaluation II (APACHE II), Pitt bacteremia scores [29], and sequential organ failure assessment score (SOFA) score [30].
Table 1. Clinical and laboratory characteristics of study patients with A. baumannii bacteremia.
| Characteristicsa | Total (n = 51) | Nonsurvivor group (n = 34) | Survivor group (n = 17) | P |
| Age | 61.5 (48.9–74.2) | 59.9 (48.0–74.6) | 61.5 (49.0–71.7) | 0.84 |
| Sex, M/F | 33/18 | 22/12 | 11/6 | 1.00 |
| Days of prior hospitalizationb | 18 (10–41) | 19.5 (10–47.3) | 18 (10–29.5) | 0.75 |
| Days of prior ICU stayb | 10 (0–18) | 10 (0.8–16.3) | 14 (0–18) | 0.80 |
| APACHE II score, ICU admission | 26 (20.5–36) | 27 (20.5–36) | 25.5 (20.3–35) | 0.73 |
| Underlying illness | ||||
| Charlson score | 3 (2–6) | 3 (2–6) | 4 (2–6) | 0.91 |
| Diabetes mellitus | 20 (39.2) | 11 (32.4) | 9 (52.9) | 0.23 |
| Congestive heart failure | 7 (13.7) | 5 (14.7) | 2 (11.8) | 1.00 |
| Chronic renal insufficiency | 16 (31.4) | 11 (32.4) | 5 (29.4) | 1.00 |
| Immunosuppressantsc | 22 (43.1) | 18 (52.9) | 4 (23.5) | 0.07 |
| Leukemia | 7 (13.7) | 7 (20.6) | 0 (0) | 0.08 |
| Lymphoma | 3 (5.9) | 2 (5.9) | 1 (5.9) | 1.00 |
| Focus of infection | ||||
| Primary bacteremia | 11 (21.6) | 7 (20.6) | 4 (23.5) | 1.00 |
| Pneumonia | 33 (64.7) | 24 (70.6) | 9 (52.9) | 0.23 |
| Intra-abdominal infection | 4 (7.8) | 2 (5.9) | 2 (11.8) | 0.59 |
| Surgical site infection | 4 (7.8) | 4 (11.8) | 0 (0) | 0.29 |
| Laboratory data on day 0b | ||||
| White blood cell count (1000/µL) | 10.29 (2.42–20.34) | 8.36 (0.55–20.64) | 12.07 (8.50–19.97) | 0.19 |
| Hemoglobin (g/dL) | 9.4 (8.5–10.4) | 9.2 (8.2–10.5) | 9.6 (8.9–10.7) | 0.35 |
| Platelet count (1000/µL) | 73 (41–134) | 56 (19–96.8) | 134 (74–275.5) | <0.001 |
| ASTd (U/L) | 50 (25–95) | 67 (30–117) | 35 (17.5–58.3) | 0.15 |
| Total bilirubin (mg/dL) | 2.56 (0.83–9.06) | 3 (0.8–25.5) | 1.9 (0.7–3.3) | 0.32 |
| Creatinine (mg/dL) | 1.8 (1.1–3.2) | 1.9 (1.1–3.8) | 1.7 (1.1–3) | 0.81 |
| C-reactive protein (mg/dL) | 9.6 (4.6–16.4) | 9.8 (5.3–21.1) | 8.7 (2.3–12.7) | 0.37 |
Data are median (interquartile range) for continuous variables and number of cases (percentage) for categorical variables, with two-tailed Mann-Whitney U test for the former and Fisher's exact test for the later.
Days of hospitalization and ICU stay prior to day 0, which was the day on which the first blood sample positive for A. baumannii by culture was drawn.
Immunosuppressants included antineoplastic therapy within 6 weeks of the onset of A. baumannii bacteremia, corticosteroids at a dose ≧ 20 mg of prednisolone daily for at least 2 weeks or 30 mg of prednisolone daily for at least 1 week before a positive blood culture for A. baumannii, and other immunosuppressants such as cyclophosphamide [15].
AST: aspartate aminotransferase.
Table 2. Antimicrobial susceptibility and usage, bacteremia score and Oxa-51 DNA level of study patients with A. baumannii bacteremia.
| Characteristicsa | Total (n = 51) | Nonsurvivor group (n = 34) | Survivor group (n = 17) | P | Multivariatef odds ratio [95% CI] | P |
| Pitt bacteremia score, on day 0b | 5 (3–7) | 5.5 (4–7.3) | 4 (2–6) | 0.02 | 1.55 [0.94–2.57] | 0.087 |
| SOFA scorec | 14.5 (11–19) | 16 (13.5–19) | 10.5 (6–13) | <0.001 | ||
| Co-infection | 16 (31.4) | 10 (29.4) | 6 (35.3) | 0.75 | ||
| Subsequent blood stream infection | 2 (3.9) | 1 (2.9) | 1 (5.9) | 1.00 | ||
| Antimicrobial resistance to | ||||||
| Anti-pseudomonas Penicillin | 43 (89.6) | 29 (87.9) | 14 (93.3) | 1.00 | ||
| Anti-pseudomonas Cephalosporin | 45 (88.2) | 31 (91.2) | 14 (82.4) | 0.39 | ||
| Carbapenem | 38 (74.5) | 25 (73.5) | 13 (76.5) | 1.00 | ||
| Aminoglycosides | 44 (86.3) | 31 (91.2) | 13 (76.5) | 0.20 | ||
| Fluoroquinolones | 48 (94.1) | 32 (94.1) | 16 (94.1) | 1.00 | ||
| Ampicillin/Sulbactam | 31 (60.8) | 25 (73.5) | 6 (35.3) | 0.01 | ||
| Tigecycline | 10 (20.8) | 8 (24.2) | 2 (13.3) | 0.47 | ||
| Colistin | 0 (0) | 0 (0) | 0 (0) | |||
| ERABd | 27 (52.9) | 22 (64.7) | 5 (29.4) | 0.04 | 1.99 [0.21–18.71]h | 0.55 |
| Antibiotics used in empirical therapy | ||||||
| Anti-pseudomonas Penicillin | 6 (11.8) | 2 (5.9) | 4 (23.5) | 0.09 | ||
| Anti-pseudomonas Cephalosporin | 16 (31.4) | 10 (29.4) | 6 (35.3) | 0.75 | ||
| Carbapenem | 26 (51.0) | 21 (61.8) | 5 (29.4) | 0.04 | ||
| Fluoroquinolones | 12 (23.5) | 8 (23.5) | 4 (23.5) | 1.00 | ||
| Ampicillin/Sulbactam | 8 (16.7) | 5 (14.7) | 3 (17.7) | 1.00 | ||
| Tigecycline | 5 (9.8) | 4 (11.8) | 1 (5.9) | 0.65 | ||
| Colistin | 5 (9.8) | 4 (11.8) | 1 (5.9) | 0.65 | ||
| Appropriate empirical therapye | 17 (33.3) | 12 (35.3) | 5 (29.4) | 0.76 | ||
| Day 3 Oxa-51 DNA (log/copies/mL)b | 1.00 (0–3.01) | 2.81 (0.59–3.48) | 0 (0–1.69) | 0.006 | 2.19 [1.18–4.07]g | 0.013g |
| Maximum Oxa-51 DNA (log copies/mL), day 0 – day 3b | 3.04 (2.30–3.62) | 3.38 (2.70–4.07) | 2.30 (1.71–2.89) | <0.001 | 10.25 [1.61–65.46] | 0.014 |
| Change of Oxa-51 DNA, day 0b – day 3 (Δ log copies/mL/day) | 0.12 (−0.73–0.76) | 0.40 (−0.09–0.97) | −0.77 (−1.25– −0.14) | <0.001 | 3.17 [1.24–8.12] | 0.016 |
Data are median (interquartile range) for continuous variables and number of cases (percentage) for categorical variables, with two-tailed Mann-Whitney U test for the former and Fisher's exact test for the later.
Day 0 was the day on which the first blood sample positive for A. baumannii by culture was drawn.
SOFA: Sequential organ failure assessment. Not included in the final multivariate regression model due to small case numbers (Text S4).
ERAB: Extensively resistant A. baumannii.
Appropriate empirical therapy is defined when A.baumannii is susceptible to at least one antibiotic used within 48 hours of day 0 except aminoglycoside monotherapy (see Methods).
Multivariate logistic regression model: n = 40, adjusted generalized R 2 = 0.634, estimated area under the ROC curve = 0.92. Deviance goodness-of-fit (GOF) test P = 0.87>0.05 (df = 36); Pearson GOF test P = 0.88>0.05 (df = 36); Hosmer and Lemeshow GOF test P = 0.98>0.05 (df = 8).
Discard “Maximum Oxa-51 DNA” and “Change of Oxa-51 DNA”, both of which require daily monitoring of Oxa-51 DNA in blood.
ERAB was considered in the predicting model of mortality based on the three independent predictors, Pitt bacteremia score, maximum Oxa-51 DNA and change of Oxa-51 DNA from day 0 to day 3.
Day 0 was the day on which the first blood sample positive for A. baumannii by culture was drawn, and the days after that were designated consecutively. Since the average time of reporting A. baumannii bacteremia was 72 h in our study, the period of empirical therapy was defined as from day 0 to day 3. Extensively resistant A. baumannii (ERAB) was defined as A. baumannii resistant to the commonly used anti-pseudomonas antibiotics (cephalosporins, extended-spectrum penicillins, carbapenems, aminoglycosides, fluoroquinolones) and ampicillin/sulbactam determined by disc diffusion method [15], [25]. Appropriate empirical therapy was defined if A. baumannii was susceptible in vitro to at least one antibiotic used within 48 hours from day 0. Aminoglycoside monotherapy without combination of other susceptible antibiotics was considered as inappropriate [9]. For ERAB, the possibly appropriate antibiotics included tigecycline and colistin. Although the MIC breakpoint for tigecycline susceptibility was 2 µg/mL according to the FDA definition for Enterobacteriaceae [26], it was considered as inappropriate therapy if used alone for A. baumannii blood stream infection with MIC ≧1 µg/mL [9]. The overall and initial changes of Oxa-51 DNA load were assessed by the summary statistics for each patient, in which the slopes of linear regression of log Oxa-51 DNA copies/mL over the whole course and from day 0 to day 3, respectively, were calculated. Co-infection was defined by the presence of pathogens other than A. baumannii in blood culture at the same time when the first blood sample positive for A. baumannii by culture was drawn. Subsequent blood stream infection was defined by the presence of pathogens other than A. baumannii in blood culture within 14 days after the onset of A. baumannii bacteremia.
Patients, who fulfilled the definition of A. baumannii bacteremia related death including (1) blood culture positive for A. baumannii at the time of death, (2) death before resolution of symptoms and signs, and (3) death within 14 days after the onset of A. baumannii bacteremia without another explanation, were classified as the nonsurvivor group. Patients who survived the episode were classified as the survivor group.
DNA extraction
One milliliter of whole blood from patients or A. baumannii-spiked whole blood was treated with 3 mL of red blood cell lysis solution (Gentra Systems, Minneapolis, MN) at room temperature for 5 min, followed by centrifugation at 5,000× g for 10 min to obtain the pellets, which were resuspended in enzyme solution (20 mM Tris [pH 8.0], 2 mM EDTA, 1.2% Triton, lysozyme 20 mg/mL) and subjected to DNA extraction using QIAamp DNA minikit (Qiagen, Hilden, Germany) [21]. The final eluate (100 µl) was stored at –20°C until use. Whole blood samples from healthy donors served as negative controls. DNA extraction and PCR were carried out in separate rooms, and precautions were taken to prevent contamination [31].
Quantitative real-time PCR
The Oxa-51-specific primers (F Oxa-51 and R Oxa-51) and probe (P Oxa-51) were designed based on the Oxa-51 sequence in GenBank and Primer Express software v2.0. Plasmid Oxa-51/pCRII-TOPO, which contained a 431-bp fragment of the Oxa-51 gene in pCRII-TOPO, was used as the standard and its copy number was calculated based on the concentration determined by spectrophotometry and molecular weight [21], [32] (Text S3). The sequences of F Oxa-51, R Oxa-51 and P Oxa-51 are 5′-TTTAGCTCGTCGTATTGGACTTGA-3′, 5′-CGGAGAACGACTCCTCAT TAAAAA-3′ and 5′-TGGCAATGCAGATA TCGGTACCCAAGTC-3′, respectively. An aliquot of extracted DNA and known copy numbers of Oxa-51 DNA standards were subjected to real-time PCR. Briefly, a 25 µl reaction mixture containing 12.5 µl of 2x TaqMan universal PCR master mix, 2.5 µl of 10 pmol/µl of each of F Oxa-51, R Oxa-51 and P Oxa-51, Oxa-51 DNA standards or extracted DNA, and nuclease free-water was subjected to real-time PCR with an ABI Prism 7000/7500 sequence detector (Applied Biosystems, Foster City, CA). The amplification conditions were 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min, as described previously [21]. A positive result was defined by the cycle number required to reach the threshold, which was 10 times the standard deviation of the mean baseline emission calculated for cycles 3 to 15 [21]. Since 5 µl of the 100 µl DNA eluate, which was derived from 1 mL of whole blood samples, was used in each reaction mixture, the Oxa-51 DNA copy number per reaction was multiplied by 20 to determine the copy numbers of Oxa-51 DNA per mL whole blood. The sensitivity of the assay was 20 copies per mL.
Statistical analysis
Mann-Whitney U test and Fisher's exact test were used to compare continuous variables and categorical variables, respectively, between two groups. Log-rank test and Kaplan-Meier survival curve were used for survival analysis, and the results were verified by multivariate Cox's proportional hazard model. Multivariate logistic regression analysis including all variables such as age, sex, underlying diseases, laboratory data, infection foci, invasive procedure (central catheter, ventilator), appropriate empirical antibiotics and bacterial load was conducted to identify predictors for mortality (Text S4). Data was analyzed using Stata software, version 10 (StataCorp, College Station, Texas).
Results
Quantitative real-time PCR assay for Oxa-51 gene
We first established a standard curve by using increasing copy numbers (5 to 5×108 copies) of the plasmid Oxa-51/pCRII-TOPO in the assay (r = 0.9992, Text S3). We next tested DNA extracted from increasing CFUs (9×101 to 9×106 CFUs) of a previously reported A. baumannii isolate 2003I053 [32], which had been spiked with whole blood from a healthy volunteer, and found a linear curve between the CFUs and Oxa-51 copy numbers (r = 0.9942), demonstrating the accuracy of this assay for quantifying A. baumannii in whole blood (Text S3). Moreover, the Oxa-51 copy numbers of DNA extracted from A. baumannii-spiked whole blood that had been stored at 4°C for up to 4 days remained stable, suggesting the feasibility of this assay to quantify A. baumannii in stored whole blood samples (Text S3).
The specificity of the Oxa-51 real-time PCR assay was verified by examining DNA extracted from 85 isolates including common nosocomial pathogens such as E. coli, K. pneumoniae, E. cloacae, P. aeruginosa, S. maltophilia, E. faecium and MRSA, as well as Acinetobacter genospecies 3, 13TU and 2 (A. baumannii) in the assay [32]–[35], which detected positive signal only in the DNA extracted from A. baumannii but not in those from other isolates.
Study Patients
Table 1 summarizes the clinical and laboratory characteristics of the 51 patients with A. baumannii bacteremia, including 34 nonsurvivors and 17 survivors. They were predominantly males with a median age of 61.5 years, and were all critically ill with high APACHE II score (median: 26). There was no difference between the two groups in the APACHE II or Charlson score, the number of days of hospitalization and of ICU stay prior to A. baumannii bacteremia, individual underlying disease such as leukemia or lymphoma, immunosuppressant, foci of infection, or initial laboratory data except lower platelet counts in the nonsurvivors. Compared with the survivors, the nonsurvivors had higher Pitt bacteremia score and SOFA score on day 0, and higher frequency of ERAB and resistance to ampicillin/sulbactam (P = 0.02 and P<0.001, Mann-Whitney U test, two tailed; P = 0.04 and 0.01, Fisher's exact test, respectively) (Table 2). There was no difference in the co-infection, subsequent blood stream infection, antimicrobial resistance to other groups of antibiotics, antibiotics used during empirical period (except carbapenem) or appropriate empirical therapy between the two groups (Table 2 and Table S1).
Higher levels of Oxa-51 DNA in nonsurvivors than survivors
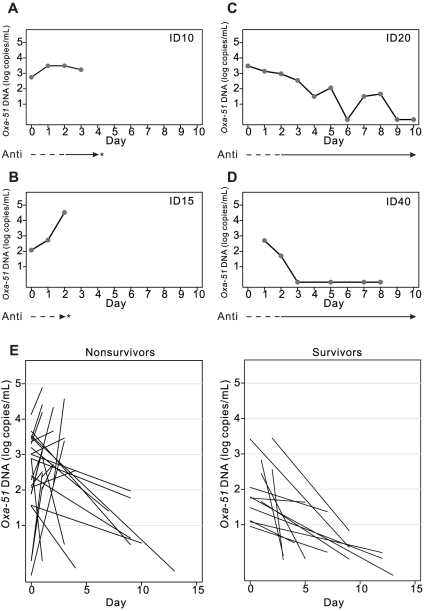
We then employed the real-time PCR assay to quantify Oxa-51 DNA in 318 sequential blood samples from 51 patients. We first examined sequential Oxa-51 DNA loads over time in 4 patients, including 2 nonsurvivors and 2 survivors. As shown in Figure 1, a decreasing trend of the Oxa-51 DNA level was found in the survivors, whereas continuously high levels of Oxa-51 DNA were found in the nonsurvivors. To investigate the overall change of Oxa-51 DNA load during the course of infection, we analyzed the slope of Oxa-51 DNA load. While all survivors had a decline in Oxa-51 DNA over time, more than half of the nonsurvivors showed an upward slope at death (P = 0.01, Mann-Whitney U test, two tailed) (Figure 1E).
Figure 1. Sequential Oxa-51 DNA loads in blood during the course of A. baumannii bacteremia.
Sequential whole blood samples from 2 nonsurvivors (A, B) and 2 survivors (C, D) with A. baumannii bacteremia were subjected to DNA extraction and the Oxa-51 real-time PCR assay. The lines beneath indicate inappropriate (—) and appropriate (—) antibiotic therapy, and the asters indicate death. (E) Overall change of Oxa-51 DNA load in blood for patients with A. baumannii bacteremia (27 nonsurvivors, 13 survivors) assessed by the slope of the linear regression of log Oxa-51 DNA copies per mL plotted against time.
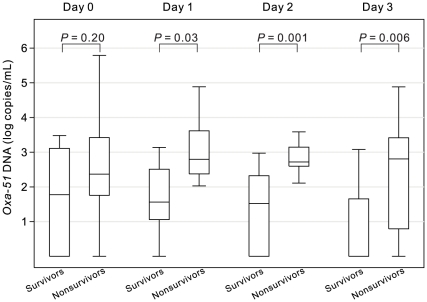
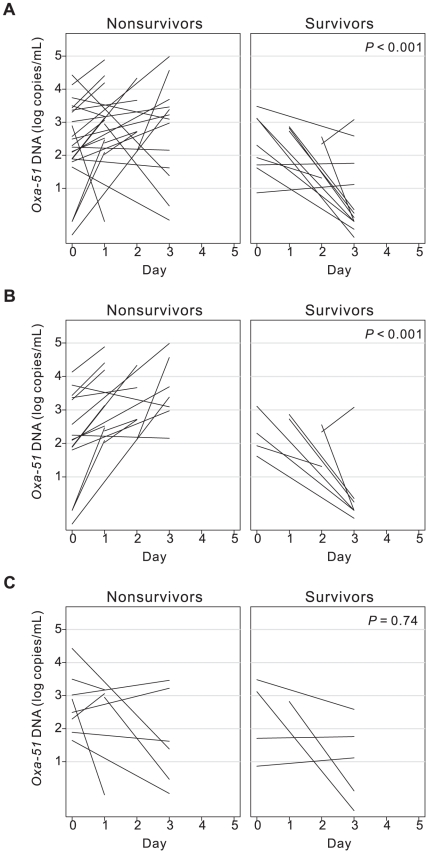
We next focused on initial Oxa-51 DNA loads from day 0 to day 3, which corresponded to the period of empirical therapy. The levels of Oxa-51 DNA were significantly higher in the nonsurvivors than survivors on day 1, day 2 and day 3 (P = 0.03, 0.001 and 0.006, respectively, Mann-Whitney U test, two tailed) (Figure 2), so was the maximum Oxa-51 DNA between day 0 and day 3 (Table 2) [20]. We then examined the change of Oxa-51 DNA load from day 0 to day 3. As shown in Figure 3A, a trend of increase was found in the majority of nonsurvivors, whereas a trend of decrease in most survivors (median 0.40 vs. −0.77 Δ log copies/mL/day, P<0.001, Mann-Whitney U test, two tailed). To further investigate whether appropriate empirical antibiotics affect the Oxa-51 DNA change during this period, we analyzed subgroups of patients. Among those receiving inappropriate empirical antibiotics, nearly all nonsurvivors showed increasing Oxa-51 DNA compared with survivors (P<0.001, Mann-Whitney U test, two tailed) (Figure 3B). Among those receiving appropriate empirical antibiotics, there was no difference in the initial Oxa-51 DNA change between the two groups (Figure 3C). Interestingly, the nonsurvivors receiving appropriate empirical antibiotics showed a trend of no or less increase in Oxa-51 DNA compared with those receiving inappropriate empirical antibiotics (P<0.001, Mann-Whitney U test, two tailed) (left panels, Figures 3B and 3C).
Figure 2. Initial Oxa-51 DNA load from day 0 to day 3 in patients with A. baumannii bacteremia (34 nonsurvivors, 17 survivors).
Data are presented as median (line), interquartile range (box) and range.
Figure 3. Initial change of Oxa-51 DNA load from day 0 to day 3 in patients, with A. baumannii bacteremia (27 nonsurvivors, 13 survivors) (A), and among subgroups including those receiving inappropriate (B) and those receiving appropriate (C) empirical antibiotics.
Predictors for mortality
Multivariate logistic regression analysis including all the variables in Tables 1 and 2 revealed that Pitt bacteremia score, maximum Oxa-51 DNA and change of Oxa-51 DNA from day 0 to day 3 were three independent predictors for mortality (Table 2). Excluding “maximum Oxa-51 DNA” and “change of Oxa-51 DNA”, which required daily monitoring, the single day measurement, Oxa-51 on day 3, was the most potent independent predictor for those who survived more than 3 days (odds ratio [OR], 2.19; 95% confidence interval [CI], 1.18–4.07; P = 0.013). The goodness-of-fit (GOF) test suggested a good fit of the multivariate logistic regression model to binary data and thus a strong prediction (Text S4). Moreover, the predicting model based on the three independent predictors had a positive predictive value of 89.3% and a negative predictive value of 83.3% with the estimated area under receiver operating characteristic (ROC) curve of 0.9202, which was higher than that of 0.6581 based on Pitt bactermia score alone.
Kaplan-Meier survival analysis
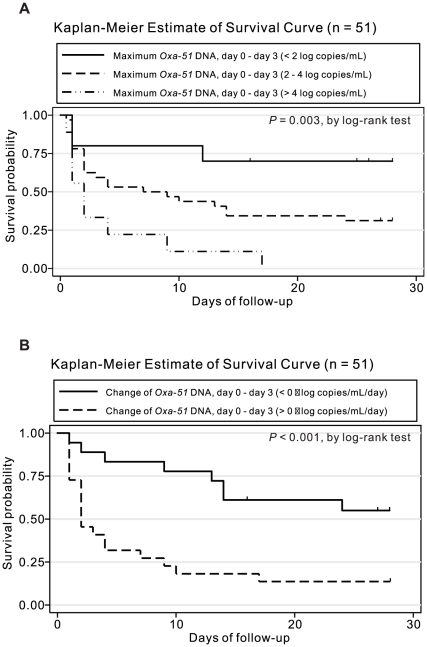
Based on the maximum Oxa-51 DNA from day 0 to day 3, three groups of patients had different survival curves. All patients with high maximum Oxa-51 DNA (>4 log copies/mL) during empirical therapy died within 17 days, whereas 70% of patients with low maximum Oxa-51 DNA (<2 log copies/mL) survived (P = 0.003, log-rank test) (Figure 4A). Moreover, based on the increase or decrease in Oxa-51 DNA from day 0 to day 3, two groups of patients had different survival curves (P<0.001, log-rank test) (Figure 4B). These were further supported by multivariate Cox's proportional hazard analysis (Table 3). Similarly, the survival curves based on a single day measurement of Oxa-51 DNA on day 2 or day 3 (>3 or <3 log copies/mL) were significantly different (data not shown).
Figure 4. Kaplan-Meier survival curves in patients with A. baumannii bacteremia.
Comparison among subgroups with different maximum Oxa-51 DNA load (A) and increased or decreased Oxa-51 DNA (B) from day 0 to day 3.
Table 3. Analysis of factors predicting mortality of study patients with A. baumannii bacteremia by multivariate Cox's proportional hazard modela.
| Covariate | P value | Adjusted hazard ratio | 95% Confidence interval |
| Pitt bacteremia score | 0.02 | 1.29 | 1.04–1.60 |
| Change of Oxa-51 DNA, day 0b – day 3(Δ log copies/mL/day) | 0.004 | 1.87 | 1.22–2.85 |
| Maximum Oxa-51 DNA, day 0b – day 3(log copies/mL) | 0.09 | 1.52 | 0.94–2.45 |
Test of proportional-hazards assumption: P = 0.14>0.05, Groennesby and Borgan GOF test P = 0.23>0.05, adjusted generalized R2 = 0.405.
Day 0 was the day on which the first blood sample positive for A. baumannii by culture was drawn.
Discussion
In light of the rapid increase in the antimicrobial resistance of A. baumannii, limited choice of antimicrobial agents and multiple complicated risk factors associated with its high mortality, it is important to identify good parameters for evaluating therapeutic responses to new or combination regimens in well controlled clinical studies [3], [4], [9]. In this study, we established an Oxa-51 quantitative real-time PCR assay to measure A. baumannii loads in blood from patients with A. baumannii bacteremia. Compared with the survivors, the nonsurvivors had higher maximum Oxa-51 DNA load and an increased trend from day 0 to day 3; both parameters and the single day measurement of Oxa-51 DNA on day 3, the day when blood culture reports were available from most clinical laboratories, were independent predictors for mortality by multivariate analysis (Table 2). To our knowledge, this is the first report demonstrating high A. baumannii loads in blood were associated with mortality. Moreover, nonsurvivors receiving appropriate antibiotics showed a trend of less increase in Oxa-51 DNA during the empirical period compared with those receiving inappropriate antibiotics, suggesting that monitoring Oxa-51 DNA load in blood could provide simple and direct parameters for evaluating therapeutic response of A. baumannii bacteremia in future clinical trials.
Several risk factors for mortality of A. baumannii bacteremia have been identified by different studies, such as septic shock [11], [14], high Pitt bacteremia score [15], immunosuppressant [11], [15], age, recent surgery, acute respiratory failure, acute renal failure [11], McCabe score, ventilator usage [14], disseminated intravascular coagulation, and inappropriate antibiotics usage [5]. With one exception [5], the majority of these were retrospective studies, which might be potentially biased by retrospective recall. We carried out a prospective study and included all factors studied in the multivariate logistic regression analysis, and three independent factors, maximum Oxa-51 DNA, its change from day 0 to day 3 and Pitt bacteremia score were identified (Table 2). These findings suggest both bacterial and host factors are important. The possible role of appropriate empirical antibiotic therapy was suggested by the trend of less increase in Oxa-51 DNA in the subgroup of nonsurvivors who received appropriate antibiotics compared with those receiving inappropriate antibiotics (Figures 3B and 3C). Analysis of this subgroup and the survivors receiving appropriate empirical antibiotics, of which both had an indistinguishable trend of Oxa-51 DNA change (Figure 3C), revealed that SOFA score and platelet counts were different (P = 0.06 and 0.03, respectively, Mann-Whitney U test, two tailed), suggesting host factors play a key role in determining mortality in this subgroup. The importance of host factors was further suggested by comparing the left panels of Figures 3B and 3C, in which appropriate empirical antibiotics were strongly associated with decreasing Oxa-51 DNA load (P<0.001, Mann-Whitney U test, two tailed), however, all patients in both groups died. This might be due to host factors; most of our patients had high APACHE II score (interquartile range [IQR], 20.5–36), Charlson score (IQR, 2–6) and Pitt bacteremia score (IQR, 3–7) (Tables 1 and 2). Another bacterial factor, antimicrobial resistance, was also carefully examined. Although ERAB was significantly associated with mortality in the univariate analysis, it was not significant in the multivariate analysis (OR, 1.99; 95% CI, 0.21–18.71; P = 0.55) (Table 2). Similarly, sulbactam resistance was not significantly associated with mortality in the multivariate analysis (Table 2).
Although SOFA score was strongly associated with mortality in the univariate analysis (Table 2), it was not included in the multivariate analysis due to small case number with SOFA score on day 0 available in our prospective observational study design. Nonetheless, including SOFA score in the multivariate logistic regression analysis revealed a similar final model, which included SOFA (OR, 1.65; 95% CI, 1.02–2.66; P = 0.04), change of Oxa-51 DNA, day 0 – day 3 (OR, 6.93; 95% CI, 1.00–48.15; P = 0.05) and maximum Oxa-51 DNA, day 0 – day 3 (OR, 14.93; 95% CI, 0.65–341.06; P = 0.09). The sensitivity, specificity, positive predictive value and negative predictive value were 100%, 87.5%, 95.2%, and 100%, respectively. The area under ROC curve was 0.9688. In multivariate Cox's proportional hazard model analysis, SOFA (Hazard ratio [HR], 1.17; 95% CI, 1.03–1.32; P = 0.01) and change of Oxa-51 DNA, day 0 – day 3 (HR, 1.63; 95% CI, 1.08–2.49; P = 0.02) were independent factors for mortality.
Of the 43 follow-up blood cultures from the 51 patients, only 3 (6.98%) were positive and 40 were negative, of which 12 had detectable Oxa-51 DNA (median 2.0 log/copies/mL, interquartitle range 1.6 to 2.6 log/copies/mL). This could be due to a drastic decrease in the sensitivity of blood culture after antibiotic usage as reported previously [2], [18]. Alternatively, the possibility that the Oxa-51 DNA detected in these samples was derived from dead or degraded bacteria rather than viable bacteria can not be completely ruled out. A recent study examining the amounts of Neisseria meningitidis in blood by quantitative PCR and simultaneous culture reported that the DNA copy numbers were more than 2 logs higher than CFU numbers, suggesting significant amounts of non-viable bacteria were present in vivo [22]. Nonetheless, the good correlation between bacterial DNA load and disease severity and/or outcome demonstrated in different bacterial infections, including Streptococcus pneumoniae, Neisseria meningitides, MRSA and A. baumanii in this study, strongly suggested bacterial DNA load, though derived from both viable and dead bacteria, could be a useful marker to predict disease outcome [19]–[22].
There are several limitations of this study. First, the sample size was small. Future study with a larger sample size is needed to verify our observations and the applicability of this method in clinical practice. Second, this study enrolled only critically ill patients in ICU. Future study enrolling patients from general wards is also needed to investigate the roles of Oxa-51 DNA load in non-critically ill patients. Moreover, the relationship of the kinetics of Oxa-51 DNA load in blood to antibiotic regimens and other factors during the course of infection require further investigation. Nonetheless, our findings that high and increased Oxa-51 DNA load can predict mortality in A. baumannii bacteremia support the notion that measuring bacterial DNA load could predict disease outcome or monitor therapeutic responses, and may have implications to our understanding of pathogenesis, particularly for those bacterial diseases with high mortality, multiple and complicated risk factors.
Supporting Information
Copy of the approval of the Institutional Review Board
(5.08 MB PDF)
Study protocol
(0.02 MB PDF)
Quantitative real-time PCR assay for Oxa-51 gene of A. baumannii
(0.08 MB DOC)
Multivariate logistic regression analysis
(0.04 MB DOC)
Antimicrobial susceptibility profile and antibiotic therapy of each study patient with A. baumannii bacteremia.
(0.09 MB DOC)
Acknowledgments
We thank Dr. Fu-Chang Hu at the Center of Excellence for General Clinical Trial and Research, National Taiwan University Hospital for his guidance in regression analysis, Su-Ru Lin, Yu-Wei Chen, Cha-Ming Wong and Rui-Juan Wu for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: National Science Council, Taiwan (NSC-95-2745-B-002-007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;4:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- 3.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 4.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 5.Cisneros JM, Reyes MJ, Pachon J, Becerril B, Caballero FJ, et al. Bacteremia due to Acinetobacter baumannii: epidemiology, clinical findings, and prognostic features. Clin Infect Dis. 1996;22:1026–1032. doi: 10.1093/clinids/22.6.1026. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, Jimenez-Jimenez FJ, Perez-Paredes C, et al. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin Infect Dis. 2001;33:939–946. doi: 10.1086/322584. [DOI] [PubMed] [Google Scholar]
- 7.Falagas ME, Kopterides P, Siempos Attributable mortality of Acinetobacter baumannii infection among critically ill patients. Clin Infect Dis. 2006;43:389; author reply 389–390. doi: 10.1086/505599. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, Jimenez-Jimenez FJ, Monterrubio-Villar J, et al. Mortality and the increase in length of stay attributable to the acquisition of Acinetobacter in critically ill patients. Crit Care Med. 1999;27:1794–1799. doi: 10.1097/00003246-199909000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Fishbain J, Peleg AY. Treatment of Acinetobacter infections. Clinical Infectious Diseases. 2010;51:79–84. doi: 10.1086/653120. [DOI] [PubMed] [Google Scholar]
- 10.Chen CH, Lin LC, Chang YJ, Huang CC, Liu CE, et al. Analysis of prognostic factors in 95 patients with Acinetobacter baumannii bacteremia. Infection. 2003;31:331–335. doi: 10.1007/s15010-003-3223-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen HP, Chen TL, Lai CH, Fung CP, Wong WW, et al. Predictors of mortality in Acinetobacter baumannii bacteremia. J Microbiol Immunol Infect. 2005;38:127–136. [PubMed] [Google Scholar]
- 12.Gomez J, Simarro E, Banos V, Requena L, Ruiz J, et al. Six-year prospective study of risk and prognostic factors in patients with nosocomial sepsis caused by Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 1999;18:358–361. doi: 10.1007/pl00015019. [DOI] [PubMed] [Google Scholar]
- 13.Poutanen SM, Louie M, Simor AE. Risk factors, clinical features and outcome of Acinetobacter bacteremia in adults. Eur J Clin Microbiol Infect Dis. 1997;16:737–740. doi: 10.1007/BF01709254. [DOI] [PubMed] [Google Scholar]
- 14.Seifert H, Strate A, Pulverer G. Nosocomial bacteremia due to Acinetobacter baumannii. Clinical features, epidemiology, and predictors of mortality. Medicine (Baltimore) 1995;74:340–349. doi: 10.1097/00005792-199511000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Tseng YC, Wang JT, Wu FL, Chen YC, Chie WC, et al. Prognosis of adult patients with bacteremia caused by extensively resistant Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2007;59:181–190. doi: 10.1016/j.diagmicrobio.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Schonheyder HC, Gottschau A, Friland A, Rosdahl VT. Mortality rate and magnitude of Staphylococcus aureus bacteremia as assessed by a semiquantitative blood culture system. Scand J Infect Dis. 1995;27:19–21. doi: 10.3109/00365549509018967. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan TD, LaScolea LJ, Jr, Neter E. Relationship between the magnitude of bacteremia in children and the clinical disease. Pediatrics. 1982;69:699–702. [PubMed] [Google Scholar]
- 18.Glerant JC, Hellmuth D, Schmit JL, Ducroix JP, Jounieaux V. Utility of blood cultures in community-acquired pneumonia requiring hospitalization: influence of antibiotic treatment before admission. Respir Med. 1999;93:208–212. doi: 10.1016/s0954-6111(99)90010-0. [DOI] [PubMed] [Google Scholar]
- 19.Carrol ED, Guiver M, Nkhoma S, Mankhambo LA, Marsh J, et al. High pneumococcal DNA loads are associated with mortality in Malawian children with invasive pneumococcal disease. Pediatr Infect Dis J. 2007;26:416–422. doi: 10.1097/01.inf.0000260253.22994.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett SJ, Guiver M, Marsh J, Sills JA, Thomson AP, et al. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch Dis Child. 2002;86:44–46. doi: 10.1136/adc.86.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho YC, Chang SC, Lin SR, Wang WK. High levels of mecA DNA detected by a quantitative real-time PCR assay are associated with mortality in patients with methicillin-resistant Staphylococcus aureus bacteremia. J Clin Microbiol. 2009;47:1443–1451. doi: 10.1128/JCM.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ovstebo R, Brandtzaeg P, Brusletto B, Haug KB, Lande K, et al. Use of robotized DNA isolation and real-time PCR to quantify and identify close correlation between levels of Neisseria meningitidis DNA and lipopolysaccharides in plasma and cerebrospinal fluid from patients with systemic meningococcal disease. J Clin Microbiol. 2004;42:2980–2987. doi: 10.1128/JCM.42.7.2980-2987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, et al. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreckenberger PC, von Graevenitz A. Acinetobacter, Achromobacter, Acaligenes, Moraxella, Methylobacterium, and other nonfermentative gram-negative rods. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. Washington, D.C: ASM Press; 2000. pp. 539–560. 7th ed. [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. Wayne, PA: CLSI; 2007. Performance standards for antimicrobial susceptibility testing: 17th informational supplement, M100-S17. [Google Scholar]
- 26.Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, et al. Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J Clin Microbiol. 2007;45:227–230. doi: 10.1128/JCM.01588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arroyo LA, Garcia-Curiel A, Pachon-Ibanez ME, Llanos AC, Ruiz M, et al. Reliability of the E-test method for detection of colistin resistance in clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:903–905. doi: 10.1128/JCM.43.2.903-905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 29.Feldman C, Alanee S, Yu VL, Richards GA, Ortqvist A, et al. Severity of illness scoring systems in patients with bacteraemic pneumococcal pneumonia: implications for the intensive care unit care. Clin Microbiol Infect. 2009;15:850–857. doi: 10.1111/j.1469-0691.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- 30.Minne L, Abu-Hanna A, de Jonge E. Evaluation of SOFA-based models for predicting mortality in the ICU: A systematic review. Crit Care. 2008;12:R161. doi: 10.1186/cc7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 32.Lin YC, Sheng WH, Chang SC, Wang JT, Chen YC, et al. Application of a microsphere-based array for rapid identification of Acinetobacter spp. with distinct antimicrobial susceptibilities. J Clin Microbiol. 2008;46:612–617. doi: 10.1128/JCM.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai TY, Chang SC, Hsueh PR, Feng NH, Wang JT. In vitro activity of isepamicin and other aminoglycosides against clinical isolates of Gram-negative bacteria causing nosocomial bloodstream infections. J Microbiol Immunol Infect. 2007;40:481–486. [PubMed] [Google Scholar]
- 34.Wang JT, Chen YC, Chang SC, Chen ML, Pan HJ, et al. Control of vancomycin-resistant enterococci in a hospital: a five-year experience in a Taiwanese teaching hospital. J Hosp Infect. 2004;58:97–103. doi: 10.1016/j.jhin.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Wang JT, Fang CT, Chen YC, Wu CL, Chen ML, et al. Staphylococcal cassette chromosome mec in MRSA, Taiwan. Emerg Infect Dis. 2007;13:494–497. doi: 10.3201/eid1303.060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Copy of the approval of the Institutional Review Board
(5.08 MB PDF)
Study protocol
(0.02 MB PDF)
Quantitative real-time PCR assay for Oxa-51 gene of A. baumannii
(0.08 MB DOC)
Multivariate logistic regression analysis
(0.04 MB DOC)
Antimicrobial susceptibility profile and antibiotic therapy of each study patient with A. baumannii bacteremia.
(0.09 MB DOC)